Abstract
Human single-strand (ss) DNA binding proteins 1 and 2 (hSSB1 and 2) are components of the hSSB1/2-INTS3-C9orf80 heterotrimeric protein complex shown to participate in DNA damage response and maintenance of genome stability. However, their roles at telomeres remain unknown. Here, we generated murine SSB1 conditional knockout mice and cells and found that mSSB1 plays a critical role in telomere end protection. Both mSSB1 and mSSB2 localize to a subset of telomeres and are required to repair TRF2-deficient telomeres. Deletion of mSSB1 resulted in increased chromatid-type fusions involving both leading- and lagging-strand telomeric DNA, suggesting that it is required for the protection of G-overhangs. mSSB1's interaction with INTS3 is required for its localization to damaged DNA. mSSB1 interacts with Pot1a, but not Pot1b, and its association with telomeric ssDNA requires Pot1a. mSSB1Δ/Δ mice die at birth with developmental abnormalities, while mice with the hypomorphic mSSB1F/F allele are born alive and display increased sensitivity to ionizing radiation (IR). Our results suggest that mSSB1 is required to maintain genome stability, and document a previously unrecognized role for mSSB1/2 in the protection of newly replicated leading- and lagging-strand telomeres.
Similar content being viewed by others
Introduction
Single-strand DNA (ssDNA) binding proteins are essential for DNA replication, DNA recombination/repair and the maintenance of genomic stability (reviewed in1). A hallmark of these proteins is that they possess oligonucleotide/oligosaccharide binding folds (OB-folds) that bind ssDNA2. In eukaryotes, the major SSB is replication protein A (RPA), a heterotrimeric protein critically important for DNA replication and repair of DNA double-strand breaks (DSBs). During the cell cycle, DSBs can be repaired either by the error-prone non-homologous end joining (NHEJ) or the error-free homologous recombination (HR) pathways, depending on the nature of the DSB and when the break occurs. Central to the DNA damage response (DDR) are the MRN (Mre11, Rad50, Nbs1) complex and ATM and ATR protein kinases. The MRN complex directly recognizes DSBs and activates ATM. Importantly, during HR-mediated repair, the MRN complex, in cooperation with Sae2/CtIP, resects the DSB to generate ssDNA3,4. RPA then binds to the ssDNA and recruits ATR to sites of DNA damage, resulting in the phosphorylation of a variety of factors, including Chk1 that then target downstream effector proteins involved in modulating HR-mediated repair and cell cycle progression5,6. DNA strand exchange catalyzed by the Rad51 protein forming a nucleoprotein filament with the ssDNA completes HR7.
ssDNA-binding proteins also play important roles in telomere homeostasis. Telomeres consist of TTAGGG repetitive sequences that terminate in a 3′ ss G-rich overhang, and invasion of the 3′ end into duplex regions of the telomere is postulated to be important to protect DNA ends from being recognized by RPA, which would otherwise activate ATR-dependent DNA damage checkpoint responses8,9. However, this protective DNA structure must be resolved when telomeres are replicated during S-phase. The proper coordination of telomere end protection and replication depends critically on a number of telomere binding proteins, in particular, the ss telomere binding protein protection of telomere 1 (POT1). All POT1 proteins contain two highly conserved OB-folds to bind to the 3′ terminus of the ss G-overhang10. The mouse genome encodes two POT1 proteins, POT1a and POT1b, each with distinct functions at telomeres. POT1a is required to repress an ATR-dependent DDR at telomeres in a cell cycle-dependent manner by out-competing RPA at newly replicated telomeres, while POT1b orchestrates the formation of newly synthesized ss telomeric G-overhangs11,12,13,14,15. These studies illustrate the importance of ssDNA-binding proteins in coordinating telomere replication with end protective functions.
Recently, two novel ssDNA-binding proteins, SSB1 and SSB2, have been identified in eukaryotes16. Both proteins contain a highly conserved OB-fold. Following ionizing radiation (IR)-induced DNA damage, human SSB1 interaction with ATM, is stabilized by ATM-dependent phosphorylation and localizes to damaged DNA as discrete foci16. Cells depleted of hSSB1 or hSSB2 become hypersensitive to IR and exhibit reduced efficiency in HR-dependent repair, suggesting that both proteins are required for proper repair of damaged DNA16,17,18,19. Recent studies revealed that hSSB1 and hSSB2 form two separate heterotrimeric complexes with the proteins INTS3 and C9orf80, termed sensor of ssDNA complex 1 and 2 (SOSS1/2)17,18,19,20. Although RPA recruitment to damaged DNA requires both CtIP and the MRN complex, localization of SOSS1 to damaged DNA in S/G2 phase requires only MRN17. In addition, SOSS1 forms damage foci independent of RPA16,17, and unlike RPA, hSSB1 does not localize to DNA replication foci17. These results reinforce the notion that RPA and SOSS1 localize to DSBs via distinct mechanisms, and likely play independent functions at DSBs.
To ascertain the in vivo functions of SSB1, we generated conditional knockout of the mouse ortholog of human SSB1, Obfc2b (henceforth termed mSSB1). mSSB1-null mice experience neonatal lethality. Surprisingly, we discovered previously unrecognized roles for both mSSB1 and mSSB2 (also called Obfc2a, the ortholog of hSSB2) in telomere end protection. Deletion of mSSB1 and removal of TPP1-POT1a/b resulted in increased chromatid fusions, which increased further when mSSB2 expression was repressed. Site-directed mutagenesis analysis revealed that localization of mSSB1 to damaged DNA requires an OB-fold essential for both DNA binding and interaction with INTS3. Our results suggest that in addition to their functions at genomic DSBs, mSSB1 and mSSB2 also play important roles in protecting newly replicated telomeres.
