Abstract
Oncogenic mutations in RAS genes are very common in human cancer, resulting in cells with well-characterized selective advantages, but also less well-understood vulnerabilities. We have carried out a large-scale loss-of-function screen to identify genes that are required by KRAS-transformed colon cancer cells, but not by derivatives lacking this oncogene. Top-scoring genes were then tested in a larger panel of KRAS mutant and wild-type cancer cells. Cancer cells expressing oncogenic KRAS were found to be highly dependent on the transcription factor GATA2 and the DNA replication initiation regulator CDC6. Extending this analysis using a collection of drugs with known targets, we found that cancer cells with mutant KRAS showed selective addiction to proteasome function, as well as synthetic lethality with topoisomerase inhibition. Combination targeting of these functions caused improved killing of KRAS mutant cells relative to wild-type cells. These observations suggest novel targets and new ways of combining existing therapies for optimal effect in RAS mutant cancers, which are traditionally seen as being highly refractory to therapy.
Similar content being viewed by others
Introduction
Activating point mutations in the genes encoding the RAS subfamily of small GTP binding proteins contribute to the formation of a large proportion of human tumors, with ∼20% of human tumor samples carrying activating mutations in the KRAS, NRAS or HRAS oncogenes. Constitutive activation of RAS proteins provides a major contribution towards the establishment of the transformed phenotype, but several other oncogenic lesions are also required in the same cells to establish a fully malignant state 1. Removal of a RAS oncogene from human tumor cell lines or mouse model systems results in reversal of transformation 2, 3, suggesting that these tumors can show RAS oncogene addiction 4 and making RAS an attractive target for tumor therapy. However, RAS proteins themselves have so far proved impossible to inhibit selectively using drugs, so attention has shifted to targeting the downstream signaling pathways controlled by RAS, which contain several more tractable enzymes, such as RAF, MEK and AKT protein kinases and the lipid phosphoinositide (PI) 3-kinases 1. Of these, the genes encoding both BRAF and the p110α PI 3-kinase catalytic subunit (PIK3CA) have been found to be frequently activated by somatic mutation in human cancer 5, with overall mutation frequencies around 10% for each, adding to their potential importance as cancer therapeutic targets. PI 3-kinase activity is further implicated in carcinogenesis by the frequent inactivation of the tumor suppressor gene PTEN, which encodes the phosphatase that reverses the incorporation of phosphate at the 3′ position of the inositol ring 6.
While targeting pathways directly controlled by RAS provides a rational approach to therapy of RAS mutant tumors, it is likely that these cancers will also show selective dependencies on activities that are not regulated by RAS, but that are needed to adapt to stresses induced by the process of transformation, a phenomenon recently referred to as “non-oncogene addiction” 7. It is possible that determining these dependencies could suggest new approaches to the therapy of RAS mutant tumors, although it is also likely that several existing chemotherapeutic regimes already act in this area.
To investigate the nature of cell functions that are needed for the survival and proliferation of cells bearing RAS mutations, but not those lacking them, we have undertaken a large-scale RNA interference screen. Initially we sought siRNAs that would kill a KRAS mutant human colon cancer cell line, HCT-116, but not an otherwise isogenic derivative lacking the mutant allele, a classic “synthetic lethal” approach 8. The top 50 or so hits from this screen were then evaluated for their ability to kill a panel of KRAS mutant but not KRAS wild-type cell lines from various cancer types. These analyses, combined with analysis of the action of a collection of drugs using the same system, identified a number of activities that are selectively required by KRAS mutant cells, albeit in quite different ways, including GATA2, CDC6, proteasome function and topoisomerase activity. Inhibiting combinations of these targets further improved the selectivity for KRAS mutant relative to wild-type cells. These findings have implications for possible new therapeutic approaches to treating cancer.
Results
Development of an RNA-interference assay for selective killing of KRAS oncogene-addicted cancer cells
To investigate possible differential effects of KRAS gene silencing in cells carrying an oncogenic KRAS mutation compared to cells with wild-type KRAS, we used HCT-116 human colon cancer cells and the isogenic derivative, HKE-3, in which the activated, but not the normal, KRAS allele has been removed by homologous recombination 2. Figure 1A shows that siRNA-mediated knockdown of KRAS expression in HCT-116 colon cancer cells leads to a rapid and profound loss of cell viability, most likely resulting from a robust induction of apoptosis. This is evident by microscopic examination (data not shown) and detection of PARP cleavage, a well-described indicator of effector caspase activation and consequent cell death. In contrast, knockdown of KRAS expression in the isogenic cell line HKE-3, which lacks the oncogenic KRAS allele, elicits a much attenuated apoptotic response (Figure 1A). PLK1 siRNA induces apoptosis to very similar levels in both cell lines, which proliferate at a comparable rate (Supplementary information, Figure S1A). Furthermore, KRAS depletion in the mutant KRAS cells is accompanied by a rapid and marked reduction in levels of phospho-ERK and phospho-AKT, two key effectors of downstream RAS signaling (Figure 1B).
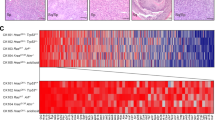
Effects of KRAS knockdown in the isogenic cell line pair and validated top hits from the large-scale KRAS differential apoptosis screen. (A) Strong apoptosis induction assessed by PARP cleavage in the KRAS mutant cell line HCT-116 as compared to an attenuated response in HKE-3 cells. Non-targeting “Scrambled” and GAPDH siRNAs serve as negative controls and knockdown of PLK1 exhibits strong apoptosis induction in both cell lines. (B) Knockdown of KRAS results in reduced levels of phospho-ERK and phospho-AKT in the KRAS mutant cell line. (C) Reproduction of the results shown in A by duplexed fluorescence microplate assay. Apoptosis induction in HCT-116 and HKE-3 is shown for the panel of control siRNAs shown in A. Calculation of an apoptosis ratio between the isogenic cell lines provides a measure of the much stronger induction of apoptosis in HCT-116 as compared to HKE-3. Data are represented as mean ± SD. (D) Validated top siRNA primary screen hits. Tabulated mean differential apoptosis-induction data from six independent validation experiments using the original siRNA pools to compare HCT-116 with HKE-3 cells together with complementary data from four independent validation experiments comparing HCT-116 with HKH-2 cells. Only three genes failed to validate across the alternative isogenic cell line comparison. Data are represented as mean ± SD. (E) STRING plot showing predicted protein-protein interactions among the top 52 hits.
The induction of apoptosis and resultant loss of cell viability can be assessed by a duplexed fluorescence microplate-based assay, suitable for high-throughput screening (HTS) applications 9. This assay employs a fluorescent DEVD effector caspase peptide substrate together with a redox dye as a monitor of cell number, allowing for direct quantitation of apoptosis levels and normalization to cell number. Figure 1C illustrates that KRAS knockdown in HCT-116 cells results in a strong induction of apoptosis whereas HKE-3 cells are much less affected. The specificity of this pro-apoptotic effect of siRNA-mediated KRAS depletion in KRAS mutant relative to KRAS wild-type cells is further illustrated in Supplementary information, Figure S1. For most of the analyses and screens described here we used pools of four siRNAs against each gene (“SMARTpools” from Dharmacon). However, deconvolution of siRNA pools into their constituent individual oligonucleotides is an important step in minimizing the potential for off-target effects to compromise the analysis of gene knockdown studies 10. Supplementary information, Figure S1B demonstrates that deconvolution of both KRAS siRNA SMARTpool and an alternative KRAS-“On Target Plus” (OTP) siRNA SMARTpool (chemically modified to reduce the potential for off-target effects), each consisting of four different oligonucleotides, clearly leads to the differential induction of apoptosis in the KRAS mutant cells. Moreover, a striking differential apoptosis in response to KRAS depletion is also observed when HCT-116 parental cells are compared to an alternative clone of mutant KRAS-deleted cells, HKH-2 (Supplementary information, Figure S1C). In addition, a parallel system of paired KRAS mutant and wild-type isogenic colon cancer cell lines, DLD-1 and DKO-4, can also be employed to demonstrate the differential induction of apoptosis in KRAS mutant cells in response to KRAS knockdown using this assay system (Supplementary information, Figure S1D). Lastly, the use of allele-specific siRNAs, designed to target preferentially either the mutant or the wild-type KRAS allele, further serves to illustrate the dependence of the KRAS mutant cells on the activated KRAS allele for the suppression of pro-apoptotic signaling (Supplementary information, Figure S1E and S1F). Mutant KRAS selective siRNA has little effect on HKE-3 cells, suggesting that the low but significant amount of apoptosis seen in HKE-3 when unselective KRAS siRNA is used may be due to the loss of expression of the wild-type KRAS allele.
