Abstract
Pathological cardiac hypertrophy is an inevitable forerunner of heart failure. Regardless of the etiology of cardiac hypertrophy, cardiomyocyte mitochondrial alterations are always observed in this context. The translocases of mitochondrial outer membrane (Tom) complex governs the import of mitochondrial precursor proteins to maintain mitochondrial function under pathophysiological conditions; however, its role in the development of pathological cardiac hypertrophy remains unclear. Here, we showed that Tom70 was downregulated in pathological hypertrophic hearts from humans and experimental animals. The reduction in Tom70 expression produced distinct pathological cardiomyocyte hypertrophy both in vivo and in vitro. The defective mitochondrial import of Tom70-targeted optic atrophy-1 triggered intracellular oxidative stress, which led to a pathological cellular response. Importantly, increased Tom70 levels provided cardiomyocytes with full resistance to diverse pro-hypertrophic insults. Together, these results reveal that Tom70 acts as a molecular switch that orchestrates hypertrophic stresses and mitochondrial responses to determine pathological cardiac hypertrophy.
Similar content being viewed by others
Introduction
Heart failure is the final stage of various cardiac diseases and is a major cause of morbidity and mortality worldwide1. Two hallmarks of chronic heart failure are cardiac hypertrophy and reduced contractility of viable myocardium2,3. As the precursor of heart failure, cardiac hypertrophy is an extremely stereotypic response of the heart to long-standing stresses, such as chronic pressure, volume overload or neurohormonal stimuli4. Cardiac hypertrophy can generally be categorized as physiological or pathological hypertrophy. An initial hypertrophic response is adopted to maintain cardiac output. After an initial phase of compensation, if hypertrophic growth leads to cardiac dysfunction, then the hypertrophy is considered pathological4. Mitochondria are central to cardiac stress responses5,6. Studies of pathological cardiac hypertrophy have reported obvious mitochondrial abnormalities, such as pronounced changes in the composition and function of the mitochondrial proteome5. However, the molecular mechanism of the mitochondrial alterations underlying pathological cardiac hypertrophy remains unclear.
Mammalian mitochondria contain approximately 1 000 different proteins. The vast majority of mitochondrial proteins are encoded by nuclear DNA, synthesized as precursor proteins in the cytosol, and imported into the mitochondria; only ∼1% of mitochondrial proteins are encoded by mitochondrial DNA. The dynamic import of these precursor proteins into mitochondria is critically required for the maintenance of mitochondrial functions ranging from cell signaling to the regulation of cell fate beyond intrinsic energy generation7.
All mitochondrial preproteins are imported via a general entry gate: the translocases of mitochondrial outer membrane (Tom) complex, which comprises a central channel (formed by Tom40, Tom5, Tom6 and Tom7) and three receptors (Tom70, Tom20 and Tom22)8. The precise targeting of mitochondrial precursor proteins requires the recognition of mitochondrial targeting signals. Tom20 and Tom70 recognize mitochondrial-targeting sequences on the precursor proteins and transfer these preproteins to Tom22 and the central channel7,8. Accumulating evidence indicates that the assembly of the Tom complex exhibits obvious plasticity in response to a variety of stressors, including enhanced contractile activity of skeletal muscle9 and elevated levels of thyroid hormones and cytosolic kinases10,11. These studies suggest that the Tom complex might incorporate a molecular toggle that fine-tunes mitochondrial biogenesis and bioenergetics to meet metabolic demands under stress. There is substantial interest in determining whether Tom complex components are altered in pathological cardiac hypertrophy.
Using various pathological cardiomyocyte hypertrophy models in vivo and in vitro, in combination with a model organism, we revealed that Tom70 functions as a molecular switch to determine pathological cardiac hypertrophy by integrating hypertrophic stresses and mitochondrial responses. The pathological overgrowth of Tom70-deficient cardiomyocytes was mediated through oxidative stress associated with the defective import of a mitochondria-shaping protein, optic atrophy-1 (Opa1). To our knowledge, this is the first study to show a link between the Tom machinery and cardiovascular diseases.
Results
Characterization of the expression of Tom machinery components in pathological hypertrophic hearts
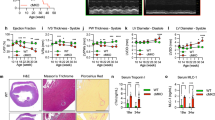
To identify mitochondrial molecular signatures underlying pathological cardiac hypertrophy, we performed mRNA screens for all known Tom complex genes that might undergo changes in end-stage failing human hearts. This screen revealed a marked decrease in the expression of Tom70 (Figure 1A).
Endogenous Tom70 is decreased in pathological hypertrophic myocardium of humans and experimental animals. (A) The expression of Tom complex genes in human cardiac tissues (n = 5 per heart failure (HF); n = 3 per non-failing control (NF)). (B) Echocardiographic analysis of heart weight/body weight (HW/BW), left ventricular posterior wall thickness (LVPWTd) and fractional shortening (FS) of rats after isoproterenol (ISO) injection. The upper panels show typical echocardiograms at the indicated times. (C, D) Western blot analysis of Tom70 protein in the hearts of rats subjected to ISO injection (C) and mice after TAC (D). GAPDH from the lysates was immunoblotted as a loading control. n = 8 per animals with pathological cardiac hypertrophy; n = 6 per control. The data represent the means ± SEM. *P < 0.05 versus the respective control, as determined using one-way ANOVA.
Given the reduction in Tom70 expression in the human pathological hypertrophic cardiac samples, we hypothesized that Tom70 expression might also be downregulated in adult animal hearts through chronic pro-hypertrophic insults. In a rat model, cardiac hypertrophy was generated through subcutaneous isoproterenol injection12. The hypertrophy developed gradually and peaked on days 14-17 (Figure 1B). Ventricular fractional shortening, an important index of cardiac function, showed a similar pattern (Figure 1B). Following the pathological progression of cardiac hypertrophy, the expression of Tom70 protein in the left ventricular myocardium was progressively downregulated (Figure 1C). A similar pattern of Tom70 expression was observed in a mouse cardiac hypertrophy model in which transverse aortic constriction was used to induce pressure overload on the heart, leading to pathological cardiac hypertrophy and dysfunction (Figure 1D). Collectively, these results show that Tom70 expression is reduced in the pathological hypertrophic hearts.