Results
mSSB1 deletion results in increased chromosomal aberrations
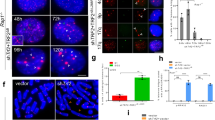
The mSSB1 gene contains seven exons, encoding a 212-amino acid protein, with the translational initiation site located within exon 2. To characterize the function of mSSB1 in vivo, we generated a conditional knockout allele of mSSB1 by introducing loxP sites flanking exons 2-6 in the mSSB1 locus (Figure 1A). Two independently targeted ES cell lines were generated to produce mSSB1F/+ and mSSB1F/F mice and mouse embryo fibroblasts (MEFs). Expression of adenoviral Cre recombinase in mSSB1F/F MEFs resulted in the efficient depletion of the mSSB1 transcript (Supplementary information, Figure S1A-S1C). We were careful not to disrupt the essential gene RNF4121 that lies only 2 kb upstream of mSSB1 (Supplementary information, Figure S1B). We crossed mSSB1F/F mice with the ZP3-Cre deleter mouse to generate mSSB1Δ/Δ MEFs. In wild-type (WT) and mSSB1Δ/+ MEFs, both mSSB1 and mSSB2 were highly expressed, with protein levels increasing following exposure to 5-Gy IR (Figure 1B). Both mSSB1 and mSSB2 localized to γ-H2AX-positive DNA damage foci following IR treatment (Figure 1C). In mSSB1Δ/Δ MEFs, mSSB1 protein was undetectable, confirming that we successfully generated a mSSB1-null allele (Figure 1B and 1C). The localization of mSSB2 to IR-induced DNA damage foci was not affected upon mSSB1 deletion, in agreement with previous reports17,18,20 (Figure 1C). Interestingly, mSSB2 protein level was upregulated in the absence of mSSB1 (Figure 1B). Time course studies of IR-induced phosphorylation of ATM, ATR, Chk1 and Chk2 revealed that deletion of mSSB1 reduced, but did not abolish global DNA damage signaling (Figure 1B). Compared with WT controls, INTS3 level was also reduced in untreated mSSB1Δ/+ and mSSB1Δ/Δ MEFs, however, increased INTS3 levels were observed in mSSB1Δ/Δ MEFs after IR exposure (Figure 1B). These results suggest that while mSSB1 is required for the complete activation of a DDR, it is dispensable for DNA damage signaling, likely due to concomitant upregulation of mSSB2. To examine whether mSSB1 has a role in DNA repair, we examined metaphase spreads of WT, mSSB1Δ/+ and mSSB1Δ/Δ MEFs by telomere-FISH. A 2-fold increase in the number of chromosome aberrations was observed in mSSB1Δ/Δ MEFs, with the majority of aberrations confined to telomeres. Specifically, increased number of telomere fragments, telomere duplication at chromatid arms and chromosome ends without telomere signals were all observed (Figure 1D and 1E). After exposure to 5-Gy IR, additional increase in the number of telomere-specific aberrations was observed in mSSB1Δ/Δ MEFs (Figure 1D and 1E). These results suggests that mSSB1 plays a role at telomeres.
Generation of a conditional mSSB1 knockout mouse and increased sensitivity of mSSB1Δ/Δ MEFs to IR exposure. (A) Schematics of the mSSB1 knockout vector. The mSSB1 gene contains seven exons, illustrated as rectangles. Red areas, protein coding regions; white areas, non-coding regions; arrowheads, loxP sites; green rectangle, FRT sites; blue rectangle, PGK-neo gene. (B) Immunoblot of phosphorylated ATM, Chk1, Chk2, ATR and SSB1, SSB2, INTS3 in lysates from SV40-immortalized mSSB1Δ/Δ, mSSB1Δ/+ and WT MEFs. Cells were harvested at the indicated time points following 5-Gy IR treatment. Total p53 was used as loading control. (C) Colocalization of endogenous mSSB1 or mSSB2 with γ-H2AX in WT and mSSB1Δ/Δ MEF exposed to 5-Gy IR. (D) Chromosome aberrations (arrows) observed in WT and mSSB1Δ/Δ MEF either untreated or after exposure to 5-Gy IR. Details of the aberrations are magnified in panels below mSSB1Δ/Δ MEF. (E) Quantification of total chromosome aberration observed in D. Mean values were derived from two independent cell lines. Error bars: standard error of the mean (s.e.m.).
mSSB1 and mSSB2 localize to telomeres and participate in the repair of uncapped telomeres
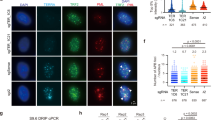
To determine whether mSSB1 and mSSB2 function at telomeres, we examined their cellular localization pattern. Endogenous mSSB1 colocalized with the telomere-binding protein TRF1 on a subset of telomeres (Figure 2A). As our mSSB2 antibody was unable to clearly detect endogenous mSSB2 by immunofluorescence, we circumvented this technical difficulty by expressing Flag-mSSB1 and Flag-mSSB2 in WT MEFs. Both mSSB1 and mSSB2 localized to telomeres in WT MEFs, with ∼30% of cells possessing >3 Flag-mSSB1- or Flag-mSSB2-positive foci at telomeres (Figure 2A and Supplementary information, Figure S2). When telomeres were rendered dysfunctional by removing endogenous TRF2 using a shRNA against TRF222, the number of telomeres containing Flag-mSSB1 and Flag-mSSB2 foci did not change significantly (Supplementary information, Figure S2). These results suggest that both mSSB1 and mSSB2 localize to telomeres regardless of their functional status. Compared with WT cells, significantly fewer telomere dysfunction-induced foci were observed in mSSB1Δ/Δ MEFs bearing dysfunctional telomeres, reinforcing the notion that complete activation of the DDR at dysfunctional telomeres decreased in the absence of mSSB1 (Supplementary information, Figure S3).
mSSB1 and mSSB2 protect telomeres. (A) Endogenous mSSB1, Flag-mSSB1 and Flag-mSSB2 colocalized with the telomeric PNA probe (telomere: Cy3-OO-(CCCTAA)4 (red)) or the telomere binding protein TRF1 on a subset of telomeres. Colocalized foci are displayed in greater detail in magnified images for Flag-mSSB1 and 2. (B) Immunoblot detection of TRF2, p-Chk2, mSSB1, mSSB2 and INTS3 in MEFs of the indicated genotypes untreated or treated with shRNA against TRF2 for 5 days. γ-tubulin is used as a loading control. (C) Examples of chromosome aberrations in metaphase spreads using Cy3-OO-(CCCTAA)4 (red) and FAM-OO-(TTAGGG)4 (green) telomere peptide nucleic acid probes. Arrows point to sites of chromosome fusions. Not all fusion sites are indicated. (D) Immunoblot detection of TPP1ΔRD, TRF2, pChk2, mSSB1, mSSB2 and INTS3 in MEFs of the indicated genotypes untreated or treated with TPP1ΔRD. γ-tubulin is used as a loading control. (E) Quantification of total telomere fusions in MEFs either untreated or treated with shRNA against TRF2. Two independent cell lines were analyzed, and a minimum of 35 metaphases and 1 500 chromosomes were scored per cell line. Mean values were derived from two independent cell lines. The one-tailed t-test was used to calculate statistical significance. Error bars: s.e.m. (F) Quantification of total telomere fusions in MEFs either untreated or treated with TPP1ΔRD and analyzed as in D. Error bars: s.e.m. (G) Immunoblot detection of p-Chk1 in WT or mSSB1Δ/Δ MEFs untreated or treated with shRNA against TRF2 or expressing TPP1ΔRD. γ-tubulin was used as a loading control.
Telomeres depleted of TRF2 only activate the ATM-Chk2-dependent DNA damage signaling pathway and are repaired by classical NHEJ (C-NHEJ)23,24. We therefore monitored the number of chromosome fusions observed in TRF2-deficient WT and mSSB1Δ/Δ MEFs. Compared with WT MEFs, mSSB1Δ/Δ MEFs depleted of TRF2 display ∼2-fold less fused chromosomes (Figure 2B, 2C and 2E). Notably, long trains of end-to-end fused chromosomes, a hallmark of TRF2 loss, were rarely observed in the setting of mSSB1 deficiency. As activation of the ATM kinase is negatively impacted when hSSB1 is depleted17,18,20, we monitored the Chk2 phosphorylation status in mSSB1Δ/Δ MEFs treated with shTRF2. Chk2 phosphorylation in mSSB1Δ/Δ MEFs appears similar to that observed in WT MEFs, although this analysis is complicated by the fact that mSSB2 levels increased following mSSB1 deletion and therefore any decline in Chk2 phosphorylation due to mSSB1 loss will not be evident (Figure 2B). In contrast, when we removed TPP1-POT1a/b from telomeres of mSSB1Δ/Δ MEFs using the dominant negative construct TPP1ΔRD, we observed significantly more chromosome fusions compared with WT cells expressing TPP1ΔRD (Figure 2C, 2D and 2F). Many of these fused chromosomes contain robust telomere signals at fusion sites, indicative of uncapped telomeres. Consistent with an overall decrease in damage signaling at dysfunctional telomeres (Supplementary information, Figure S3), Chk1 phosphorylation was reduced by mSSB1 deletion in MEFs treated with shTRF2 or TPP1ΔRD (Figure 2G). Taken together, our results suggest that mSSB1 promotes C-NHEJ-mediated DNA repair while repressing alternative-NHEJ (A-NHEJ) mediated repair at dysfunctional telomeres, as chromosome fusions in cells devoid of TPP1-POT1a/b arise from A-NHEJ-mediated repair24.