A large-scale siRNA screen identifies candidate genes selectively essential for the survival of cells bearing an oncogenic KRAS allele
Despite considerable efforts over a number of years, the specific targeting and inactivation of RAS proteins for therapeutic purposes has proved largely unsuccessful 1. Thus, in an effort to identify novel targets that may perform functions that are critical to the maintenance and survival of tumor cells carrying an activating KRAS mutation, but not to cells lacking this oncogene, we performed a large-scale siRNA differential apoptosis screen. HCT-116 and HKE-3 cells were screened in parallel, each in quadruplicate, using a library of more than 7 000 siRNA pools targeting the druggable human genome, employing the activated effector caspase activity and cell number assays in a HTS format (Supplementary information, Figure 2A). Following data normalization, Z-scores were calculated for each cell line (Supplementary information, Table S1) and, for differential hit calling, a ΔZ-score cut-off value of 3.3 was selected to generate a primary hit list of 89 genes (∼ 1.2% of the total number of genes screened). KRAS itself was placed ninth in this ΔZ-score ranking list, scoring very highly in HCT-116 and weakly in HKE-3 cells and thereby serving as an important internal control (Supplementary information, Table S1). Of the remaining 88 genes, 18 with a Z-score in excess of 2.0 in the HKE-3 cell line alone were eliminated from further analysis, following the reasoning that high levels of apoptosis resulting from siRNA-mediated silencing of these genes would likely constitute an undesirably strong cytotoxic effect in wild-type KRAS cells (Supplementary information, Figure S2B and S2C). Thus, our starting list for further validation comprised 70 candidate genes.
As an initial hit-validation step we performed six independent repeat apoptosis assays in the HCT-116/HKE-3 cell system using siRNA pools measured against a series of positive and negative controls including KRAS siRNA as an HCT-116-specific apoptosis inducer and CDC2L2 siRNA, which scored lowest in the ΔZ-score ranking list (Supplementary information, Table S1), as an HKE-3-specific apoptosis inducer. As a consequence of this analysis, by adopting a KRAS mutant/wild-type threshold cut-off of 1.25, we eliminated 10 relatively weak differential pools (Supplementary information, Figure S2B and S2C). Next, we performed pool deconvolution experiments with the 60 consistently strong differential pools that remained. The four individual siRNAs comprising each pool were assessed in two independent assays for their ability to induce differential apoptosis in the mutant/wild-type KRAS assay. Of the 60 pools assayed in this fashion, eight failed to pass the threshold of three or more positive oligonucleotides out of four per pool (Supplementary information, Figure S2B and S2C), leaving a validated differential apoptosis list of 52 genes.
As an additional approach to assess the wider applicability of the validated hit list in distinguishing between isogenic KRAS mutant and wild-type cells, we compared HCT-116 cells to the alternative mutant KRAS-deleted isogenic clone HKH-2. As anticipated, the vast majority of siRNA pools in the list also induced differential apoptosis in this comparison. Figure 1D displays the list of 52 validated genes, ranked according to original differential apoptosis screen data, with mean differential apoptosis-induction scores in response to gene silencing for both cell line pairings. Supplementary information, Figure S3A shows representative pool deconvolution experiments targeting CDC6, MMP7, GATA2, PSMA1, PSMB6 and PSMD14 in the HCT-116/HKE-3 cell line pairing. The use of three additional individual siRNA duplexes per gene, targeting CDC6, PSMA1, PSMB6 and PSMD14, provides further support for the view that the KRAS mutant cells exhibit a greater degree of dependence on these genes than their KRAS wild-type counterparts (Supplementary information, Figure S3B). Analysis of mRNA levels by Q-PCR established effective knockdown of the top screen hits by the appropriate siRNA pools in both cell lines (Supplementary information, Figure S3C). In addition, effective knockdown of CDC6 upon pool deconvolution was confirmed by Q-PCR (Supplementary information, Figure S3C) and by western blotting (data not shown). Interestingly, KRAS siRNA also clearly decreases CDC6 expression in both cell lines (Supplementary information, Figure S3C, S3D); this may reflect changes in the cell cycle distribution of cells when KRAS function is ablated.
A strikingly high number of proteasome components appear among the top 52 genes. Indeed, from a total of 13 proteasome components present in the screen, eight rank within the top 300 genes and four in the final 52 selected genes. Analyzing the list of top 52 hits for protein-protein interactions using “STRING” 11 places a number of genes into two separate networks (Figure 1E), clearly emphasizing the prevalence of proteasome components.
Screening a panel of KRAS mutant and wild-type cell lines identifies genes broadly required by KRAS mutant cells
To test the impact of silencing hit-list genes from this screen in other cell types, we used the validated collection of 52 siRNA pools, together with a collection of positive and negative controls, to screen a panel of 28 tumor cell lines comprising 14 that carry an activating KRAS mutation and 14 that do not (Figure 2A). Many of the cell lines in the panel were selected from the NCI60 tumor cell line collection and are derived from cancer types that frequently harbor RAS mutations. We excluded the cell lines used in the primary differential apoptosis screen. Since the magnitude of an apoptosis response elicited by any given pro-death insult might be expected to vary considerably across such a wide range of cell lines, we chose to exploit the duplexed nature of the cell viability/apoptosis assay and evaluated these secondary screen data as distinct cell viability and cell apoptosis datasets, in the expectation of deriving maximum information from the assay results. Figure 2B illustrates the output from gene-based hierarchical clustering analysis of both viability and apoptosis datasets across the panel of KRAS mutant and KRAS wild-type cells. Positive control siRNAs that tend to produce a strong impact on both cell viability and apoptosis read-outs, such as UBB, UBC, PLK1 and NDC80, cluster closely in both analyses. More importantly, both of the siRNA pools that target KRAS also cluster tightly in both datasets together with a small number of other genes.
Use of a cell line panel for secondary screening of KRAS selective hits. (A) Cell line panel used in a secondary screen with siRNAs targeting the validated 52 genes identified by the primary screen together with a panel of control siRNAs: 14 KRAS mutant and 14 KRAS wild-type cell lines from various cancer types. (B) Hierarchical cluster analysis of both viability and apoptosis assays. Negative controls cluster closely together, as do strong pan-positive controls such as UBB/UBC and PLK1/NDC80. KRAS clusters tightly with a small set of genes including GATA2. (C) Unpaired t-test analysis of both viability and apoptosis datasets to identify genes whose knockdown has a statistically significant differential effect in KRAS mutant versus KRAS wild-type cells. In addition to KRAS, significant differences are found for a total of ten genes, four of which are common to both viability and apoptosis datasets.