Tom70 is downregulated during pathological cardiomyocyte hypertrophy
Ventricular myocyte enlargement is a hallmark of cardiac hypertrophy4. To characterize the expression of Tom70 in hypertrophic cardiomyocytes, several well-established experimental models of pathological cardiomyocyte hypertrophy employing cultured neonatal rat ventricular myocytes (NRVMs)13 were adopted. The application of isoproterenol (ISO, 1 μM), angiotensin-II (Ang-II, 100 nM) or endothelin-1 (ET-1, 100 nM) alone to the culture medium of spontaneously beating NRVMs for 48 h induced a characteristic hypertrophic effect, as evidenced by an obvious physical signature of hypertrophy, changes in cell surface area, and alterations of the expression of principal biomarkers atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP) and alpha-myosin heavy chain (α-MHC). As expected, these pathological provocations led to a significant increase in the size of the NRVMs (Figure 2A and 2B), elevations in ANP and BNP mRNA expression and a reduction in α-MHC expression (Figure 2C). Immunoblot analyses revealed a time-dependent reduction in Tom70 protein in response to these pro-hypertrophic stimulators (Figure 2D). These results provide further support for the potential link between Tom70 reduction and pathological cardiac hypertrophy.
Cardiomyocyte Tom70 is downregulated during pathological hypertrophy. (A) Representative images of NRVMs treated with ISO, angiotensin-II (AngII) and endothelin-1 (ET-1) for 48 h. Immunohistochemistry using α-actinin and DAPI was followed by cell area quantifications, as described in the Materials and Methods. (B) Pooled data from A. (C) mRNA expression of pathological hypertrophic markers. Atrial natriuretic peptide, ANP; brain natriuretic peptide, BNP; alpha-myosin heavy chain, α-MHC. (D) Western blot analysis of Tom70 protein in NRVMs subjected to ISO, AngII or ET-1 treatment. GAPDH from the lysates was immunoblotted as a loading control. #P < 0.05. *P < 0.05 versus the control using one-way ANOVA.
Knockdown of Tom70 induces pathological cardiomyocyte hypertrophy in vitro
The reduced expression of Tom70 in cardiomyocytes during pathological cardiac hypertrophy suggested a potential functional role for Tom70 in this process. To examine this hypothesis, we knocked down Tom70 in intact NRVMs using siRNA. The decrease in Tom70 levels by siRNA induced a clear increase in cell size in the absence of pro-hypertrophic stimulators (Figure 3A and 3B). The reduction in Tom70 expression was also sufficient to trigger changes in the expression of ANP, BNP and α-MHC (Figure 3C) without affecting cell viability (Figure 3D).
Reduction in Tom70 in primary cardiomyocytes results in hypertrophic growth. Primary NRVMs were infected with Tom70 siRNA-expressing adenovirus and compared with the corresponding controls. All the experiments were performed 48 h after transfection. (A) Immunohistochemistry against α-actinin and DAPI was followed by cell area quantifications, as described in the Materials and Methods. At least 100 cells were quantified in all the groups. (B) Pooled data from A. (C) mRNA expression of pathological hypertrophic markers. Atrial natriuretic peptide, ANP; brain natriuretic peptide, BNP; alpha-myosin heavy chain, α-MHC. (D) TUNEL staining assay for cell viability. Left, typical images for the TUNEL-staining cardiomyocytes. Right, pooled data. All the data represent the means ± SEM. *P < 0.05 versus control as determined using one-way ANOVA.
To exclude a potential role of cardiac fibroblast transdifferentiation in the Tom70 reduction-related cardiac hypertrophy, we also studied the effects of Tom70 knockdown on fibroblast transdifferentiation into cardiomyocytes. Our results showed that Tom70 reduction did not affect the expression of cardiogenesis markers, including MEF2C, GATA4, Nkx2.5, actinin, β-MHC and Troponin T, in the fibroblasts (Supplementary information, Figure S1). Thus, Tom70 deficiency-induced cardiomyocyte hypertrophy did not involve cardiac fibroblast transdifferentiation.
Tom70 reduction results in pathological cardiac hypertrophy in vivo
To confirm the link between Tom70 reduction and pathological cardiac hypertrophy, we next examined whether Tom70 modulated the in vivo growth of cardiomyocytes in zebrafish, an excellent model organism for obtaining insights into various cellular aspects of vertebrate cardiac development14. First, we identified the protein sequence of the zebrafish ortholog of Tom70, which has overall identity to the human (79%), mouse (81%) and rat (78%) Tom70 orthologs (Supplementary information, Figure S2A). Tom70 is expressed in the heart, liver, brain and skeletal muscle tissues of adult zebrafish (Supplementary information, Figure S2B).
To inactivate zebrafish tom70, morpholino-modified antisense oligonucleotides directed against either the translational start site (MO-Tom70ATG) or the splice donor site of exon 1 (MO-Tom70) were injected into single-cell zebrafish embryos carrying a transgene for specific cardiac myosin light chain-2 (cmlc2)-GFP. MO-Tom70 was designed to block splicing at the exon 1-intron 1 boundary, leading to either the inclusion of intron 1 into the mature transcript, resulting in the introduction of a premature stop codon, or the skipping of exon 1, resulting in a shift in the reading frame and the premature termination of protein translation. We detected both aberrant splice products in the injected embryos, whereas wild-type tom70 mRNA was completely absent (Figure 4A). A western blot analysis confirmed the effective knockdown of the Tom70 protein using MO-Tom70 or MO-Tom70ATG (Figure 4B).
Inactivation of Tom70 leads to pathological cardiac hypertrophy in zebrafish. (A) RT-PCR amplification of RNA from MO-ctrl- and MO-Tom70-injected zebrafish showing altered splicing of Tom70 with either the integration of intron 1(*) or the partial skipping of exon 1 (+). (B) Western blot analysis of MO-ctrl-, MO-Tom70- and MO-Tom70ATG-injected embryos at 2 dpf. GAPDH was used as a loading control. The data are representative of three independent experiments. (C) Dissection of beating hearts at 2 dpf followed by integrin staining (blue = DAPI, green = GFP and red = Zn-8). (D) Quantification of cardiomyocyte size and number from a total of 10 hearts per group in C. (E) Top, lateral close-up view of zebrafish hearts at 5 dpf. MO-Tom70ATG-injected embryos with hypertrophic hearts (yellow dotted line) were compared with control (MO-ctrl)-injected zebrafish. Bottom, H&E staining of zebrafish heart sections of MO-ctrl- and MO-Tom70ATG-injected embryos. The ventricular (v) chambers of MO-Tom70ATG-injected zebrafish are constricted. n = 10 embryos. (F) Quantification of ventricular fractional shortening (FS) in MO-Tom70ATG-injected zebrafish embryos at the indicated stages. The data represent the means ± SEM. n = 10 embryos per time point. *P < 0.05 versus the respective control using one-way ANOVA.