The mSSB1 OB-fold is required for both ssDNA binding and interaction with INTS3
To determine mechanistically how mSSB1 participates in telomere end protection, we used site-directed mutagenesis to interrogate the functions of a group of evolutionarily conserved amino acid (residues i.e., W55, Y74, F78, K79, Y85 and F98) within its N-terminal OB-fold (Figure 3A)20. To determine whether these residues indeed interact with ssDNA or with INTS3, we mutated them to alanines individually. Epitope-tagged 293T and mSSB1Δ/Δ MEF or mutant mSSB1 constructs were overexpressed in WT MEFs and then subjected to an oligo-dT DNA binding assay (Figure 3B and 3C)16. We found that the residues W55 and Y74 were essential for mSSB1's interaction with ssDNA, while mutant mSSB1Y85A bind ssDNA with reduced affinity. In contrast, both mSSB1WT and mSSB1F98A are efficiently bound to ssDNA (Figure 3C). Coimmunoprecipitation experiments between mSSB1 WT or mutant proteins and INTS3 revealed that only the mSSB1F98A mutant failed to interact with INTS3 (Figure 3D). This result suggests that F98 is essential for promoting mSSB1's interaction with INTS3.
The mSSB1 OB-fold is required for ssDNA binding and interaction with INTS3. (A) Schematics of conserved amino acid residues postulated to be important for DNA binding and interaction with INTS3 (red box). Residues were mutated individually to alanines. (B) Detection of mSSB1 WT and mutant proteins in 293T cells and mSSB1Δ/Δ MEFs. γ-tubulin served as loading control. (C) ssDNA binding assay. Cell lysates extracted from mSSB1Δ/Δ MEFs expressing mSSB1 WT and mutant proteins were incubated with biotin-oligo-dT40-coated streptavidin-beads. ssDNA-bound mSSB1 proteins were eluted and detected by immunoblotting. (D) Interaction between WT or mSSB1 mutants with endogenous INTS3 in mSSB1Δ/Δ MEFs was assayed by coimmunoprecipitation assays. (E) Morphology of mSSB1Δ/Δ MEFs reconstituted with WT or mutant mSSB1. (F) Immunoblot detection of mSSB1Δ/Δ MEFs reconstituted with the indicated constructs, probed with antibodies to detect phosphorylated Chk2, mSSB1 and mSSB2. γ-tubulin served as loading control. (G) Co-localization of 53BP1 with WT or mSSB1 mutants reconstituted in mSSB1Δ/Δ MEF, following exposure to 5Gy IR. 0.5% Triton treatment of mSSB1Δ/Δ MEFs expressing SSB1F98A reduced nuclear staining.
We next addressed the in vivo functions of these residues by reconstituting WT or mSSB1 mutants in mSSB1-null MEFs. While all mutants expressed well in 293T cells, efficient expression in mSSB1Δ/Δ MEFs was observed for the mSSB1WT, mSSB1Y85A and mSSB1F98A cDNAs (Figure 3B). The remaining mutants were either unable to express in mSSB1Δ/Δ MEFs (double mutant mSSB1F78A,K79A) or their expression resulted in rapid cell death (mutants mSSB1W55A and mSSB1Y74A) (Figure 3B and 3E). Therefore, all subsequent analyses were confined to mSSB1Δ/Δ MEFs reconstituted with mSSB1WT, mSSB1Y85A or mSSB1F98A. Importantly, we found that expression of mSSB1WT and mSSB1Y85A, but not mSSB1F98A, in mSSB1Δ/Δ MEFs efficiently repressed mSSB2 expression (Figure 3F). In addition, unlike reconstituted mSSB1WT controls, the reconstituted mSSB1Y85A and mSSB1F98A mutants were not able to repress chromosomal aberrations induced by increased IR exposure (Supplementary information, Figure S4). Indirect immunofluorescence following 5-Gy IR exposure revealed that mSSB1Y85A formed tiny, discrete foci, only a few of which colocalized with 53BP1 (Figure 3G). In contrast, mSSB1F98A completely failed to localize to DNA damage foci and instead displayed a diffuse pattern of nuclear and cytoplasmic staining (Figure 3G). Treatment with 0.5% Triton X-100 almost completely removed mSSB1F98A from the nucleus (Figure 3G). Taken together, these results suggest that localization of mSSB1 to sites of DNA damage requires not only an intact DNA binding domain, but also the ability to associate with INTS3.
Increased chromatid fusions in mSSB1-null MEFs
Previous studies suggest that hSSB1 and hSSB2 both form complexes with INTS3 and possess overlapping functions in the DNA damage response (DDR)17,18. The elevated expression of mSSB2 observed when mSSB1 is deleted (Figures 1C, 2B, 2C and 3F) suggests that upregulation of mSSB2 likely compensates for mSSB1 loss, thereby complicating the analysis of mSSB1 function. We found that shRNA-mediated depletion of mSSB2 in mSSB1-null MEFs resulted in rapid cell death (data not shown), precluding the further analysis of mSSB1 function. This result also suggests that at least one functional mSSB1/2 protein is essential for cell viability. To circumvent this technical limitation, we utilized the mSSB1WT and mSSB1Y85A constructs, as reconstitution of these cDNAs into mSSB1Δ/Δ MEFs was able to almost completely suppress mSSB2 expression without affecting cellular proliferation (Figures 3E, 3F and 4A). Therefore, these constructs allow us to interrogate the role of the mSSB1 OB-fold in telomere end protection without concomitant upregulation of mSSB2. We examined chromosome fusions observed in reconstituted mSSB1-null MEFs devoid of TRF2 (Supplementary information, Figure S5). Reconstitution of mSSB1Δ/Δ MEFs with mSSB1WT restored end-to-end chromosome fusions to near WT levels, while mSSB1Y85A reconstitution led to significantly less fused chromosomes observed (Figure 4B and 4C). These results confirm that mSSB1 plays a role in promoting C-NHEJ-mediated repair at telomeres when devoid of TRF2, and suggest that mSSB2 is also required in this process. In addition, compared with mSSB1WT reconstituted cells, the level of phosphoylated Chk2 was attenuated in mSSB1Y85A reconstituted MEFs, correlating with the decline in mSSB2 protein levels in this cell line (Figure 4A). This result suggests that both mSSB1 and mSSB2 are required to promote the full activation of the ATM-Chk2-dependent DDR, a result masked in mSSB1-null MEFs by the dramatic upregulation of mSSB2 (Figure 2B and 2D).