The separate datasets were also analyzed for statistical significance using the unpaired t-test to compare individual gene-silencing effects across KRAS mutant and wild-type cell lines. Strikingly, both of the different KRAS siRNA pools are clearly able to distinguish between the mutant and wild-type cell lines in the panel when either dataset is assessed (Figure 2C and Supplementary information, Figure S4A-S4D). Moreover, nine additional genes in the viability dataset and five genes in the apoptosis dataset achieve statistical significance in discriminating between KRAS mutant and wild-type cells. In addition to KRAS, four targets, MMP7, CDC6, GATA2 and LAMB3, score as statistically significant in both datasets (Figure 2C), with PSMD14 significant in the apoptosis dataset and NHP2L1, MAP4K1, P11, POLR2B and E2F1 in the viability dataset. Additional analysis approaches, using Mann-Whitney and Kolmogorov-Smirnov statistical tests (Supplementary information, Figure S4A-S4D), also identify MMP7, CDC6 and GATA2 together with KRAS as targets that are most closely associated with the survival of mutant KRAS cells.
Further analysis of target knockdown by our top siRNA hits in HCT-116 and HKE-3 cells reveals that, although the intended targets are successfully downregulated by their cognate siRNA pools, transfection with MMP7 siRNA also leads to a striking reduction in the levels of expression of KRAS protein and mRNA (Supplementary information, Figure S3D). This effect is also apparent when several other cell lines were tested in a similar manner (data not shown). Deconvolution of the MMP7 siRNA pool pinpoints one individual duplex as being responsible for this KRAS-specific off-target effect (Supplementary information, Figure S3D). Thus, the appearance of MMP7 in our validated hit list is due predominantly to a profound off-target knockdown of KRAS itself and MMP7 is discounted from further consideration.
Selective induction of apoptosis in KRAS mutant cells in response to CDC6 silencing and to proteasome inhibition
An in-depth characterization of the role of the GATA2 transcription factor in KRAS mutant cancer is the subject of a separate manuscript 12. In addition, we sought to further assess the effects of CDC6 knockdown and of proteasome inhibition in KRAS mutant cells.
Given the established role of CDC6 in the control of DNA replication initiation, we were interested in determining whether acute loss of CDC6 expression is characterized by an increase in DNA damage signaling with concomitant effects on cell cycle distribution and whether any such effects would be more apparent in RAS mutant cells. Consistent with its role in DNA synthesis, Figure 3A illustrates that CDC6 silencing in both KRAS mutant and wild-type cells leads to a marked reduction in the number of cells in S phase that is accompanied by an increase in the number of cells in G1. The less evident increase in cells accumulating in G2/M occurs only in HCT-116 cells. To investigate these cell cycle effects in more detail, we employed a BrdU pulse-chase experiment to follow the fate of cells labelled in S phase, with or without CDC6 knockdown, over a period of 6 h. Figure 3B illustrates that S phase-labelled HKE-3 cells fail to progress and remain stably arrested in response to CDC6 depletion, whereas S phase-labelled HCT-116 cells continue to cycle even in the absence of CDC6 expression.
CDC6 knockdown leads to increased DNA damage in RAS mutant cells. (A) CDC6 knockdown for 48 h leads to loss of S phase cells in both HCT-116 KRAS mutant and HKE-3 KRAS wild-type cells, as assessed by flow cytometry. CDC6-depleted HCT-116 cells also exhibit an increase in the proportion of G2/M phase cells that is not evident in CDC6-depleted HKE-3. Non-targeting “Risc-free” siRNA serves as a negative control. (B) A pulse of BrdU, used to label S phase cells after a 48-h CDC6 depletion, followed by a 6-h chase, indicates that whereas HKE-3 cells are stably arrested, HCT-116 cells continue to move through S phase. Non-targeting “Risc-free” siRNA serves as a negative control to monitor the normal S phase traverse of each cell line over the same time period. (C) Strong induction of DNA damage assessed by γH2AX, p53 and p21 accumulation in response to depletion of CDC6 over two and three days in HCT-116 as compared to an attenuated response in HKE-3 cells. Non-targeting “Risc-free” siRNA serves as a negative control. (D) In contrast to GATA2 knockdown, CDC6 knockdown produces no synergistic synthetic lethal pro-apoptotic effect when combined with proteasome inhibition. 24 h after siRNA transfection cells were treated with a wide titration range of the proteasome inhibitor bortezomib and apoptosis induction was measured 24 h later.
Parallel western blotting analysis (Figure 3C) reveals that the CDC6-depleted KRAS mutant cells contain elevated levels of the tumor suppressor p53 and correspondingly increased levels of the product of the p53 transcriptional target gene CDKN1A, the cyclin-dependent kinase inhibitor, p21WAF1/CIP1. In contrast, CDC6 knockdown in wild-type KRAS cells over the same time period does not elicit upregulation of p53, although p21WAF1/CIP1 levels are modestly increased from a relatively high steady-state level. In keeping with the robust p53 response to CDC6 loss-of-expression, HCT-116 KRAS mutant cells also strongly induce expression of γH2AX, a C-terminally phosphorylated histone variant localized to sites of DNA damage, whereas CDC6-depleted HKE-3 cells exhibit a considerably diminished response. Taken together, these data suggest that, while both HCT-116 and HKE-3 cells upregulate p21WAF1/CIP1levels in response to CDC6 depletion, the RAS mutant cells exhibit a much stronger DNA damage response, most likely as a consequence of overriding cell cycle checkpoint control.
The differential apoptosis/viability siRNA screen (Figure 1) identified several components of the proteasome among the validated top 52 genes hit list (PSMA1, PSMA2, PSMB6, PSMD14) and functional enrichment analysis of this list compared to the rest of the library indicates the proteasome complex as being significantly overrepresented. In addition, PSMD14 (also known as POH1, a Lys63-specific deubiquitinase of the 26S proteasome) shows significant selectivity for KRAS mutant cell lines in the apoptosis assay with the larger panel of 28 cancer cell lines. In light of the fact that the connectivity map (Figure 1E) shows that proteasome components functionally interact with CDC6, we sought to determine whether CDC6 knockdown would result in additional synthetic lethal effects when combined with proteasome inhibition. We elected to compare the induction of apoptosis in response to the depletion of CDC6 with the pro-apoptotic response to depletion of GATA2, another highly significant screen hit, against a background of chemical inhibition of proteasome activity. Figure 3D demonstrates that combining CDC6 siRNA with the small-molecule proteasome inhibitor bortezomib leads to an apparently additive effect in both HCT-116 and HKE-3 cells, with respect to apoptosis induction, with each treatment alone showing selectivity towards the RAS mutant cells. Interestingly, whereas the combination of GATA2 siRNA and bortezomib also produces an additive effect in HKE-3 cells, combined GATA2 silencing and proteasome inhibition in HCT-116 cells leads to a striking increase in apoptosis with increasing drug dosage, indicating a degree of synergy for this combination in RAS mutant cells.