During the first 2 days of development, the MO-Tom70ATG- or MO-Tom70-injected embryos exhibited normal heart morphogenesis, such as heart tube jogging, looping and heart chamber demarcation, compared with the MO-ctrl-injected embryos. However, we observed an evident increase in cardiomyocyte size and concentric cardiac hypertrophy in the Tom70 morphants (Supplementary information, Figure S2C and Movie S1) 2 days post fertilization (dpf). Figure 4C shows zebrafish hearts dissected at 2 dpf and subjected to nuclear (DAPI-blue) and plasma membrane (Zn-8-red) staining. The quantification of these images revealed cardiomyocyte enlargement in the Tom70 morphants; however, no significant change in cardiomyocyte number was observed (Figure 4D). At 5 dpf, the thickness of the ventricular wall was markedly increased and the ventricular cavity was constricted in the morphants, as demonstrated by a high-magnification observation of the heart and immunological staining of cardiac sections in the diastole phase (Figure 4E). The analysis of cardiac function revealed reduced ventricular contractility in the morphants after 2 dpf, as determined by ventricular fractional shortening (Figure 4F), a typical feature of pathological cardiac hypertrophy. This pathological growth behavior of the cardiomyocytes was almost fully counteracted by the cardiac-specific overexpression of human Tom70 under the control of the zebrafish cmlc-2 promoter region (Supplementary information, Figure S2D and S2E and Movie S2). In another set of experiments, we analyzed the potential role of Tom70 in early heart development by profiling the expression of cardiogenesis markers, including GATA4, Hand2 and NKx2.5 (Supplementary information, Figure S2F). The results revealed that Tom70 reduction had no effect on these markers. Thus, Tom70 might play an essential role in regulating cardiomyocyte growth, and reduced Tom70 expression leads to pathological cardiac hypertrophy without affecting early cardiogenesis or cardiomyocyte proliferation.
Oxidative stress mediates Tom70 deficiency-induced cardiomyocyte hypertrophy
Substantial evidence has indicated that oxidative stress might be the common mechanism underlying the pathological cardiac hypertrophy stimulated by hemodynamic overload and G protein-coupled receptor agonists15. We observed that Tom70 reduction induced an overt elevation of reactive oxygen species (ROS) in NRVMs, which resembled the effects of pathological provocation using ET-1 for 48 h (Figure 5A). The application of the antioxidant N-acetyl-cysteine (NAC, 2 mM) and catalase (100 U/ml) greatly ameliorated cardiomyocyte enlargement (Figure 5B and 5C) and the expression change of ANP, BNP and α-MHC (Figure 5D-5F), further supporting an important role for oxidative stress in the Tom70 deficiency-induced cardiomyocyte hypertrophy.
Tom70 mediates pathological hypertrophy of cardiomyocytes by regulating intracellular oxidation states. (A) Measurements of superoxide generation in primary cardiomyocytes treated with ET-1 and Tom70 siRNA using a MitoSOX Red probes. Effects of an antioxidant NAC (2 mM) and catalase on cardiomyocyte size (B-C) and the expression levels of ANP, BNP and α-MHC (D-F). (G) MitoSOX Red staining indicates high ROS generation in the Tom70-deficient hearts of zebrafish (white dotted line) compared with MO-ctrl-injected embryos at 2 dpf. The same exposure intensity was used in different experimental groups. (H) qPCR analysis of the indicated genes in zebrafish hearts injected with MO-Tom70ATG for 48 h with or without NAC (2 mM). NS: nonsignificance. *P < 0.05 versus the respective control using one-way ANOVA. #P < 0.05, δP < 0.05.
To investigate the potential in vivo effects of oxidative stress induced by Tom70 deficiency on pathological cardiac hypertrophy, we analyzed the levels and effects of ROS in the heart. The in vivo staining of zebrafish embryos at 2 dpf with MitoSOX Red revealed the presence of high levels of ROS in the hearts of Tom70 morphants compared with MO-ctrl (Figure 5G). We also measured the expression levels of a regulated set of genes involved in cardiac hypertrophy, including ANP, α-MHC and GATA416,17, in the morphant hearts. As shown in Figure 5H, the inactivation of Tom70 increased the expression of ANP and reduced the expression of α-MHC and GATA4. These effects were mitigated by the administration of NAC. Thus, the combined in vitro and in vivo data identified oxidative stress as a conserved pro-hypertrophic mediator of the effects of Tom70 deficiency.
Abnormal mitochondrial fusion caused by defect in a mitochondria-shaping protein, optical atrophy-1, underlies oxidative stress caused by Tom70 deficiency
ROS are primarily generated in mitochondria, and mitochondrial abnormalities induce ROS overproduction18. Using transmission electron microscopy, we observed that mitochondria in the ventricular myocytes of Tom70 morphants exhibited blurred cristae, swelling and fragmentation (Figure 6A). This abnormal organelle morphology was also observed in the pathological hypertrophic hearts of experimental animals and Tom70-deficient NRVMs (Supplementary information, Figure S3A and S3B). Using MitoTracker, a mitochondria-localizing fluorescent probe, we next analyzed the dynamics of mitochondria in primary cardiomyocytes. The cytoplasm of control NRVMs contained a mitochondrial network with a long filamentous shape; however, this network was fragmented into small organelles in Tom70-deficient cells (Figure 6B and 6C). These observations suggest that Tom70 is crucial for the maintenance of mitochondrial morphology, and reduced Tom70 expression impairs mitochondrial fusion in cardiomyocytes.
Mitochondrial disorder contributes to oxidative stress and hypertrophic growth of Tom70-deficient cardiomyocytes. (A) Transmission electron microscopic examination of mitochondria from Tom70-deficient zebrafish hearts. (B) Effects of Tom70 knockdown on mitochondrial dynamics in primary cardiomyocytes. The cells were treated with Tom70 siRNA or scrambled siRNA for 48 h and were subsequently loaded with MitoTracker red. Images were obtained, and the boxed areas show magnifications (right). (C) Mitochondrial morphological change was analyzed as the mean roundness and fusion/fission ratio. (D) Western blot analysis of the mitochondria-shaping protein Opa1 in the Tom70-deficient cardiomyocytes. (E) Pooled data from five separate experiments in D. (F) Co-immunoprecipitation of Tom70 and Tom20 with Opa1 in normal primary cardiomyocytes. IP: immunoprecipitation. IB: immunoblotting. (G) In vitro import analysis of Opa1 in cardiomyocyte mitochondria. Top, representative western blots showing the time-dependent import of biotin-labeled Opa1 proteins. Bottom, pooled data. All the data were obtained from five separate experiments. (H) Inactivation of Opa1 resulted in hypertrophic growth of cardiomyocytes in zebrafish. Images of dissection of beating hearts followed by staining (blue = DAPI, green = GFP and red = Zn-8) at the indicated times. Bar: 10 μm. Upper right, western blot examination of Opa1 knockdown by MO-opa1ATG injection at 2 dpf. Lower right, quantification of the cardiomyocyte size from a total of 6-8 hearts per group. All the data were obtained from three independent experiments. *P < 0.05 versus the respective control using one-way ANOVA.