Increased chromatid fusions in mSSB1Δ/Δ MEFs. (A) Expression of HA-TPP1ΔRD, phospho-Chk2, mSSB1, mSSB2 and INTS3 in mSSB1Δ/Δ MEFs reconstituted with WT or mSSB1 mutants. Reconstituted MEFs were either untreated (vector) or treated with shTRF2 or HA-TPP1ΔRD for 5 days before immunoblot analysis. γ-tubulin served as loading control. (B) mSSB1Δ/Δ MEFs reconstituted with the indicated constructs were treated with shTRF2 or TPP1ΔRD for 5 days and metaphase spreads were analyzed by CO-FISH. Cy3-OO-(CCCTAA)4 (red) and FAM-OO-(TTAGGG)4 (green) telomere peptide nucleic acid probes were used to monitor chromosome ends. White arrows point to chromosome fusions and red arrows point to chromatid fusions. (C, D) Quantification of telomere fusions treated with shTRF2 (C) or TPP1ΔRD (D). Mean values were derived from three independent experiments from over 3 500 chromosome ends per genotype. P values were calculated by one-tailed t-test. Error bars: s.e.m.
While the majority of the chromosome fusions observed in TRF2-depleted mSSB1Δ/Δ MEFs occurred primarily in G1 or G2 phase of the cell cycle, a small percentage of chromatid fusions was observed, especially when both mSSB1 and mSSB2 functions were compromised in mSSB1Y85A reconstituted MEFs (Figure 4B and 4C). Chromatid fusions represent postreplicative repair of newly synthesized telomere ends and occur exclusively in G2 phase of the cell cycle25. We have previously shown that the TPP1-POT1a/b complex plays a critical role in the protection of newly replicated telomere ends25, and hypothesize that mSSB1/2 might also have a role in the protection of newly replicated telomeres. To test this hypothesis, we removed TPP1-POT1a/b from reconstituted mSSB1-null MEFs. The number of chromatid fusions increased significantly in mSSB1Δ/Δ MEFs, accounting for 30% of all fusions observed (Figure 4B and 4D). Chromatid fusions further increased in mSSB1Y85A reconstituted MEFs, suggesting that together with TPP1-POT1a/b, both mSSB1 and mSSB2 protect newly replicated telomere ends (Figure 4B and 4D).
mSSB1 and mSSB2 protect both leading- and lagging-strand telomeres
Newly replicated leading-strand telomeres are initially blunt ended, and must be processed by the 5′ to 3′ nuclease Apollo to generate the protective 3′ ss overhang preventing the initiation of NHEJ-mediated chromatid fusions15,25. In contrast, lagging-strand telomeres already possess the 3′ overhang due to placement of the RNA primer15,25. To ascertain whether mSSB1 and mSSB2 preferentially protect leading- or lagging-strand telomeres, we utilized chromosome-orientation FISH (CO-FISH), a technique that allows the differentiation between telomeres generated by leading- and lagging-strand DNA synthesis26, to analyze the types of chromatid fusions generated by removal of TPP1-POT1a/b in the absence of mSSB1. Random chromatid-type telomere fusions would be expected to yield a 1:2:1 ratio of leading-leading, leading-lagging and lagging-lagging chromatid fusions. Analysis of over 40 000 chromatid ends indicated that unlike ApolloΔ/Δ MEFs, which displayed mainly leading-leading chromatid fusions, deletion of mSSB1 resulted in a 1:2:1 ratio of chromatid fusions, indicating that it is required to protect both leading- and lagging-strand telomeres (Figure 5A and 5B). In support of this notion, sister chromatid fusions, the joining of leading- and lagging-strand telomeres, predominate in the absence of mSSB1, accounting for ∼70% of all chromatid fusions (Figure 5A and 5B). A significant increase in the number of chromatid fusions was observed in mSSB1Δ/Δ MEFs reconstituted with the mSSB1Y85A mutant, suggesting that mSSB2 also plays a role in protecting newly replicated leading- and lagging-strand telomeres (Figure 5C).
mSSB1 and mSSB2 protect both leading and lagging G-overhangs. (A) Examples and schematics illustrating the various chromosome and chromatid fusions observed in the absence of mSSB1 or both mSSB1 and mSSB2. (B, C) Quantification of chromatid fusions (treated with TPP1ΔRD) in WT or mSSB1Δ/Δ MEF (B) or mSSB1Δ/Δ MEFs reconstituted with WT or mSSB1 mutants (C). Mean values were derived from three independent experiments. P values were calculated by one-tailed t-test. Error bars: s.e.m. (D) Detection of G-overhang (native) and total telomere length (denature) in reconstituted mSSB1Δ/Δ MEFs by TRF Southern blot. Numbers under the native gel indicate the relative ratios of G-overhang to total telomere signal. γ-32P- (CCCTAA)4 telomere probe was used for both native and denature gel hybridizations. Molecular weight markers are indicated.
The increased number of chromatid fusions observed after deletion of mSSB1 could arise from a failure to protect the 3′ G-overhang from nucleolytic degradation. To examine this possibility, we monitored the status of the 3′ G-overhang in mSSB1Δ/Δ MEFs and reconstituted cell lines using an in-gel hybridization assay14,25. The ratio of the ss G-overhang to total telomere did not differ appreciably in mSSB1Δ/Δ MEFs expressing vector, mSSB1WT, mSSB1Y85A or mSSB1F98A (Figure 5D). Taken together, these results suggests that mSSB1 and mSSB2 are required to protect both newly replicated leading- and lagging-strand telomeres from engaging in inappropriate DNA repair, and their removal from telomeres does not result in the nucleolytic degradation of the 3′ G-overhang.
Binding of mSSB1 to ss telomeric DNA requires Pot1a
The observation that mSSB1/2 and Pot1a/b cooperate to prevent fusions of newly replicated telomeres prompted us to examine whether these two protein complexes interact with each other on telomeric ssDNA. We performed coimmunoprecipitation experiments with epitope-tagged mSSB1, Pot1a and Pot1b proteins and demonstrated that robust protein-protein interaction occurs between mSSB1 and Pot1a, but not Pot1b, even in the absence of ss (TTAGGG)6 (G) or (CCCTAA)6 (C) DNA (Figure 6A). The observation that the Pot1aF62A OB-fold mutant incapable of binding to telomeric ssDNA14 was able to interact with mSSB1 confirmed that interactions between mSSB1 and Pot1a could occur independently of telomere binding by Pot1a (Figure 6A). As mSSB1 by itself bound poorly to telomeric ssDNA (Figure 6B), we asked whether Pot1a is required to recruit mSSB1 to telomeres. Using a pull-down assay with biotinylated telomeric ssDNA oligos, we showed the recruitment of mSSB1 to telomeric ssDNA in a Pot1a-, but not Pot1b-, dependent manner (Figure 6B and 6C). Efficient recruitment of mSSB1 to telomeric ssDNA requires the Pot1a OB-fold, as the Pot1aF62A mutant was incapable of facilitating mSSB1 localization to telomeric oligos (Figure 6B). Finally, overexpression of Pot1a in WT MEFs resulted in a 2.5-fold increase in the localization of endogenous mSSB1 to telomeres (Figure 6D and 6E). Our results suggest that Pot1a interacts with mSSB1, and facilitates its localization to telomeric ssDNA.