To address in greater detail whether mutant KRAS cells are selectively sensitive to proteasome inhibition, we assessed the impact of several small-molecule chemical inhibitors of proteasome function on cell viability and apoptosis in the HCT-116 isogenic cell line system. Figure 4A demonstrates that chemical inhibition of proteasome function using the licensed cancer drug bortezomib results in a selective loss of cell viability associated with the induction of apoptosis in KRAS mutant cells. Flow cytometric analysis provides additional evidence of the differential cytotoxic impact of bortezomib treatment upon KRAS mutant versus wild-type cells, as shown by a relatively increased number of Annexin V-positive apoptotic cells and a relatively reduced residual live cell number. The specificity and selectivity of this differential response is supported by the additional use of two alternative proteasome inhibitors, MG-132 and proteasome inhibitor I (Z-Ile-Glu(OtBu)-Ala-Leu-CHO), both of which clearly elicit stronger pro-apoptotic and loss of viability responses in HCT-116 cells in comparison to either HKE-3 or HKH-2 cells (Figure 4B, Supplementary information, Figure S5A).
Differential killing of mutant RAS cells by proteasome inhibitors and by topoisomerase inhibitors. (A) Bortezomib preferentially impairs viability of HCT-116 as compared to HKE-3 or HKH-2 cells across a wide range of drug concentration (top left panel). Parallel differential apoptosis induction can also be monitored, indicated by apoptosis ratios (bottom left panel) and apoptosis-induction data for individual cell lines (top right panel – relative to control vehicle treatment). Data are represented as mean ± SD. FACS analysis to monitor annexin V staining following bortezomib treatment shows preferential induction of cell death in HCT-116 cells (bottom right panel). (B) An alternative proteasome inhibitor (MG-132) also produces a preferential viability loss in HCT-116 as compared to HKE-3 or HKH-2 cells across a wide range of drug concentrations (top panel). Parallel differential apoptosis induction is indicated by apoptosis ratios (bottom panel). (C) Topotecan preferentially impairs viability of HCT-116 as compared to HKE-3 or HKH-2 cells across a wide range of drug concentration (top left panel). Parallel differential apoptosis induction can also be monitored, indicated by apoptosis ratios (bottom left panel) and apoptosis-induction data for individual cell lines (top right panel – relative to control vehicle treatment). Data are represented as mean ± SD. FACS analysis to monitor annexin V staining following topotecan treatment shows preferential induction of cell death in HCT-116 cells (bottom right panel). (D) An alternative topoisomerase inhibitor (doxorubicin) also produces a preferential viability loss in HCT-116 as compared to HKE-3 or HKH-2 cells across a wide range of drug concentrations (top panel). Parallel differential apoptosis induction is indicated by apoptosis ratios (bottom panel).
Selective loss of viability and induction of apoptosis in KRAS mutant cells in response to chemotherapeutic drugs
To provide a complementary approach to the large-scale siRNA differential apoptosis screen, we also performed a loss-of-viability screen with the same isogenic colon cell lines using a collection of commercially available chemotherapeutic drugs, excluding proteasome inhibitors (Supplementary information, Table S2). Several of these agents induced a selective loss of viability in the KRAS mutant cells, most notably drugs that are known to inhibit DNA topoisomerases, either directly or indirectly, such as camptothecin, daunorubicin and doxorubicin (Supplementary information, Figure S5B). Both apoptosis induction and viability loss, measured in response to treatment with either topotecan, a topoisomerase I inhibitor derived from camptothecin (Figure 4C), or doxorubicin, an anthracycline intercalator that inhibits the progression of the topoisomerase II and stabilizes its complex with cleaved DNA (Figure 4D), are considerably more marked in HCT-116 cells as compared to either of the mutant KRAS-deleted derivative lines, HKE-3 and HKH-2. Furthermore, similar effects are seen with the use of other topoisomerase inhibitors such as camptothecin and etoposide (a topoisomerase II poison) and with other drugs that inhibit DNA replication and induce a DNA damage response, such as mitomycin (a DNA intercalator and crosslinker), hydroxyurea (a ribonucleotide reductase inhibitor) or aphidicolin (a DNA polymerase inhibitor) (Supplementary information, Figure S5C).
To assess the broader impact of both proteasome inhibition and topoisomerase inhibition on RAS mutant cells, we sought to determine loss of viability in response to these agents in a panel of lung cancer cells. Mutations in KRAS are relatively frequent in lung cancer and we reasoned that testing cell lines derived from a single tissue of origin would help avoid some of the heterogeneity of response likely to be manifest across a wider range of cancer types. Thus, we analyzed the response of a panel of 13 lung cancer cell lines, seven KRAS mutant and six KRAS wild-type, across a titration range of bortezomib and doxorubicin. We find that, although there is considerable variability of response across both genotypes (Supplementary information, Figure S5D, lower panels), each drug shows selectivity toward the RAS mutant cells (Supplementary information, Figure S5D, upper panels).
Effects of proteasome inhibition and topoisomerase inhibition on DNA damage, cell cycle arrest and accumulation of reactive oxygen species
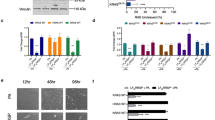
To investigate the increased sensitivity of RAS mutant cells to proteasome inhibition, we employed a cell-based proteasome activity assay, which determines the chymotrypsin-like activity associated with intact proteasomes toward a luminogenic peptide substrate. At steady state, HCT-116 and HKE-3 cell lysates contain very similar absolute chymotrypsin-like activity, with HKE-3 cells generally possessing modestly higher levels on a per-cell basis (data not shown). However, in response to proteasome inhibition across a range of concentrations of bortezomib (Figure 5A) or MG-132 (Supplementary information, Figure S5E), the chymotrypsin-like activity of HCT-116 cells is preferentially inhibited as compared to HKE-3 cells. The differential response toward proteasome inhibition is also manifest by the accumulation of ubiquitinated proteins at lower doses of bortezomib when comparing RAS mutant to wild-type cells (Figure 5B).
Analysis of the effects of proteasome and topoisomerase inhibition on RAS mutant and wild-type cells. (A) Steady-state chymotrypsin-like proteasome activity is more strongly inhibited in HCT-116 as compared to HKE-3 cells across a titration range of bortezomib. 24 h after seeding, cells were treated with bortezomib for 90 min before exposure to luminogenic substrate for 15 min. Data are represented as mean ± SD. (B) 24-h bortezomib treatment leads to a pronounced accumulation of ubiquitinated proteins at lower drug doses in HCT-116 as compared to HKE-3 cells. (C) Treatment of HCT-116 cells with a range of doses of bortezomib or topoisomerase inhibitors for 24 h leads to a stronger G2/M arrest when compared with HKE-3 cells, as assessed by flow cytometry. (D) 24-h treatment with a range of doses of topoisomerase inhibitors used in C leads to a more pronounced DNA damage response in HCT-116 when compared with HKE-3 cells, as monitored by γH2AX and p53 accumulation. (E) Steady-state ROS levels are elevated in HCT-116 cells as compared to HKE-3 counterparts, as determined by flow cytometry. (F) Topoisomerase inhibition for 24 h, using 600 nM topotecan, further increases ROS levels in both HCT-116 and HKE-3 cells, as determined by flow cytometry. Hydrogen peroxide treatment for 90 min serves as a positive control to assess the response of each cell line to oxidative stress.