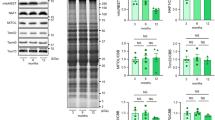
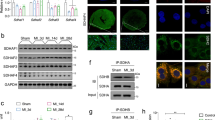
Based on the observations of mitochondrial morphology described above, the proteins responsible for mitochondrial fusion and fission were expected to be altered in Tom70-deficient cardiomyocytes. First, we isolated and purified total mitochondrial proteins from the Tom70-deficient NRVMs to perform a proteome analysis. When we compared the two-dimensional gel electrophoresis protein patterns in mitochondria from control (scramble-treated) and Tom70-deficient cells, we observed a reduction in the expression of optical atrophy-1 (Opa1; Supplementary information, Table S1), a protein located in the inner mitochondrial membrane that is critically responsible for mitochondrial fusion19. In separate experiments, an immunoblot analysis verified that a long isoform of mitochondrial Opa1 protein was reduced, but other well-known proteins involved in the regulation of mitochondrial morphology, such as dynamin-related protein-1 (Drp1) and mitofusion-1 and mitofusion-2 (Mfn1/2), were not reduced (Figure 6D, 6E and Supplementary information, Figure S4A). These results suggest that the abnormality in mitochondrial morphology caused by Tom70 reduction is associated with deficiency in mitochondrial Opa1.
We also explored the potential causes of mitochondrial Opa1 deficiency. Using real-time PCR, we measured the Opa1 mRNA expression in cardiomyocytes. We observed that the Opa1 transcript levels did not differ between Tom70-deficient NRVMs and intact cardiomyocytes or between rat HF hearts and related controls (Supplementary information, Figure S4B and S4C), suggesting that the change in Opa1 protein levels is post-transcriptionally regulated. We further assessed the potential association between Tom70 and Opa1 using immunoprecipitation-based assays. The immunoprecipitation of endogenous Tom70 pulled down endogenous Opa1 from lysates of freshly isolated intact cardiomyocytes, and vice versa (Figure 6F). As Tom20 is an alternative receptor that is important for mitochondrial preprotein recognition and transport, the possibility of Opa1 binding to Tom20 was also analyzed. Coimmunoprecipitation assays showed that Opa1 did not physically interact with Tom20 in myocytes (Figure 6F). Moreover, we performed a cell-free import assay to investigate whether Tom70 plays a role in the mitochondrial import of Opa1. Cell-free translated biotin-labeled Opa1 was incubated with mitochondria isolated from intact and Tom70-deficient NRVMs, and the results showed that the import efficiency of Opa1 was considerably reduced upon Tom70 knockdown (Figure 6G). Collectively, defects in Tom70-targeting import contribute to the reduction in mitochondrial Opa1 and the consequent disruption of mitochondrial morphology in pathological hypertrophic cardiomyocytes.
As Opa1 deficiency is critically responsible for Tom70 deficiency-induced cardiomyocyte hypertrophy, we further tested whether loss of Opa1 function in zebrafish embryos would produce a similar cardiac phenotype. A translation-blocking morpholino directed against Opa1 (MO-opa1ATG) was used to downregulate Opa1 in zebrafish. Western blot analysis confirmed that the injection of MO-opa1ATG reduced the expression of Opa1, whereas the controls were unaffected. The inactivation of Opa1 obviously enlarged the ventricular myocytes at 2 dpf compared with MO-control-injected zebrafish embryos (Figure 6H). Ultrastructural observations revealed fragmented mitochondria with blurred cristae in the hearts of the opa1 morphants (Supplementary information, Figure S5), similar to the morphology observed in the Tom70-morphant hearts (Figure 6A). Thus, deficiency in mitochondrial Opa1 mimics the cellular features of pathological cardiac hypertrophy induced by Tom70 deficiency.
Enforced expression of Tom70 provides potent cardioprotection against the pathological hypertrophy caused by pro-hypertrophic insults
To confirm the causative effects of Tom70 reduction on pathological cardiomyocyte hypertrophy, we further examined whether cardiomyocyte Tom70 might provide inherent protection against the pathological growth induced by multiple pro-hypertrophic insults. We observed that the adenovirus-mediated overexpression of Tom70 did not alter cell size under basal conditions, but it fully ablated the hypertrophic responses of NRVMs to ISO, Ang-II or ET-1 treatment, as evidenced by the restoration of cell size and ANP expression (Figure 7A-7C). These data demonstrate that multiple hypertrophic stresses might converge on the mitochondrial receptor Tom70 and that the increased expression of Tom70 is sufficient to abolish the pathological hypertrophic growth of cardiomyocytes caused by pathological provocations.
Cardiomyocytes with Tom70 overexpression are resistant to prohypertrophic insults. (A) Representative images of primary cardiomyocytes treated with ISO, AngII and ET-1 in the presence or absence of Tom70 overexpression. (B) Pooled data from A. (C) mRNA expression of the hypertrophic marker ANP. All the data were obtained from three independent experiments. (D) Western blot analysis of Tom70 proteins in mouse hearts at the indicated times following the direct injection of Adv-Tom70 into the ventricular cavity. (E) Analysis of left ventricular posterior wall thickness (LVPWTd) of mice after ISO injection for 7 days. Top, typical echocardiographic images. Bottom, representative H&E staining. (F) Protein expression analysis of the cardiac hypertrophy marker ANP in mice treated with ISO for 7 days. GAPDH expression was used as a loading control. (G) Superoxide generation of mouse hearts with or without Tom70 overexpression was analyzed 7 days after ISO infusion. n = 12 per ISO treatment group, n = 11 per ISO treatment with Tom70 overexpression group, n = 8 per vector injection group, n = 7 per control group. *P < 0.05 versus the respective control using one-way ANOVA. #P < 0.05.
Next, we evaluated whether a systemic effect resulting from Tom70 overexpression in the heart also has a significant influence on pathological hypertrophy in a mouse model of cardiac hypertrophy. An adenoviral vector expressing Tom70 was administered through direct injection into the ventricular cavity of mice. Following adenoviral injection, the expression of Tom70 in the ventricular myocardium was confirmed using western blot analysis. The overexpression of Tom70 was detected 5 days after vector delivery, and levels above baseline persisted for at least 14 days (Figure 7D). Echocardiographic studies revealed overt cardiac hypertrophy in response to ISO-mediated adrenergic stress, whereas hypertrophy was reversed in the mice with Tom70 overexpression in this context (Figure 7E). Histological sectioning and hematoxylin-eosin (H&E) staining confirmed that enforced Tom70 expression resulted in a marked improvement in left ventricular posterior wall thickness (Figure 7E). The administration of Tom70 in the heart also reversed the prototypical gene expression pattern associated with pathological cardiac hypertrophy, as demonstrated by the normalized expression of ANP (Figure 7F).