mSSB1 interacts with Pot1a on telomeric ssDNA. (A) Detection of interaction between Pot1a, Pot1b and mSSB1 by coimmunoprecipitation. Epitope-tagged proteins were expressed in 293T cells and whole-cell lysates (WCL) were probed for proein-protein interactions. After binding of Flag-Pot1b, Myc-Pot1a or Myc-Pot1aF62A to anti-Flag or anti-Myc beads, the beads were incubated first with ssDNA (Tel-C (CCCTAA)6 or Tel-G (TTAGGG)6) and then with HA-SSB1 or Flag-SSB1. Anti-Flag or anti-Myc antibodies were used to detect bound Pot1b and Pot1a; anti-HA and anti-Flag antibodies were used to detect coimmunoprecipitated SSB1. (B) The schematic of the experiment is illustrated. Streptavidin (SA) beads coated with biotinylated (B) telomeric ssDNA (Tel-G: (TTAGGG)6) were incubated with 293T WCLs containing different ratios of Myc-Pot1a and Flag-SSB1. Proteins bound to the biotinylated ssDNA were detected by western blot analysis with anti-Myc (to detect Pot1a) or anti-Flag (to detect mSSB1) antibodies. Pot1aF62A was used to determine whether the Pot1a ssDNA-binding ability was required for mSSB1 interaction with biotinylated ssDNA. The OB-fold of Pot1a or Pot1b is illustrated as a red box. (C) Similar to B, a biotinylated telomeric ssDNA was used to show that mSSB1 interacts preferentially with Pot1a but not Pot1b. Pot1a* denotes the use of Pot1aF62A as a substitute for WT Pot1a. (D) Overexpression of Pot1a in WT MEFs resulted in increased colocalization of endogenous mSSB1 with telomeres. Telomere localization was assayed with PNA-probe (Cy3-OO-(CCCTAA)4 (red)). (E) Quantification of endogenous SSB1 colocalized to telomere in D. Cells with more than three colocalized SSB1 to telomere foci were scored positive.
mSSB1 is essential for mouse development and genome stability
The increased chromosomal aberrations observed in mSSB1Δ/Δ MEFs suggest that mSSB1Δ/Δ might be required to maintain genome stability in vivo. Live mSSB1Δ/Δ mice from mSSB1F/Δ intercrosses were never found. However, E13.5 to E17.5 mSSB1Δ/Δ embryos were viable and found at Mendelian ratios (Figure 7A). 100% of P0 mSSB1Δ/Δ pups were found dead and displayed developmental abnormalities, including growth delay and abnormal hindlimb development, in agreement with a recently published report27 (Figure 7B). This neonatal lethality phenotype suggests that mSSB1 is essential for embryonic development, and that increased mSSB2 expression cannot compensate for mSSB1 loss (Figure 7C). In support of this notion, examination of the expression profiles of mSSB1 and mSSB2 during embryonic stem (ES) cell differentiation and various embryonic stages revealed that mSSB1 was highly expressed during ES cell differentiation and during embryogenesis from E7 to E17, while mSSB2 expression remained low throughout all stages of embryo development (Supplementary information, Figure S6A). mSSB2 expression was robust only in certain adult tissues, including the testes, lung and spleen (Supplementary information, Figure S6B). These results suggest that mSSB1 expression is essential for normal murine development.
mSSB1 deletion results in neonatal lethality and increased genomic instability. (A) mSSB1 deletion results in neonatal lethality. Table of mSSB1Δ/+ intercrosses. (B) Images of neonatal mSSB1Δ/Δ pup and WT littermate. Arrow points to abnormal hindlimb. (C) Detection of mSSB1 and mSSB2 in lungs from P0 WT or mSSB1Δ/Δ pups. (D) Immunoblot detection of mSSB1 and mSSB2 in adult testis and lungs from WT or mSSB1F/F mice. (E) Immunoblot detection of mSSB1, mSSB2, pATM, pChk2 in WT or mSSB1F/F MEFs with or without exposure to 5-Gy IR. (F) Quantification of total chromosome aberrations in mSSB1F/F and WT MEFs untreated or exposed to 5-Gy IR. (G) Kaplan-Meier survival analysis of mSSB1F/F and mSSB1F/+ controls. (H) Significant difference in the ratio of spleen to body weight of age-matched female WT or mSSB1F/+ mice (others) and mSSB1F/F mice. (I) mSSB1F/F;p53−/− female mice display disseminated lymphomas, with invasion into livers (top) and kidneys (bottom). Scale bar, 25 μm.
To circumvent the prenatal lethality phenotype observed in mSSB1Δ/Δ newborns, we generated mSSB1F/F mice, which expressed mSSB1 protein at very low levels due to insertion of the Neo gene within the mSSB1 promoter (Figures 1A and 7D). This hypomorphic mSSB1F/F mouse was viable, generated at normal Mendelian ratios and appeared outwardly normal, with increased expression of mSSB2 in all tissues examined (Figure 7D and data not shown). However, mSSB1F/F MEFs were sensitive to IR exposure. Even after exposure to 5-Gy IR, mSSB1 protein expression remained low (Figure 7E). mSSB1F/FMEFs treated with 2-and 5-Gy IR displayed increased chromosomal aberrations, reinforcing earlier observations that mSSB1 is required to repair damaged DNA (Figure 7F and Supplementary information, Figure S7A).
Survival analysis revealed that mSSB1F/F mice die prematurely, with male mice dying earlier than females (median survival of male mSSB1F/F mice was 8.6 months vs 13 months for females) (Figure 7G). While the exact cause of premature death remains unclear, 50% of mSSB1F/F mice developed splenomegaly at time of death (Figure 6H, Supplementary information, Figure S7B and data not shown). These spleens developed red pulp hyperplasia but still displayed normal appearing germinal centers without any overt signs of tumor (data not shown). To further interrogate this phenotype, we generated mSSB1F/F;p53−/− mice to ascertain whether the increased genomic instability observed in mSSB1F/F mice would promote lymphomagenesis. While cancer incidence was not significantly elevated in mSSB1F/F;p53−/− mice, several mice developed disseminated lymphoma into the kidney and liver which were never found in p53−/− animals (Figure 7I and data not shown). Taken together, these results suggest that mSSB1 deletion results in elevated genome instability that is tumor-promoting in the setting of p53 deficiency.