A number of studies have shown that proteasome and topoisomerase inhibition can promote cell cycle arrest and we therefore evaluated the consequences of bortezomib, topotecan and doxorubicin treatments on the cell cycle profiles of HCT-116 and HKE-3. Flow cytometric analysis (Figure 5C) illustrates that in response to all three agents, mutant RAS cells generally exhibit a stronger G2/M arrest than their wild-type counterparts (see also Supplementary information, Figure S5F). In addition, bortezomib and topotecan treatments provoke a more evident depletion of G1 phase cells from HCT-116, whereas doxorubicin tends to lead to an accumulation of cells in G1 in HKE-3. Figure 5D demonstrates that in response to the same topoisomerase inhibitors, levels of p53 increase much more strongly in HCT-116 cells than in HKE-3 cells. Moreover, both inhibitors elicit a striking dose-dependent induction of γH2AX in HCT-116 in contrast to HKE-3. Given that oncogenic RAS expression is frequently associated with elevated levels of reactive oxygen species (ROS), a known potential source of endogenous DNA damage, we were interested to determine whether ROS levels might play a role in the differential response of RAS mutant cells to proteasome and topoisomerase inhibitors. Figure 5E demonstrates that HCT-116 cells harbor considerably more ROS at steady state as compared to HKE-3 cells. Upon treatment with topotecan levels of ROS increase strikingly in both cell lines (Figure 5F). Taken together, these data suggest that DNA damage signaling produced in response to chemotherapeutic drugs may be augmented by existing ROS in mutant RAS cells above a critical threshold beyond which the ability of existing cellular detoxifying mechanisms may become overwhelmed.
Inducible oncogenic Ras signaling sensitizes cells to topoisomerase inhibition but not proteasome inhibition
To further investigate the role of oncogenic RAS signaling in the differential induction of cell death in response to DNA damage or proteasome inhibition, we used a 4-hydroxytamoxifen (4OHT)-inducible oncogenic RAS fusion protein with estrogen receptor hormone binding domain that allows rapid and titratable induction of downstream signals emanating directly from the mutant RAS protein 13. We stably expressed this inducible RAS fusion protein in the HKE-3 cell line, allowing the re-activation of oncogenic RAS in an acute fashion 14. This provides a means to assess whether the enhanced sensitivity of cells with activated RAS to the chemotherapeutic drugs described above represents an acute synthetic lethal response, such as might be expected from direct conflict of the cell's response to drug-induced damage with the activated signaling pathways emerging from RAS. Alternatively, the selective disadvantage of the mutant RAS-expressing cells could be due to a more chronic addiction phenotype that might reflect the result of selective pressures acting over prolonged periods in vivo during the evolution of the tumor and not readily mimicked by short-term treatments in vitro. Strikingly, acute re-activation of oncogenic RAS in HKE-3.ER-RAS cells produces no discernible effect on the induction of apoptosis in response to bortezomib treatment (Figure 6A, left panel), with these cells showing the same sensitivity to proteasome inhibition whether or not RAS signaling is activated. In stark contrast, topotecan-induced apoptosis is strongly enhanced by acute RAS activation (Figure 6A, right panel) to similar levels as those seen in the HCT-116/HKE-3 or HCT-116/HKH-2 isogenic cell line pairings (Figure 6B, left panel and compare Figure 6B with Figure 4A and 4C). The use of alternative proteasome inhibitors and DNA topoisomerase inhibitors unequivocally support these data (Figure 6B, right panel). RAS-dependent enhancement of camptothecin-induced apoptosis in HKE-3.ER-RAS cells can be modulated across a broad range of drug concentration by titrating 4OHT in order to control the intensity of oncogenic RAS activation (Figure 6C).
Inducible oncogenic RAS signaling sensitizes cells to topoisomerase inhibition. (A) In contrast to the comparison between HCT-116 and HKE-3 cells, 4-hydroxy-tamoxifen-inducible oncogenic RAS in an HKE-3 cell background does not confer preferential induction of apoptosis in response to bortezomib treatment (left panel). Conversely, topotecan treatment does still elicit a clear differential apoptosis response when RAS-induced cells are compared to uninduced cells (right panel). Data are relative to control vehicle-treated cells and are represented as mean ± SD. (B) Ratio of apoptosis induced in mutant versus wild-type or ER-RAS-induced versus uninduced cells treated with bortezomib or topotecan (left panel). HKE-3 cells carrying a 4-hydroxy-tamoxifen-inducible oncogenic RAS construct fail to show a preferential induction of apoptosis following treatment with two additional proteasome inhibitors (MG-132 and PI-I) when comparing RAS-induced with uninduced cells. Conversely, treatment with two alternative topoisomerase inhibitors (doxorubicin and camptothecin) still results in a strong preferential induction of apoptosis when comparing RAS-induced with uninduced cells (right panel). (C) Modulation of inducible ER-RAS fusion protein activity, by titration of 4-hydroxy-tamoxifen, shows a gradual decline in apoptosis induction upon treatment with a wide range of camptothecin concentrations to baseline uninduced levels. Differential apoptosis induction is represented by plotting the induced/uninduced ratio. Data are represented as mean ± SD. (D) 24 h induction of oncogenic RAS signaling produces an increase in ROS levels that is elevated further in response to topotecan treatment.
In light of our observations that DNA topoisomerase inhibitors elicit an increase in ROS and that RAS mutant cells contain elevated levels of ROS at steady state in comparison to wild-type cells (see Figure 5E and 5F), we were prompted to evaluate the production of ROS in HKE-3.ER-RAS cells in response to 4OHT. Figure 6D demonstrates that ROS levels increase markedly upon acute oncogenic RAS activation and that this effect is further exacerbated by topotecan treatment, with ROS production reaching levels comparable to those of topotecan-treated HCT-116 cells (see Figure 5F). Taken together, these data suggest that the mechanisms for selective sensitivity of RAS mutant cells to proteasome inhibitors and DNA damaging agents are fundamentally different. Proteasome inhibitors show no sign of acute synthetic lethality with RAS activation: selective toxicity in RAS mutant cancer cells is likely to reflect non-oncogene addiction of the tumor cells to proteasome function to deal with proteotoxic stress 7. By contrast, activation of RAS shows rapid synthetic lethal effects with DNA damaging agents, likely due to the prevention of an appropriate adaptive response of the cells to the DNA damage signals, including an inability to detoxify ROS efficiently, due to overriding proliferative signals from the oncogenic RAS.
Drug combinations enhance selective killing of KRAS mutant cells
The data presented above show that RAS mutant cells are preferentially sensitive to inhibition of both the DNA replication machinery and proteasome function relative to cells lacking this activated oncogene, and that this sensitization has a fundamentally different basis for the two classes of drugs. We therefore assessed the possibility of increasing these differential sensitivity effects by combining drugs from each class of inhibitor in the hope that improved selectivity might be achieved whilst using lower drug concentrations. Figure 7A (left panel) illustrates the differential effects on cell viability as a consequence of combining the proteasome inhibitor bortezomib across a wide range of topotecan concentrations. When employed at the low doses used here, the single agents have a minimal impact on apoptosis induction and cell viability (Supplementary information, Figure S6B and see also Figure 4). However, the combination of topoisomerase and proteasome inhibition leads to a marked increase in selective apoptosis induction and viability loss in KRAS mutant as compared to wild-type cells (Figure 7A, bar-charts). These effects can be replicated when using an alternative proteasome inhibitor, MG-132 (Supplementary information, Figure S6A).
Drug combinations enhance killing of mutant RAS cells. (A) The combination of low (sub-lethal) doses of bortezomib and topotecan leads to a preferential loss of viability in HCT-116 as compared to HKE-3 cells. These effects can be seen across a wide topotecan titration range in combination with several bortezomib concentrations (left panel). Selected viability ratios are displayed (middle panel) and parallel differential apoptosis induction can also be monitored, indicated by apoptosis ratios (right panel). Data are represented as mean ± SD. (B) Different scheduling of drug treatment influences the effect of drug combinations: Adding bortezomib together with topotecan results in an elevated apoptosis induction in HCT-116 cells in comparison to HKE-3 cells as shown in A. The addition of bortezomib for 24 h prior to topotecan treatment does not elicit this response whereas addition of proteasome inhibitor 24 h post induction of DNA damage leads to an even greater differential effect. Apoptosis induction in the individual cell lines is displayed (left panel) and differential apoptosis induction is indicated by apoptosis ratios (right panel). Data are represented as mean ± SD. (C) Enhanced differential apoptosis induction effects can also be produced in response to scheduling the combination of gemcitabine and bortezomib. Selected apoptosis ratios are displayed. Data are represented as mean ± SD. (D) The application of low doses of bortezomib 24 h post induction of DNA damage can enhance the preferential loss of viability in KRAS mutant lung cancer cells as compared to KRAS wild-type cells. These effects can be seen across a wide doxorubicin titration range in combination with several bortezomib concentrations (curves representing the ratios of average values for each KRAS genotype, left panel; single data points representing individual cell lines, middle and right panels).