Whether the cardioprotection of Tom70 was linked to the mitigation of myocyte oxidative stress was also examined. As shown in Figure 7G, the intracellular ROS levels of heart tissues were increased in the ISO-injected mice, but this was significantly attenuated in the Tom70-overexpressing mice. Together, these results indicate that the increased expression of Tom70 results in resistance to pathological cardiac hypertrophy and the improvement of cardiac function.
Discussion
The mitochondrial protein import machinery has been studied mainly in yeast and fungal model organisms7,8. Two major pathways for importing precursor proteins into the mitochondrial outer membrane have been characterized in detail: the presequence pathway for the import of matrix proteins and the carrier pathway for the import of hydrophobic inner membrane proteins7,8. Tom20 and Tom22 recognize the presequences of cleavable precursor proteins, and Tom70 typically recognizes hydrophobic precursor proteins with several internal targeting signals, many of which are inner membrane metabolite carriers7. Tom70 also functions as a docking site for cytosolic chaperones, such as Hsp70 and Hsp90, to receive mitochondrial proteins20, implying a distinct biological feature of Tom70 in cellular functions. Recently, mammalian counterparts of Tom70 have been identified21, but their functions in the heart remain unknown. Herein, we identified a marked reduction in Tom70 among the Tom complex proteins in pathological hypertrophic cardiomyocytes from different species and discovered that Tom70 knockdown induced the pathological hypertrophic growth of mammalian primary cardiomyocytes and zebrafish hearts without affecting fibroblast transdifferentiation into cardiomyocytes (Supplementary information, Figure S1) or the expression of early cardiogenesis markers (Supplementary information, Figure S2F). However, the Tom70-mediated cardiac hypertrophy does not involve Tom20/22 receptors: Tom20 defect caused a lethal developmental defect (Supplementary information, Figure S6), and Tom22 knockdown did not induce obvious abnormalities in the heart22. Thus, our study provides evidence that Tom70 exerts an evolutionarily conserved and specific function in regulating cardiomyocyte growth, and deficiency in this protein may directly promote the pathological hypertrophy of cardiomyocytes.
A balanced activity of the fusion and fission machineries shapes the mitochondrial compartment, and the behavior of mitochondria allows the cell to respond to ever-changing stressful conditions23. The dynamic regulation of mitochondrial morphology is mediated by large guanosine triphosphatase proteins19,23. These proteins include Drp1 and Fis1, which are responsible for mitochondrial fission, and Mfn1, Mfn2 and Opa1, which mediate fusion. Multiple lines of evidence highlighted Opa1 as a downstream effector of the Tom70-dependent pathway underlying the development of pathological cardiac hypertrophy. First, Tom70 downregulation induced mitochondrial fragmentation and swelling in cardiomyocytes. Second, among the above-mentioned mitochondria-shaping proteins, only Opa1 was found to be reduced in response to Tom70 deficiency. Third, Tom70 directly bound to Opa1, and a marked reduction in mitochondrial Opa1, resulted from the defects in Tom70-targeting import. Fourth, the Opa1 reduction greatly mimicked the phenotypes caused by Tom70 deficiency (Figure 6). Finally, Opa1 reduction tightly links to the chronic pressure overload-induced cardiac hypertrophy and the concurrently fragmented and swollen mitochondria24. Additionally, it has been described that an increase in the short cytoplasmic fragments of Opa1 links to cell apoptosis25. The finding of the change in the long, but not the short, isoform of Opa1 (Figure 6) may further account for the Tom70 deficiency-related cellular phenotypes.
Oxidative stress is the common mechanism of pathological cardiac hypertrophies induced by multiple etiologies18. Our results revealed that Tom70 reduction induced a widely changed expression of the components in the mitochondrial respiratory chains and the accumulation of ROS in the cardiomyocytes. NAC is an effective scavenger of free radicals and a major contributor to the maintenance of cellular glutathione status (antioxidative defense) in cardiac cells26. The beneficial effects of the antioxidative NAC (Figure 7 and Supplementary information, Figure S7) suggest that Tom70 reduction induced the hypertrophic growth of cardiomyocytes mainly by promoting ROS generation. Mitochondria are an important source of ROS. Reduced NADH:ubiquinone oxidoreductases (also called complex I) and/or increased NADPH oxidase leads to oxidative stress18. A previous study showed that mitochondrial Opa1 reduction induced complex I deficiency through the disruption of mitochondrial morphology, leading to the accumulation of mitochondrial ROS27. Herein we identified Opa1 as a downstream effector of Tom70, and Tom70 reduction caused the significant downregulation of complex I activity (Supplementary information, Table S1 and Figure S7). ROS scavenger NAC did not reconcile the reduced expression of Opa1 by Tom70 reduction and the Tom70 expression in hypertrophic cell models (Supplementary information, Figure S8), indicating that Opa1 acts as an upstream regulator of ROS generation in Tom70-deficient cardiomyocytes. Moreover, the collapse in mitochondrial membrane potential drives partial Opa1 degradation28. It is likely that the prior changes in mitochondrial membrane potential by Tom70 defect may further facilitate the reduction of Opa1. Taken together, these results lead to the reasonable deduction that impaired Tom70-targeted Opa1 import causes abnormal mitochondrial fusion and, consequently, a reduction in complex I activity, resulting in oxidative stress and pathological cardiomyocyte hypertrophy.
Our current findings indicated that multiple pro-hypertrophic signals converged on Tom70 and reduced its expression. The transcript levels of Tom70 were reduced in hypertrophic hearts and cardiomyocytes (Supplementary information, Figure S9), suggesting a defect in the transcriptional regulation of Tom70. A screen identified early growth response protein-1 (EGR1) as a transcriptional regulator of Tom70. EGR1 was downregulated in hypertrophic hearts, and its knockdown markedly reduced the protein level of Tom70 in cardiomyocytes (Supplementary information, Figure S9). Thus, the lower synthesis of Tom70 may account for the reduction in Tom70 protein in hearts with pathological hypertrophy.
Recent reports describe that Tom70 is essential for Pink1 translocation to mitochondria29, and Pink1 is required for mitophagy and subsequently ROS production30. To elucidate whether Pink1 was also involved in the development of cardiac hypertrophy caused by Tom70 reduction, we conducted knockdown experiments using Pink1 siRNA in cardiomyocytes. Our data showed that Pink1 knockdown could not induce a significant enlargement of cardiomyocytes (Supplementary information, Figure S10), precluding the contribution of Pink1 to the Tom70 reduction-mediated cardiomyocyte hypertrophy.