Discussion
mSSB1/2 and the protection of newly replicated telomeres
In this report, we demonstrate previously unrecognized roles for mSSB1 and mSSB2 in telomere end protection. mSSB1Δ/Δ MEFs show a significant decrease in the number of end-to-end chromosome fusions after removal of TRF2, due to decreased activation of the ATM-Chk2 signaling pathway essential for C-NHEJ-mediated repair of telomeres24,28. In addition, mSSB1 and mSSB2 function to protect newly replicated G-overhangs. Immediately after DNA replication, leading-strand telomeres are blunt ended, while lagging-strand telomeres possess a 3′ ss telomeric overhang. We have previously shown that protection of leading-strand telomeres requires the Apollo nuclease, which mediates 5′ to 3′ nucleolytic processing of the C-strand, generating an ss G-overhang that protects from classic NHEJ-mediated DNA repair25. Along with the ss overhang already generated at lagging-strand telomeres, both overhangs are then bound and protected by the TPP1-POT1a/b complex15,25. Our current data expand on this scenario and implicate mSSB1 and mSSB2 as additional factors that participate in the protection of newly replicated ss G-overhangs. Several lines of evidence support this assertion. Whereas deletion of TPP1-POT1a/b resulted in a minimal number of chromatid fusions observed11,14,25,29,30, deletion of both mSSB1 and TPP1-POT1a/b significantly increased these fusions (Figures 4B and 5B). However, unlike the leading-leading chromatid fusions that occurs almost exclusively in Apollo−/− MEFs, leading-leading, leading-lagging and lagging-lagging chromatid fusions were characterized in MEFs lacking both mSSB1 and mPOT1a/b. This is expected if mSSB1 is involved in the protection of both leading- and lagging-strand telomeres. Suppression of mSSB2 expression with the mSSB1Y85A mutant further increased the number of chromatid fusions, suggesting that mSSB2 is also required to protect the 3′ G-overhang at both leading- and lagging-strands. Our results suggest that both Pot1a and Pot1b are required to fully protect newly replicated telomeric ends: Pot1b for the modulation of the length of the newly synthesized telomeric ss overhangs15,25, while Pot1a confers end protection in part by facilitating the interaction of mSSB1 with telomeric ssDNA. mSSB1 directly interacts with Pot1a and its affinity to telomeric ssDNA increases with increasing Pot1a concentration, suggesting that Pot1a directly recruits mSSB1 to telomeres. However, we cannot exclude the possibility that alteration in DNA structure after binding to Pot1a31 promotes mSSB1's interaction with telomeric ssDNA.
Coordination of telomere end protection and replication by ss telomere binding proteins
The localization of three distinct sets of ssDNA-binding proteins at newly replicated telomeres underscores the importance of protecting nascent ss G-overhangs from inappropriately engaging the DNA repair machinery. A model has emerged on how ssDNA-binding proteins POT1a/b and RPA coordinate telomere end protection and DNA replication, respectively13,32. After DNA replication of lagging-strand telomeres, POT1a/b binds to and protects the 3′ overhang, while at leading-strand telomeres 5′ to 3′ nucleolytic processing by Apollo generates the 3′ overhang for POT1a/b to load on15,25. During S-phase, POT1a/b is displaced transiently from telomeres by RPA, possibly as a result of the arrival of the replication machinery at telomeres32,33. The subsequent RPA to POT1a/b switch on telomeres in turn depends on additional factors, including telomere repeat-containing RNA and heterogeneous nuclear ribonucleoproteins A1 and A2/B132. The discovery that mSSB1 and mSSB2 are also required to protect newly replicated telomeres adds additional complexities to this scenario. As mSSB1 and mSSB2 are highly expressed and their levels on telomeres or damaged DNA do not vary appreciably during the cell cycle (16 and data not shown), we speculate that some mSSB1/2 are always present on telomeric ssDNA, conferring telomere end protection when Pot1a/b are displaced from telomeres. In addition, we speculate that the presence of mSSB1/2 at telomeres might function to limit RPA from binding to telomeric ssDNA. RPA does not colocalize with hSSB1 at DNA damage sites and functions independently from hSSB1 in HR-mediated DNA repair16,17. Reducing RPA accumulation on telomeric DNA by mSSB1/2 would promote TPP1-POT1a/b binding to newly replicated telomeric ssDNA, thereby promoting telomere end protection by facilitating the recruitment of additional mSSB1/2 to telomeres.
Localization of mSSB1 to damaged DNA requires both interaction with INTS3 and an intact DNA-binding domain
siRNA knockdown experiments suggest that INTS3 links hSSB1/2 and C9orf80 into heterotrimeric complexes17,18,20. As INTS3 controls the levels of hSSB1 through transcriptional regulation20, it is unclear whether hSSB1 is able to localize to damaged DNA independent of INTS3. By performing mutational analyses, we defined residues within the OB-fold that interact with ssDNA and INTS3. Reconstitution of mSSB1 mutants into mSSB1-null MEFs revealed that the mSSB1F98A mutant was not able to localize to IR-induced DNA damage sites, even though its DNA-binding domain remains intact. In contrast, a point mutation within the mSSB1 DNA-binding domain (Y85A) was able to reduce mSSB1 interaction with ssDNA in vitro. These results suggests that localization of mSSB1 to DSBs requires both its interaction with INTS3 and an intact DNA-binding domain. Interestingly, expression of mSSB1W55A and mSSB1Y74A mutants incapable of binding to ssDNA in mSSB1Δ/Δ MEFs resulted in rapid cell death. We speculate that at least one functional mSSB1/2-INTS3-C9orf80 complex is required for cell survival. Overexpression of mSSB1 DNA-binding mutants likely titrated INTS3 away from endogenous mSSB2, leading to cell death. This notion is supported by our observation that overexpression of the mSSB1F98A mutant that cannot bind INTS3 does not adversely impact cell survival.
shRNA-mediated depletion of mSSB2 in mSSB1Δ/Δ MEFs results in rapid cell death, prompting us to utilize mSSB1Δ/Δ MEFs reconstituted with the mSSB1Y85A mutant to repress mSSB2 expression without affecting cellular proliferation. Using this approach, we found that both mSSB1 and mSSB2 play roles in regulating fusions of telomeres devoid of TRF2. Chk2 phosphorylation was markedly decreased in shTRF2-treated mSSB1Δ/Δ; mSSB1Y85A MEFs (Figure 4A). As end-to-end chromosome fusions following TRF2 depletion depends strictly on an intact ATM-Chk2 pathway23,24, this result suggests that like hSSB1, both mSSB1 and mSSB2 activate the ATM-Chk2 pathway following DNA damage16,17,18,20. Our results contrast with a recent report documenting that mSSB1 and mSSB2 do not play roles in ATM-dependent DDR in primary lymphocytes and MEFs27. We speculate that the small amount of residual mSSB1 and mSSB2 proteins following conditional deletion of mSSB1 and shRNA-mediated knockdown of mSSB2 might be able to support ATM checkpoint activation in primary cells. In support of this notion, the very low levels of mSSB1 protein in our mSSB1F/F mice was able to fully support embryonic development to bypass neonatal lethality observed in mSSB1Δ/Δ mice. Finally, we cannot exclude the possibility that ATM-dependent checkpoint activation by dysfunctional telomeres might be sensitized in the SV40-immortalized mSSB1Δ/Δ MEFs utilized in our studies.
mSSB1 is essential for maintenance of genome stability
Deletion of mSSB1 results in neonatal lethality. mSSB1Δ/Δ mice display increased skeletal abnormalities due to impaired bone formation and increased apoptosis27. However, we and others found that compound deletion of both p53 and mSSB1 does not rescue this lethal phenotype (data not shown)27. While we generated CAG-CreER;mSSB1F/F mice to enable tamoxifen-mediated deletion of mSSB1 in adult tissues, we found that residual levels of mSSB1 complicated data interpretation (data not shown). To circumvent this technical limitation, we generated mSSB1F/F mice, which expressed mSSB1 at very low levels that still enabled these animals to survive. mSSB1F/F mice appeared grossly normal but died prematurely from as yet unknown causes, although splenomegaly was found in ∼50% of mice at time of death. Interestingly, mSSB1F/F;p53−/− mice displayed disseminated lymphomas never observed in age-matched p53−/− mice (Figure 7I). We postulate that in mSSB1F/F;p53−/− mice, the formation of chromosomal rearrangements as a result of incorrectly repaired telomeres likely results in increased chromosome instability to facilitate progression to cancer.