As an additional factor in the use of drug combinations we sought to assess whether alternative scheduling of drug treatments could have an impact on differential sensitivity of KRAS mutant and wild-type cells to the induction of apoptosis by topoisomerase I and proteasome inhibitors. Figure 7B demonstrates that whereas low-dose simultaneous treatment with bortezomib together with topotecan can enhance differential killing in a RAS-dependent fashion (as shown also in Figure 7A), pre-treatment of these cells with a proteasome inhibitor for 24 h prior to topotecan addition abrogates any differential apoptosis induction. However, reversal of this regime, by pre-treatment with topotecan for 24 h prior to proteasome inhibition results in a marked enhancement of selective killing with a substantial increase in apoptosis induction observed in RAS mutant cells (Figure 7B left panel showing apoptosis in individual cell lines, right panel showing apoptosis ratios). These effects can be replicated when using the alternative proteasome inhibitor, MG-132 (Supplementary information, Figure S6C, left panel). Moreover, this staging effect is not restricted to the specific combination of proteasome inhibitor together with an anti-topoisomerase drug as Figure 7C additionally demonstrates that inhibiting proteasome function following a prior treatment with a low dose of the anti-metabolite gemcitabine also leads to a marked increase in the induction of differential apoptosis. Gemcitabine is licensed for use in a number of tumor types, including lung and pancreas cancers where KRAS mutations are common. Further, these effects can also be replicated when using the alternative proteasome inhibitor, MG-132 (Supplementary information, Figure S6C, right panel).
To assess how the drug scheduling effects might be explained, we examined HCT-116 and HKE-3 cells for possible differences in cell cycle, DNA damage or ROS levels in response to staged treatments with the same low doses of topotecan and bortezomib. However, we observed no clear positive alteration to any individual parameter (Supplementary information, Figure S6D). Thus, although pre-treatment of RAS mutant cells with a DNA damaging agent prior to inhibition of proteasome function exacerbates the differential induction of apoptosis as compared to wild-type cells, this regime provides no obvious augmentation of ROS production beyond levels elicited by simultaneous treatment or by topoisomerase inhibition alone. Likewise, we see no clear enhanced differential effect on cell cycle arrest or DNA damage signaling, as monitored by p53 activation and γH2AX induction. One possible explanation for these observations is that the effects produced by low-dose topoisomerase treatment for 24 h on cell cycle, ROS and DNA damage (Supplementary information, Figure S6E) collectively prime the cell, rendering it more susceptible to subsequent proteasome inhibition, leading to increased cell death.
In an effort to assess the wider impact of staged topoisomerase inhibition with proteasome inhibition on mutant RAS cells, we evaluated loss of viability in response to single and staged combination treatment with doxorubicin and bortezomib in the panel of KRAS mutant and wild-type lung cancer cell lines described above. Figure 7D illustrates that despite the variability in responses across the panel (also see Supplementary information, Figure S5D), the evident trend toward selective loss of viability of mutant KRAS cells across a range of doxorubicin concentration is improved by secondary proteasome inhibition.
Discussion
Probing the vulnerabilities of RAS-mutant cancer cells with large-scale screens
The coming together of our complete knowledge of the sequence of the human genome and the development of RNA interference technology has enabled the rapid probing of the biological roles of genes on a large scale in cell-based assays. A number of screens have been reported, which aimed to identify genes whose inhibition is selectively deleterious to cells with activated mutant RAS. Scholl et al. used a library of RNAi vectors targeting about a thousand human genes, including all those known to encode kinases, to determine which of these is specifically required for the survival of tumor cells expressing an activated KRAS oncogene 15. Using a panel of cancer cell lines, the authors observed that inhibiting the expression of the protein kinase STK33 proved to be most selective of these thousand genes for killing KRAS mutant cells, leading to the suggestion that STK33 might be an attractive novel drug for treating cancers with mutant KRAS. STK33 did not feature as a hit in our screen although it should be noted that our primary screen measured apoptosis, whereas that of Scholl et al. measured cell survival. Others have reported failure to see synthetic lethality of STK33 loss with KRAS activation 16.
A second screen for selective killing of KRAS mutant cancer cell lines was reported by Luo et al., in this case using an RNAi library that targeted the entire human genome 17. This screen, carried out in colon cancer cells and followed up in a panel of lung cancer cell lines, revealed a more complex set of dependencies, with KRAS mutant cells being particularly reliant on genes in pathways related to mitotic functions, such as the Polo-like kinase PLK1, components of the anaphase-promoting complex (APC) and the proteasome. As most of these are essential cellular functions that are not directly controlled by RAS, it is likely that many of these hits reflect non-oncogene addiction 7. By way of comparison with the screen that we report here, we did not see selective effects of APC/C components and found that loss of PLK1 was toxic to both KRAS mutant and wild-type cells, perhaps due to very efficient knockdown by this siRNA pool. However, in common with the work of Luo et al., we did also clearly see the selective requirement of KRAS mutant cells for proteasome function; indeed, four of the top 52 hits from the HCT-116 cell screen were proteasome components. This provides reassurance that screens probing the same question but carried out by different methodologies in different systems can lead to at least partially overlapping results.
The Luo et al. screen used isogenic cell pairs – a cancer cell line with an activating KRAS mutation and a derivative from which the mutant KRAS allele has been removed – whereas the Scholl et al. screen used a large panel of naturally occurring cancer cell lines, with or without KRAS mutation. Our approach involved aspects of both, starting with the isogenic cell pair for a near genome-scale screen, then following up candidate hits across a large 28 cancer cell line panel. A concern with the isogenic system is that, since the HCT-116 cells are KRAS addicted, the deletion of mutant KRAS during the generation of the isogenic derivatives may have resulted in clonal selection of cells that were not representative of the parental cell line. It is certainly possible that the isogenic cell pair is not suitable for finding all genotype selective targets. However, the most prevalent model of oncogene addiction 18 suggests that on withdrawal of oncogene signaling from addicted cancer cells, the more rapid decay of survival signals relative to damage signals leads to a period of “oncogenic shock” in which the cells are particularly vulnerable, but stochastically some will survive this and will then return to a stable equilibrium after a few days. It is thus possible that the surviving clones may not be fundamentally different from the cells that died, although clearly there may be some tendency to favor rare cells with stronger pro-survival pathways.
Previously, a smaller scale siRNA screen was reported using the isogenic colon cancer cell line DLD-1 and a mutant KRAS-deleted derivative 19. The only statistically significant hits found in this report were survivin (BIRC5), CDC2 and C20ORF18, none of which were hits in the HCT-116 system that we used. PLK1 was reported to be toxic to both DLD-1 cells and mutant KRAS-deleted derivatives. Another kinome-focused RNA interference screen for KRAS synthetic lethality yielded the TANK-binding kinase 1, TBK1 20. In KRAS mutant cells, the RAS effector RalB activated TBK1 21, which maintained survival through activation of the NF-κB pathway. The NF-κB signaling cascade has been found to be required for KRAS-induced lung cancer formation in a mouse model 22, 23. We also previously carried out a smaller scale screen investigating the differential requirement of KRAS mutant cells for ∼ 2 500 genes in the HCT-116 isogenic system using a pooled inducible shRNA library 14. There was relatively little overlap between the results of this screen and the one reported here, no doubt in part due to the differing knockdown efficiencies of the synthetic siRNAs and the retroviral shRNAs, different time frames of the assays and different choice of RNA interference sequences.