Several potential mechanisms may account for the cardioprotective effect of Tom70 overexpression against hypertrophic stress. On one hand, ROS are central for cardiac functioning under pathophysiological conditions. High levels of ROS have been found to promote the development of cardiac hypertrophy by regulating the expression of pro-hypertrophic factors5. Given that Tom70 overexpression inhibited ROS generation in cardiomyocytes subjected to hypertrophic stress (Supplementary information, Figure S7), it is plausible that Tom70 may protect the heart against hypertrophic stress by targeting ROS production. On the other hand, the starvation of intracellular bioenergetics contributes to the progression of cardiac hypertrophy and heart failure4,5,6. In view of our finding of the beneficial effects of Tom70 on ATP production in cardiomyocytes (Supplementary information, Figure S7), Tom70 overexpression may protect the heart from hypertrophic stress by maintaining bioenergetic homeostasis.
Pre-existing cardiac hypertrophy is a unique risk factor for developing heart failure. Pathological cardiac hypertrophy has been recognized as a new therapeutic target for heart failure and an independent predictor of cardiovascular mortality3. Despite the broad use of angiotensin-converting enzyme inhibitors, β-adrenoceptor blockers and aldosterone antagonists, heart failure remains a leading cause of mortality worldwide1,2. Our data showed that overexpression of Tom70 provided cardiomyocytes with full resistance to diverse pro-hypertrophic insults, such as the G protein-coupled receptor agonists ISO, AngII and ET-1. Importantly, an in vivo study also confirmed that enhanced Tom70 levels protected the heart against pathological hypertrophy. These findings convincingly indicate that Tom70 may serves as a molecular switch to determine pathological cardiac hypertrophy.
In conclusion, Tom70 critically governs the development of pathological cardiac hypertrophy via the Tom70-Opa1-ROS signaling pathway. Tom70 modulates hypertrophic stresses and mitochondrial responses to dramatically abolish the hypertrophic effects induced by diverse pro-hypertrophic insults. The identification of a correlation between Tom machinery defects and cardiac hypertrophy may provide a novel therapeutic target for the prevention of pathological cardiac hypertrophy and heart failure.
Materials and Methods
Human heart samples
Approval for this study was obtained from the ethics committee of the Tongji University School of Medicine. This study complied with the principles governing the use of human tissues outlined in the Declaration of Helsinki. Informed consent was obtained from all the subjects.
Left ventricular samples were obtained from five explanted human hearts obtained at the time of cardiac transplantation and from the hearts of three healthy controls with no cardiovascular pathology. The controls were prospective multiorgan donors whose organs could not be transplanted because of technical reasons.
Animal models
This study conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (Publication No. 85-23, revised 1996) and the policies of the Animal Care and Use Committee of the Tongji University School of Medicine.
Heart failure models were established through the administration of ISO to Sprague Dawley (SD, male, 180-200 g) rats and by performing transverse aortic constriction in mice, as previously reported12,31. Animal groupings and randomization were performed to ensure a balanced distribution of animals across all the study groups. The animal studies were not blinded: the same investigators performed the grouping and analyses, rendering the blinding of the studies unfeasible.
NRVM, culture and measurement of hypertrophic growth
NRVMs were isolated and cultured as previously described13. Briefly, neonatal rat pups (1-2 days old) were killed through decapitation, and their hearts were isolated and rinsed 2-3 times with PBS to remove debris. The ventricles were minced with scissors and placed in a small volume of PBS at 4 °C. After digestion with collagenase (2% w/v) and 0.5 ml of trypsin (2% w/v), the cells were centrifuged at 146× g at 4 °C for 1 min. The supernatant was discarded, and the pellet was resuspended in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and incubated at 37 °C in a 60-mm culture dish. As non-myocardiocytes attach to the substratum more quickly than myocardiocytes, the harvested cells were first incubated for 2 h to allow the attachment of non-myocardiocytes. The majority of myocardiocytes remained in the culture medium. The suspended cells were collected and plated at a density of 2 × 105/ml into a new cell culture dish, and BrdU (0.1 mmol/l) was added to the culture medium for 72 h to prevent the proliferation of non-myocardiocytes. Twenty-four hours after seeding, the culture medium was changed to serum-free DMEM.
A solvent carrier (control), 10 μM ISO (Sigma), 100 nM ET-1 (Sigma) or 100 nM AngII (Sigma) were added to the culture medium to induce cardiomyocyte hypertrophy. Forty-eight hours after the addition of drugs, the cells were stained with anti-α-actinin antibody (Sigma). Images of cultured cardiomyocytes were collected with laser confocal microscope (SP5, Leica) and were analyzed using the ImagePro Plus software (Media Cybernetics) to measure the cell surface area. In each group, the surface area was measured for >100 cells.
Zebrafish maintenance, morpholino microinjection, cell size measurements and quantification, and functional assessment
Zebrafish care and breeding were performed as previously described32. A morpholino-modified oligonucleotide from Gene Tools was directed against the translational start site (MO-Tom70: 5′-CTACGGGCTTCGATGCAGCCATCAT-3′) and the splice donor site of exon 1 (Tom70a-splice1-MO: 5′-AAATTAGGCTGATTACCAGGTTCTC-3′) of Tom70. The morpholinos were injected into wild-type embryos at the one-cell stage. As a negative control, a standard control oligonucleotide (5′-CTACcGcCTTCGATcCAcCgATCAT-3′) was injected at identical doses. Photographs and Movies were recorded at the indicated time points after the microinjection treatments. The dissection of beating hearts transgenic for a specific cmlc2 promoter at 48 h was followed by integrin staining (blue = DAPI, red = zn-8 and green = cmlc2:GFP), and the staining with both DAPI and Zn-8 per area of cardiomyocytes was assessed using confocal imaging. The cardiac output and stroke volume of the Tom70 morphants were monitored using a high-speed confocal laser line-scanning microscope (SP5, Leica).
Plasmid construction
To drive the cardiac-specific expression of Tom70 in zebrafish hearts, the Tom70-coding sequence was amplified from a cDNA clone (7391, from the laboratory of Han, Xiamen University) using the following Tom70 primers: HsaTOM70-F: 5′-CTCGAGATGGCCGCCTCTAAACCTGT-3′ and HsaTOM70-R: 5′-GGTACCGTTAATGTTGGTGGTTTTAATCCGT-3′. The coding sequences were separately subcloned into the peGFP-N1 vector (Clontech) under the control of the zebrafish cmlc2 promoter. The plasmids were pressure-injected into one-cell-stage fish embryos. The GFP-expressing cells were identified in vivo using fluorescence Movie microscope (Leica) and were used to analyze cardiac function.
H&E staining
Embryos and adult tissue samples were fixed in 4% paraformaldehyde in PBS for 48 h at 4 °C. The samples were dehydrated in ethanol, embedded in paraffin and sectioned at 5 μm using a rotary microtome (Leica). The histological H&E staining of the sections was subsequently performed using standard protocols (Beyotime).