Materials and Methods
Generation of mSSB1 conditional knockout mice
A long-range PCR strategy was used to obtain three correct genomic fragments for generating a conditional targeting vector. A 2.87 kb KpnI-BglII fragment (5′ arm), a 5.24 kb HindIII-SalI fragment (3′ arm) and a 1.9 kb HindIII-SacII fragment containing exons 2-6 and the second loxP at 3′ end were inserted into pKOII vector34. The targeting vector was linearized with SalI and electroporated into AB2.1 ES cells. ES cells were screened by long-range PCR and two selected ES cell candidates (1C6, 2C3) with correct recombination were injected into C57BL/6 blastocysts. Fourteen Chimeric mice (12 males and 2 females) were mated with female or male WT C57BL/6 to generate mSSB1F/+ offsprings. mSSB1F/+ mice were intercrossed to obtain mSSB1F/F homozygous mice. Female mSSB1F/F mice were mated with zona pellucida 3 (ZP3)-Cre male mice to give rise to mSSB1F/+;ZP3-Cre mice. Female mSSB1F/+;ZP3-Cre mice were crossed with WT mice to obtain mSSB1Δ/+ heterozygous mice. The homozygous SSB1Δ/Δ mice were generated by intercrossing SSB1Δ/+ mice. p53 knockout mice were obtained from the Donehower lab35. All mice were maintained according to Yale University IACUC-approved protocols.
Generation of mSSB1Δ/Δ and mSSB1F/F MEFs
mSSB1Δ/Δ MEFs were isolated from E13.5 embryos by intercrossing mSSB1Δ/+ mice. mSSB1F/F MEFs were obtained from E13.5 embryos by intercrossing mSSB1F/+ mice. Primary MEFs at passage one were immortalized with SV40 large-T antigen retrovirus. To completely delete mSSB1, mSSB1F/F MEFs were infected with adeno-Cre virus twice for 2 days and deletion efficiency was checked by RT-PCR. Total RNA was extracted with Trizole reagent (Invitrogen) and reverse transcription was performed with Super transcriptase III Kit (Invitrogen). RT-PCR was performed with SybrGreen PCR Kit (BioRad) and reactions were run on an ABI PCR machine. All MEFs were cultured in DMEM with 10% serum.
Expression plasmids and antibodies
Full-length mouse mSSB1 or mSSB2 cDNA expression vectors were generated by inserting N-terminal Flag- or HA- tagged cDNAs into pQCXIP retrovirus vectors. Anti-human SSB1 or SSB2 antibodies were kindly provided by Dr Junjie Chen19. Anti-mouse TRF1 was a gift from Dr Jan Karlseder. Commercial antibodies used were: anti-Phospho-Chk1 (Ser345) (# 2348), anti-Phospho-ATR (Ser428) (# 2853), anti-Phospho-ATM (Ser1981) (# 4526) from Cell signaling; anti-Chk2 (# 611570) from BD Pharmingen; anti-Flag, ant-HA, anti-γ-tubulin from Sigma; anti-γ-H2AX (# 05-636) from Upstate; anti-INTS3 (FLJ21919) from Bethyl.
Reconstitution of mSSB1Δ/Δ MEFs with mSSB1 mutants and telomere uncapping
mSSB1 point mutants were generated by site-directed mutagenesis (Stratagene). mSSB1Δ/Δ MEFs were infected with retrovirus expressing mSSB1 WT and mutants and selected by puromycin for at least 1 week. To disrupt telomere end protective function, WT, SSB1Δ/Δ MEFs or SSB1Δ/Δ MEFs reconstituted with WT or mSSB1 mutants were infected with shTRF2 or HA-mTPP1ΔRDretrovirus twice, and chromosomes were harvested 5 days after first infection.
DNA binding assay and co-IP assays
Streptavidin-sepharose beads (Invitrogen) coated with Biotin-oligodT40 or Biotin-Tel-G (TTAGGG)6 was used for the ssDNA binding assay. Protein A/G-sepharose beads (GE Healthcare) was used for co-IP. Both beads were incubated with crude cell lysates in TEB150 buffer (50 mM Hepes, pH 7.3, 150 mM NaCl, 2 mM MgCl2, 5 mM EGTA, 0.5% Triton-X-100, 10% Glycerol, proteinase inhibitors) overnight at 4 °C. After washing with the same buffer, the beads were analyzed by immunoblot assay. In Figure 6A, antibody-crosslinked agarose beads were used for co-IP. Anti-Myc agarose beads (A7470) and anti-M2-flag agarose beads (A2220) were purchased from Sigma.
IR treatment of MEFs
For chromosome aberration assay, 2 × 106 of cells were seeded in 100-mm culture dish at 30% confluency overnight. After irradiation at various IR levels, cells were incubated at 37 °C for 24 h. Chromosomes were harvested after additional 4-h colcemid treatment. For immunoblot detection, IR-irradiated cells in 60-mm dishes were harvested at 0, 1, 3 or 8 h. For IR-induced foci assay, 5 × 104 of cells were seeded in 8-well chambers overnight, irradiated, allowed to recover for 2 h. Cells were pretreated with hypotonic solution (20 mM Hepes, pH 7.3, 20 mM NaCl, 5 mM MgCl2, 0.5% NP40) for 5 min on ice to reduce background and fixed with 4% paraformaldehyde solution before immunostaining.
PNA-FISH, CO-FISH and TIF assays
Cells were treated with 0.5 μg/ml of colcemid for 4 h before harvest. Trypsinized cells were treated with 0.06 M KCl, fixed with methanol:acetic acid (3:1) and spread on glass slides. Metaphase spreads were hybridized with 5′-Cy3-OO-(CCCTAA)4-3′ probe. For CO-FISH, cells were treated with BrdU for 14 h before addition of colcemid. Metaphase spreads were sequentially hybridized with 5′-PAM-OO-(TTAGGG)4-3′ and 5′-Cy3-OO-(CCCTAA)4 probes. For the TIF assay, cells were seeded in eight-well chambers and immunostained with primary antibodies and FITC-secondary antibodies, then hybridized with the 5′-Cy3-OO-(CCCTAA)4-3′ probe to detect telomeres.