GATA2 and CDC6 as novel targets in RAS mutant cells
The screen presented here yielded two novel hits that were extensively validated to be selectively lethal when targeted in KRAS mutant tumor cells relative to cells lacking this mutation. GATA2 is a transcription factor that has been traditionally considered a master regulator of the hematopoietic, and also adipocyte, cell lineages 24, 25. It is, however, much more broadly expressed than just these lineages. Knocking down GATA2 was particularly associated with the induction of apoptosis in KRAS mutant cells, while effects on wild-type cells were generally minor, making this an interesting possible target for selective killing of KRAS mutant tumors. Further investigation in our laboratory of the role of GATA2 in KRAS mutant tumors in vivo has shown that deletion of GATA2 gene causes regression of mutant KRAS-induced lung tumors in a mouse model 12. While transcription factors are generally considered undruggable, through analysis of the transcriptional networks controlled by GATA2 we have identified druggable pathways that appear to be involved in the GATA2 dependency of KRAS mutant cells and have shown that these provide an effective combination therapy against KRAS-induced lung tumors in our mouse model 12. This work showed that one of the transcriptional pathways controlled by GATA2 in lung cancer cells is the production of the proteasome, tying in with the identification of proteasome dependency of KRAS mutant cell lines in our screen.
CDC6 is a critical regulator of DNA replication. The CDC6 protein is an essential component of the multicomponent pre-replicative complexes (pre-RCs), which are assembled at origins of DNA replication during G1 26. In addition, CDC6 may also be important after DNA replication, during mitosis 27. Deficiency in CDC6 leads to failure of replication origin firing and incomplete DNA replication due to reduced licensing of replication origins. The resulting cell cycle checkpoint triggered by low CDC6 levels may be overridden by signals from oncogenic RAS, leading to accumulation of DNA damage and ultimately loss of cell survival. CDC6 silencing by siRNAs affects ATR-dependent inhibition of mitotic entry elicited by modest replication stress 28 and suppression of ATR function in oncogenic RAS-expressing cells synergistically increases genomic instability and DNA damage-induced cell death 29, suggesting a common axis in CDC6 and ATR dependency of KRAS mutant cancer cells.
As might be expected from a critical cell cycle regulator, targeting CDC6 in cells lacking KRAS mutation also had significant growth inhibitory effects, although not as great as for the KRAS mutant cells. This suggests that any possible beneficial effects of targeting CDC6 in cancer might show a rather narrow therapeutic window.
Exploiting synthetic lethality versus oncogene and non-oncogene addiction in combinatorial therapy of cancer
The results of the screens reported here clearly showed a requirement for a number of activities that are inhibited by existing licensed anti-cancer drugs. The proteasome scored well for mutant RAS selectivity both in the HCT-116 system and in the broader panel of cancer cell lines. Since there is no evidence that the proteasome itself can have oncogenic function or is regulated by oncogenic RAS, this is presumably an example of non-oncogene addiction 7: RAS mutant cells are selectively dependent on proteasome function to deal with proteotoxic stress resulting from RAS-driven transformed growth, possibly quite indirectly. The proteasome inhibitor bortezomib has been approved for use in multiple myeloma, a malignancy of antibody-producing plasma cells with a high rate of KRAS and NRAS mutation 30.
A striking aspect of the dependency of cells with activated RAS expression on proteasome function is that it requires sustained RAS activation. Short-term activation of RAS over a period of up to several days using an inducible system in mutant KRAS-deleted HKE-3 cells failed to reproduce this sensitization to proteasome inhibitors. By contrast, sensitivity to several other inhibitory drugs identified in our small chemical library screen was rapidly established in HKE-3 cells following RAS activation, with differential sensitivity seen within a few hours. These drugs include most prominently a number of DNA damaging agents, most of which are direct or indirect inhibitors of topoisomerases. Some of these have been reported previously to be selectively toxic for RAS-transformed cells 31.
The fact that short-term activation of RAS sensitizes cells to these chemotherapeutic drugs, such as camptothecin, topotecan, doxorubicin and daunorubicin, suggests that this represents an acute synthetic lethal effect, likely due to direct conflict of the cells' response to drug-induced DNA damage with the activated signaling pathways driven by RAS. Following treatment with DNA damaging agents, KRAS mutant cells show more DNA damage than KRAS wild-type cells, also more p53 induction, ROS and a greater G2/M arrest, indicating that KRAS mutant cells have less ability to deal with DNA damage stress, perhaps reflecting increased underlying replicative and oxidative stress driven directly by the activated RAS protein.
In the case of the selective sensitivity of RAS mutant cells to bortezomib inhibition of the proteasome, as the effect cannot be reproduced by short-term activation of RAS, this may reflect addiction acquired as a consequence of prolonged selective pressures operating in vivo during the evolution of the tumor, which might be hard to mimic by short-term treatments in vitro. The KRAS mutant cells appear to have less ability to deal with proteotoxic stress, possibly due to long-term increased basal levels of such stress and therefore reduced capacity of the proteasome to handle the effects of pharmacological proteasome inhibition.
These observations point to a significant distinction between addiction of RAS mutant cancer cells, either to the continued activity of oncogenic signaling pathways or to the (non-oncogenic) stress response pathways that enable the cells to tolerate them, and acute synthetic lethal interactions between RAS signaling and certain forms of cellular damage or loss of gene function. One possible conclusion that can be drawn from this is that the dependencies of cancer cells on damaging agents or loss of gene function may be better studied in cells derived from tumors rather than from cells in which the transformed phenotype has been recreated in vitro by the expression of defined combinations of oncogenes 32. Tumor cells may display additional vulnerabilities, and possibly strengths, that reflect the natural history of their evolution in the host that are not seen in cells that have been transformed in tissue culture, where they have not been subject to such rigorous selective pressure.
One practical implication of the fundamentally different mechanistic basis for the response of RAS mutant cancer cells to DNA damaging agents and proteasome inhibitors is to suggest the possibility that combinations of these drugs may show improved selectivity for cancer cells relative to normal cells. This indeed appears to be the case, with low, sub-lethal doses of each class of drug combining to give selective killing of RAS mutant cells, especially when the proteasome inhibitor treatment is given a day after the DNA damaging agent. The mechanism underlying the dependence of the KRAS genotype selectivity on timing of administration of these two drug classes is the subject of ongoing investigations.
One synergistic drug combination observed in this study, bortezomib plus gemcitabine, has been the subject of a phase II trial in non-small cell lung cancer, where it was found to provide significant benefits in terms of overall survival 33; the KRAS mutational status of the patients responding was not determined, but might be of interest in the light of our findings. It is possible that other combinations of drugs targeting the proteasome and causing DNA damage, such as bortezomib with topotecan, might show particular clinical benefit against tumors bearing RAS mutations, especially when proteasome inhibition occurs subsequent to topoisomerase inhibition.