Transmission electron microscopy
Embryos were fixed in 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.4) overnight at 4 °C and were post-fixed in 1% OSO4 in PBS (pH 7.2), followed by standard infiltration, embedding and sectioning procedures. The sections were analyzed using a JEM-1230 transmission electron microscope.
Adenovirus construction and transfection
Recombinant adenoviral vectors expressing CMV-Tom70-eGFP and CMV-Tom70-siRNA-eGFP under the control of a CMV promoter were created at Shanghai R&S Biotechnology Co, Ltd. NRVMs were isolated, cultured and infected with adenoviruses (multiplicity of infection = 100).
In vivo administration of adenoviral vectors
CD-57 mice (6 weeks) were anesthetized with a ketamine (100 mg/kg)-xylazine (8 mg/kg) mixture, intubated and ventilated with room air. Adenovirus (2 × 109 plaque-forming units/ml) was administered by direct injection into the left ventricular cavity using an insulin syringe with a 29-gauge needle. At the indicated times after the injection of adenoviruses expressing Tom70 or an empty vector control, the mice were killed. Cardiac tissue samples were collected for immunological blotting analysis and were subsequently embedded, counterstained with H&E, and cut into 10-μm sections for histological analysis.
Mitochondrial membrane potential (ΔΨm)
The mitochondrial membrane potential-selective dye TMRE (Molecular Probes) was used to label the cells. At 48 h, TMRE was added to NRVMs at a final concentration of 600 nM. The cells were incubated for 20 min at 37 °C and washed with PBS. The images were collected by laser confocal microscopy (SP5, Leica) and analyzed using the Leica AF lit software. Before the experiments, carbonyl cyanide m-chlorophenyl hydrazone (CCCP; 5 μM; Sigma), which depolarizes mitochondria by increasing their permeability to protons, was employed to confirm the dye behavior.
Measurement of ATP concentrations
The NRVMs were cultured in white 96-well flat-bottom plates at 30 000 cells per well. The ATP content was measured using the Apoglow luciferin-luciferase bioluminescence kit (Cambrex, East Rutherford, NJ, USA) according to the manufacturer's protocol.
Quantification of mitochondrial fusion and fission in living cells
The mitochondrial morphology of the cells was observed after culturing for 48 h. The cells were stained with 600 nM MitoTracker Red FM for 30 min to visualize the mitochondrial morphology. As NRVMs have two different types of mitochondrial structure (center: round, pseudopod area: tubular), we observed and analyzed morphological change only in the pseudopod area. Images were captured using laser confocal microscope (SP5, Leica). The morphological changes are described using ImageProPLUS software (Media Cybernetics, Silver Spring, MD, USA). Fusion/fission ratio was calculated by number of elongated mitochondria (roundness ≥ 3)/number of fragmented mitochondria (roundness < 3).
In vivo and in vitro detection of ROS generation
For in vivo ROS detection, live 2- to 4-day-old embryos (2-4 dpf) were incubated with 5 μM MitoSOX Red (Molecular Probes) for 20 min at 28.5 °C and were washed 3 × 5 min with embryo medium. The fluorescence was observed under a 488-nm wavelength excitation. To measure the intracellular ROS generation in vitro, myocytes with or without transfection were preincubated with 5 μM MitoSOX Red in DMEM for 30 min at 37 °C before treatment. Experiment was repeated at least five times to avoid transfection variations.
In vitro mitochondrial import assay
Rat cardiomyocyte mitochondria were purified. The in vitro mitochondrial import assay was performed using the freshly isolated mitochondria and biotin-labeled precursor proteins (synthesized with a TNT-coupled reticulocyte lysate system; Promega, Madison, WI, USA). The biotin-labeled proteins (Opa1) were incubated at 22-25 °C for various time periods with 25 μg of mitochondria in 50 μl of import buffer (10 mM Hepes-KOH, pH 7.4, containing 1 mM ATP, 20 mM sodium succinate, 5 mM NADH, 0.5 mM magnesium acetate, 220 mM mannitol, 70 mM sucrose and 0.5 mM PMSF). The mitochondria were subsequently isolated through centrifugation and were analyzed by SDS-PAGE. The intensity of bands representing imported proteins was visualized with fluorography and was quantified using a Bioimage Analyzer FLA5000 (Fuji, Tokyo, Japan).
RT-PCR
Heart, liver, muscle and brain tissues were separated from adult zebrafish, and mRNA was extracted from the pooled tissues using TRIzol reagent (Invitrogen). The following RT-PCR primer sequences were used: DaTOM70-RT-F (5′-GCAGCTGGCACTCCTGGTGG-3′) and DaTOM70-RT-R (5′-CAGTGCCTTGGCACGCCTGA-3′).
Western blot analysis and antibodies
Ventricular tissues or myocytes were lysed in radioimmunoprecipitation (RIPA) buffer, and the lysates were heated to 95 °C, size-fractionated in NuPAGE gels (Invitrogen) and transferred to polyvinylidene fluoride (PVDF) membranes. The intensities of the immunoblots were analyzed using Quantity One imaging software (Bio-Rad).
The mouse monoclonal antibodies Tom70 (sc-390545, Santa Cruz), Tom20 (sc-17764, Santa Cruz) and Opa1 (sc-393296, Santa Cruz), and the goat polyclonal antibody ANP (sc-18811, Santa Cruz), were used to detect the relative protein expression levels and physical interactions. The mouse monoclonal antibody against Zn-8 (1:1 000) was from the Developmental Studies Hybridoma Bank.
Statistical analysis
The analysis was performed using GraphPad Prism (version 6.00 for Mac, GraphPad Software). An unpaired t-test with Welch's correction or a one-way ANOVA test was used to compare the experimental groups with their appropriate controls, unless otherwise specified. Results were considered significant when P < 0.05 and are expressed as the means ± SEM.
References
WRITING COMMITTEE MEMBERS, Yancy CW, Jessup M, et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American college of cardiology foundation/American heart association task force on practice guidelines. Circulation 2013; 128:e240–e319.
Jessup M, Brozena S . Heart failure. N Engl J Med. 2003; 348:2007–2018.
Frey N, Katus HA, Olson EN, Hill JA . Hypertrophy of the heart: a new therapeutic target? Circulation 2004; 109:1580–1589.
Frey N, Olson EN . Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol 2003; 65:45–79.
Abel ED, Doenst T . Mitochondrial adaptations to physiological vs pathological cardiac hypertrophy. Cardiovasc Res 2011; 90:234–242.
Rosca MG, Tandler B, Hoppel CL . Mitochondria in cardiac hypertrophy and heart failure. J Mol Cell Cardiol 2013; 55:31–41.
Schmidt O, Pfanner N, Meisinger C . Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol 2010; 11:655–667.
Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N . Importing mitochondrial proteins: machineries and mechanisms. Cell 2009; 138:628–644.