TRF Southern blot
Twenty micrograms of total genomic DNA were separated by pulse-field gel electrophoresis (BioRad). The gels were dried at 50 °C and prehybridized at 58 °C in Church mix (0.5 M NaH2PO4, pH 7.2, 7% SDS) and hybridized with γ-32P-(CCCTAA)4 oligonucleotide probes at 58 °C overnight. Gels were washed with 4× SSC, 0.1% SDS buffer at 55 °C and exposed to Phosphorimager screens. After in-gel hybridization for the G-overhang under native conditions, gels were denatured with 0.5 N NaOH, 1.5 M NaCl solution and neutralized with 3 M NaCl, 0.5 M Tris-Cl, pH 7.0, then reprobed with (TTAGGG)4 oligonucleotide probes to detect total telomere DNA. To determine the relative G-overhang signals, the signal intensity for each lane was scanned with Typhoon (GE) and quantified by ImageQuant (GE) before and after denaturation. The G-overhang signal was normalized to the total telomeric DNA and compared between samples.
References
Richard DJ, Bolderson E, Khanna KK . Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Crit Rev Biochem Mol Biol 2009; 44:98–116.
Flynn RL, Zou L . Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit Rev Biochem Mol Biol 2010; 45:266–275.
Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P . Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 2007; 28:134–146.
Sartori AA, Lukas C, Jackson SP, et al. Human CtIP promotes DNA end resection. Nature 2007; 450:509–514.
Bartek J, Lukas J . DNA repair: Damage alert. Nature 2003; 421:486–488.
Zou L, Elledge SJ . Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300:1542–1548.
Moynahan ME, Jasin M . Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 2010; 11:196–207.
Griffith JD, Comeau L, Rosenfield S, et al. Mammalian telomeres end in a large duplex loop. Cell 1999; 97:503–514.
Murti KG, Prescott DM . Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc Natl Acad Sci USA 1999; 96:14436–14439.
Baumann P, Cech TR . Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 2001; 292:1171–1175.
He H, Wang Y, Guo X, et al. Pot1b deletion and telomerase haploinsufficiency in mice initiate an ATR-dependent DNA damage response and elicit phenotypes resembling dyskeratosis congenita. Mol Cell Biol 2009; 29:229–240.
He H, Multani AS, Cosme-Blanco W, et al. POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J 2006; 25:5180–5190.
Flynn RL, Chang S, Zou L . RPA and POT1: friends or foes at telomeres? Cell Cycle 2012; 11:652–657.
Wu L, Multani AS, He H, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 2006; 126:49–62.
Wu P, Takai H, de Lange T . Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 2012; 150:39–52.
Richard DJ, Bolderson E, Khanna KK, et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 2008; 453:677–681.
Huang J, Gong Z, Ghosal G, Chen J . SOSS complexes participate in the maintenance of genomic stability. Mol Cell 2009; 35:384–393.
Li Y, Bolderson E, Kumar R, et al. HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J Biol Chem 2009; 284:23525–23531.
Zhang F, Wu J, Yu X . Integrator3, a partner of single-stranded DNA-binding protein 1, participates in the DNA damage response. J Biol Chem 2009; 284:30408–30415.
Skaar JR, Richard DJ, Pagano M, et al. INTS3 controls the hSSB1-mediated DNA damage response. J Cell Biol 2009; 187:25–32.
Abdullah JM, Li X, Nachtman RG, Jurecic R . FLRF, a novel evolutionarily conserved RING finger gene, is differentially expressed in mouse fetal and adult hematopoietic stem cells and progenitors. Blood Cells Mol Dis 2001; 27:320–333.
Deng Y, Guo X, Ferguson DO, Chang S . Multiple roles for MRE11 at uncapped telomeres. Nature 2009; 460:914–918.
Dimitrova N, de Lange T . Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol 2009; 29:5552–5563.
Rai R, Zheng H, He H, et al. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J 2010; 29:2598–2610.
Lam YC, Akhter S, Gu P, et al. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J 2010; 29:2230–2241.
Bailey SM, Cornforth MN, Goodwin EH, et al. Strand-specific postreplicative processing of mammalian telomeres. Science 2001; 293:2462–2465.
Feldhahn N, Ferretti E, Nussenzweig MC, et al. The hSSB1 orthologue Obfc2b is essential for skeletogenesis but dispensable for the DNA damage response in vivo. EMBO J 2012; 31:4045–456.
Celli GB, de Lange T . DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 2005; 7:712–718.
Hockemeyer D, Daniel JP, Takai H, de Lange T . Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell 2006; 126:63–77.
Hockemeyer D, Palm W, Wang RC Couto SS, de Lange T . Engineered telomere degradation models dyskeratosis congenita. Genes Dev 2008; 22:1773–1785.
Lei M, Podell ER, Cech TR . Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol 2004; 11:1223–1229.
Flynn RL, Centore RC, Zou L, et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 2011; 47:532–536.
Verdun RE, Crabbe L, Haggblom C, Karlseder J . Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell 2005; 20:551–561.
Gu P, Min JN, Wang Y, et al. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J 2012; 31:2309–2321.
Donehower LA, Harvey M, Bradley A, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992; 356:215–221.
Acknowledgements
We would like to thank Dr Jan Karlseder (Salk Institute for Biological Studies, USA) for anti-TRF1 antibodies and Dr Junjie Chen (MD Anderson Cancer Center, USA) for anti-hSSB1 and anti-hSSB2 antibodies. This work was supported by the NCI (RO1 CA129037) to SC.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
(A) Genotyping of WT and mSSB1F/F MEFs untreated or treated with Adeno-Cre recombinase. (PDF 111 kb)
Supplementary information, Figure S2
Quantification of Flag-mSSB1 and Flag-mSSB2 localization to functional and uncapped telomeres devoid of TRF2. (PDF 40 kb)
Supplementary information, Figure S3
TIF analysis in WT and mSSB1Δ/Δ MEFs. (PDF 331 kb)
Supplementary information, Figure S4
Percentage of IR-induced chromosomal aberrations in WT MEFs and mSSB1Δ/Δ MEFs reconstituted with mSSB1 WT or mSSB1 mutant cDNAs. (PDF 53 kb)
Supplementary information, Figure S5
TRF2 mRNA levels detected by real-time RT-PCR in mSSB1Δ/Δ MEFs reconstituted with WT or mSSB1 mutants. (PDF 65 kb)
Supplementary information, Figure S6
Expression profiles of mSSB1 and mSSB2 transcripts during mouse development A and in different tissues from adult mice B. (PDF 33 kb)
Supplementary information, Figure S7
(A) Elevated number of fused chromosomes in mSSB1F/F MEFs treated with 5Gy IR. (PDF 109 kb)
Rights and permissions
About this article
Cite this article
Gu, P., Deng, W., Lei, M. et al. Single strand DNA binding proteins 1 and 2 protect newly replicated telomeres. Cell Res 23, 705–719 (2013). https://doi.org/10.1038/cr.2013.31
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2013.31
Keywords
This article is cited by
-
hSSB2 (NABP1) is required for the recruitment of RPA during the cellular response to DNA UV damage
Scientific Reports (2021)
-
Backbone 1H, 13C and 15N resonance assignments of the OB domain of the single stranded DNA-binding protein hSSB2 (NABP1/OBFC2A) and chemical shift mapping of the DNA-binding interface
Biomolecular NMR Assignments (2018)
-
Pot1 OB-fold mutations unleash telomere instability to initiate tumorigenesis
Oncogene (2017)
-
The Integrator complex controls the termination of transcription at diverse classes of gene targets
Cell Research (2015)
-
Mouse models uncap novel roles of SSBs
Cell Research (2013)