Joseph AM, Ljubicic V, Adhihetty PJ, Hood DA . Biogenesis of the mitochondrial Tom40 channel in skeletal muscle from aged animals and its adaptability to chronic contractile activity. Am J Physiol Cell Physiol 2010; 298:C1308–C1314.
Colavecchia M, Christie LN, Kanwar YS, Hood DA . Functional consequences of thyroid hormone-induced changes in the mitochondrial protein import pathway. Am J Physiol Endocrinol Metab 2003; 284:E29–E35.
Schmidt O, Harbauer AB, Rao S, et al. Regulation of mitochondrial protein import by cytosolic kinases. Cell 2011; 144:227–239.
Grimm D, Elsner D, Schunkert H, et al. Development of heart failure following isoproterenol administration in the rat: role of the renin-angiotensin system. Cardiovasc Res 1998; 37:91–100.
Simpson P, McGrath A, Savion S . Myocyte hypertrophy in neonatal rat heart cultures and its regulation by serum and by catecholamines. Circ Res 1982; 51:787–801.
Bakkers J . Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res 2011; 91:279–288.
Seddon M, Looi YH, Shah AM . Oxidative stress and redox signalling in cardiac hypertrophy and heart failure. Heart 2007; 93:903–907.
Heineke J, Molkentin JD . Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol 2013; 7:589–600.
van Berlo JH, Maillet M, Molkentin JD . Signaling effectors underlying pathologic growth and remodeling of the heart. J Clin Invest 2013; 123:37–45.
Takimoto E, Kass DA . Role of oxidative stress in cardiac hypertrophy and remodeling. Hypertension 2007; 49:241–248.
Liesa M, Palacín M, Zorzano A . Mitochondrial dynamics in mammalian health and disease. Physiol Rev 2009; 89:799–845.
Young JC, Hoogenraad NJ, Hartl FU . Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 2003; 112:41–50.
Suzuki H, Maeda M, Mihara K . Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J Cell Sci 2002; 115:1895–1905.
Curado S, Ober EA, Walsh S, et al. The mitochondrial import gene tomm22 is specifically required for hepatocyte survival and provides a liver regeneration model. Dis Model Mech 2010; 3:486–95.
Youle RJ, van der Bliek AM . Mitochondrial fission, fusion, and stress. Science 2012; 337:1062–1065.
Piquereau J, Caffin F, Novotova M, et al. Down-regulation of OPA1 alters mouse mitochondrial morphology, PTP function, and cardiac adaptation to pressure overload. Cardiovasc Res 2012; 94:408–417.
Duvezin-Caubet S, Jagasia R, Wagener J, et al. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J Biol Chem 2006; 281:37972–37979.
Tanaka K, Honda M, Takabatake T . Redox regulation of MAPK pathways and cardiac hypertrophy in adult rat cardiac myocyte. J Am Coll Cardiol 2001; 37:676–685.
Ramonet D, Perier C, Recasens A, et al. Optic atrophy 1 mediates mitochondria remodeling and dopaminergic neurodegeneration linked to complex I deficiency. Cell Death Differ 2013; 20:77–85.
Ishihara N, Fujita Y, Oka T, et al. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J 2006; 25:2966–2977.
Kato H, Lu Q, Rapaport D, et al. Tom70 is essential for PINK1 import into mitochondria. PLoS ONE 2013; 8:e58435.
Vincow ES, Merrihew G, Thomas RE, et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci USA 2013; 110:6400–6405.
Arany Z, Novikov M, Chin S, et al. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc Natl Acad Sci USA 2006; 103:10086–10091.
Westerfield M . The zebrafish book: A guide for the laboratory use of zebrafish (Danio rerio). Eugene, University of Oregon Press. Paperback 2007.
Acknowledgements
This work was supported by the National Key Basic Research Program of China (2013CB531100, to Y-H C), the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (81221001, to Y-H C), the Major International Joint Research Program Fund of China (81120108004, to Y-H C), the National Natural Science Foundation of China (81170224 and 81270313, to JL; 31271214, to Y-H C), and the Shanghai Natural Science Fund (10JC1414700, to Y-H C).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Analysis of fibroblast transdifferentiation into cardiomyocytes under prohypertrophic stimulation and Tom70 reduction. (PDF 176 kb)
Supplementary information, Figure S2
Deficiency of Tom70 in zebrafish induces cardiac hypertrophy and dysfunction but does not affect early heart development. (PDF 239 kb)
Supplementary information, Figure S3
Ultrastructural characterization of cardiomyocyte mitochondria. (PDF 280 kb)
Supplementary information, Figure S4
Effects of Tom70 knockdown on mitochondria-shaping protein expression. (PDF 201 kb)
Supplementary information, Figure S5
Ultrastructural analysis of cardiomyocyte mitochondria in zebrafish. (PDF 168 kb)
Supplementary information, Figure S6
Effects of Tom20 on the biological development of zebrafish embryos. (PDF 181 kb)
Supplementary information, Figure S7
Tom70 overexpression stabilizes mitochondrial bioenergetics and morphology of cardiomyocytes. (PDF 344 kb)
Supplementary information, Figure S8
Opa1 modulates mitochondrial morphology and mediates the generation of ROS in cardiomyocytes. (PDF 229 kb)
Supplementary information, Figure S9
Early growth response-1 (EGR1) acts as a transcriptional regulator of Tom70. (PDF 214 kb)
Supplementary information, Figure S10
Effects of Pink1 reduction on the growth of primary cardiomyocytes. (PDF 231 kb)
Supplementary information, Movie S1
Knockdown of Tom70 by MO-Tom70 caused concentric growth of the myocardium in zebrafish. (MOV 238 kb)
Supplementary information, Movie S2
Cardiac-specific overexpression of Tom70 under the control of a zebrafish cmlc2 promoter. (MOV 179 kb)
Rights and permissions
About this article
Cite this article
Li, J., Qi, M., Li, C. et al. Tom70 serves as a molecular switch to determine pathological cardiac hypertrophy. Cell Res 24, 977–993 (2014). https://doi.org/10.1038/cr.2014.94
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2014.94
Keywords
This article is cited by
-
Transmembrane protein 117 knockdown protects against angiotensin-II-induced cardiac hypertrophy
Hypertension Research (2023)
-
Irisin Attenuates Apoptosis Following Ischemia–Reperfusion Injury Through Improved Mitochondria Dynamics and ROS Suppression Mediated Through the PI3K/Akt/mTOR Axis
Molecular Neurobiology (2023)
-
Targeting NAD+: is it a common strategy to delay heart aging?
Cell Death Discovery (2022)
-
Tom70 protects against diabetic cardiomyopathy through its antioxidant and antiapoptotic properties
Hypertension Research (2020)
-
Human cytomegalovirus-encoded US9 targets MAVS and STING signaling to evade type I interferon immune responses
Nature Communications (2018)