Materials and Methods
Cell lines and culture
HCT-116, HKE-3, HKH-2, DLD-1, DKO-4 and DKS-8 were obtained from Prof S Shirasawa (Fukuoka University). SW837, LoVo, T84, SW620, HCC2998, KM12, HT-29, SW48, NCI-H23, NCI-H727, NCI-H358, NCI-H460, EKVX, NCI-H322M, NCI-H520, NCI-H522, NCI-H2170, CFPAC-1, HPAF-II, BXPC3, OVCAR-5, OVCAR-4, SKOV-3, AGS, MKN-45, Calu-1, A549, A427 and SK-MES-1 were obtained from the CRUK Central Cell Services facility and all cell lines were maintained in RPMI/10% FCS or DMEM/10% FCS as appropriate. HKE3.ER-RAS cells were constructed by transducing HKE-3 cells with a bleocin-resistant retrovirus carrying the murine ecotropic receptor. Selected cells were subsequently infected with puromycin-resistant ER-Ras retrovirus 13.
siRNA reagents and assays
The Protein Kinase (G003500), GPCR (G003600) and Druggable (G004650) siGENOME siRNA SMARTpool collections and all individual siRNAs were obtained from Dharmacon, except KRAS-MUT, KRAS-WT siRNAs and additional siRNAs for Allstars (negative control), CDC6, PSMD14, PSMA1 and PSMB6, which were from Qiagen. Further information relating to individual siRNAs is contained in the accompanying Supplementary information, Data S1. siRNA transfection experiments were performed in 96-well format in antibiotic-free medium, using a reverse transfection employing 25 nM siRNA and 0.15 μl Dharmafect1 (Dharmacon) per well together with a starting cell density optimized to produce an 80% confluent monolayer in mock-treated cells at the conclusion of the experiment. Cell lines were assayed at 72 h or 96 h post transfection. Cell viability was determined using Cell Titer Blue (Promega) by incubation with the cells for 1.5 h. Apoptosis induction was recorded using a caspase 3/7 consensus site peptide (Z-DEVD) conjugated to rhodamine 110 (Invitrogen). Cells were incubated for 5 h with the substrate and lysis buffer mix. Fluorescence was read on an EnVision 2102 Plate-reader (Perkin-Elmer).
Antibodies
Antibodies directed against cleaved PARP, Phospho-ERK (Thr202/Tyr204), Phospho-AKT (Ser473), Pan-ERK, Pan-AKT, Phospho-Histone H2AX (Ser139) and CDC6 were obtained from Cell Signaling Technology. Antibodies against KRAS and Tubulin (loading control) were obtained from Santa Cruz Technology. Antibodies against p21, p53 and ubiquitin were obtained from Abcam.
Flow cytometry
FACS analysis was used as an additional means to quantitate apoptotic, dead and live cell fractions, as well as to monitor cell cycle. Cells were seeded 24 h prior to drug treatment. After a 48-h incubation period, cells were harvested and stained with Annexin V-FITC to detect apoptotic cells. Additionally, treatment with ribonuclease A and propidium iodide was used to identify dead cells. For cell cycle analysis 5-bromo-2-deoxyuridine (BrdU) staining was used to label S phase cells. Cells were seeded 24 h prior to drug treatment. After a 24 or 48 h treatment, cells were incubated with 10 μM BrdU for 1 h, harvested, fixed with cold 70% ethanol and stained using propidium iodide in order to measure DNA content. To assess the duration of the phases of the cell cycle, cells were incubated with 10 μM BrdU for 30 min, washed and incubated in fresh medium, followed by harvesting and fixation at various time points as appropriate. ROS levels were detected using the redox-sensitive dye CM-H2DCFDA (Invitrogen). 24 h after drug treatment, cells were incubated with CM-H2DCFDA for 30 min. Cells were harvested and ROS levels were measured on an LSRII flow cytometer. As a positive control, cells were incubated with 1 mM hydrogen peroxide for 60 min prior to CM-H2DCFDA addition. Results were analyzed using FlowJo 9.4.7 software.
Proteasome activity
Proteasome activity was measured using Proteasome-Glo™ Chymotrypsin-Like Cell-Based Assay (Promega). Cells were seeded 24 h prior to drug treatment. Proteasome activity was measured after 2 h incubation with proteasome inhibitors following the manufacturer's protocol.
Reagents
MG-132, proteasome inhibitor I, camptothecin, doxorubicin, etoposide, mitomycin and aphidicolin were from Calbiochem. Bortezomib was from LC Laboratories, topotecan was from TopoGen, hydroxyurea was from Sigma and gemcitabine was from Discovery Fine Chemicals. Unless otherwise stated, cells were treated with these drugs for 48 h. Biolog Phenotype Microarrays panel 11-14 (Anti-Cancer Agents) were screened by seeding 7 000 HCT-116 and HKE-3 cells per well for 72 h onto the plates. Biolog Redox Dye Mix MA was used as a cell viability readout.
Data analysis
Quadruplicate data points from the primary HCT-116 and HKE-3 apoptosis screens underwent plate and position normalization and Z-score calculation using cellHTS software 34. Differential Z-scores between the two screens were subsequently used to create a gene hit list.
Data from the multi cell line screen were clustered using Gene Cluster 3.0. A centric correlation similarity metric was applied followed by average linkage. Results are displayed using Treeview software 35.
Prism Graphpad software was used for two-tailed unpaired t-test analysis to identify genes showing a significant difference upon knockdown in KRAS mutant versus wild-type cells. Statistical differences in siRNA-mediated cell viability and apoptosis induction between KRAS mutant versus wild-type cells were determined with two additional non-parametric tests, the Mann-Whitney U test and the more stringent Kolmogorov-Smirnov test. A two-tailed P-value < 0.05 was considered significant.
References
Downward J . Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 2003; 3:11–22.
Shirasawa S, Furuse M, Yokoyama N, Sasazuki T . Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science 1993; 260:85–88.
Chin L, Tam A, Pomerantz J, et al. Essential role for oncogenic Ras in tumour maintenance. Nature 1999; 400:468–472.
Sharma SV, Gajowniczek P, Way IP, et al. A common signaling cascade may underlie “addiction” to the Src, BCR-ABL, and EGF receptor oncogenes. Cancer Cell 2006; 10:425–435.
Weir B, Zhao X, Meyerson M . Somatic alterations in the human cancer genome. Cancer Cell 2004; 6:433–438.
Cully M, You H, Levine AJ, Mak TW . Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer 2006; 6:184–192.
Luo J, Solimini NL, Elledge SJ . Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 2009; 136:823–837.
Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH . Integrating genetic approaches into the discovery of anticancer drugs. Science 1997; 278:1064–1068.
Niles A . Apo-ONE™ homogeneous caspase-3/7 assay: robust, high-throughput apoptosis detection. Promega Notes 2002; 81:6–8.
Echeverri CJ, Beachy PA, Baum B, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods 2006; 3:777–779.
von Mering C, Jensen LJ, Snel B, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 2005; 33:D433–D437.
Kumar MS, Hancock DC, Molina-Arcas M, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell 2012; 149:642–655.
Dajee M, Tarutani M, Deng H, Cai T, Khavari PA . Epidermal Ras blockade demonstrates spatially localized Ras promotion of proliferation and inhibition of differentiation. Oncogene 2002; 21:1527–1538.
Wang Y, Ngo VN, Marani M, et al. Critical role for transcriptional repressor Snail2 in transformation by oncogenic RAS in colorectal carcinoma cells. Oncogene 2010; 29:4658–4670.
Scholl C, Frohling S, Dunn IF, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 2009; 137:821–834.
Babij C, Zhang Y, Kurzeja RJ, et al. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res 2011; 71:5818–5826.
Luo J, Emanuele MJ, Li D, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 2009; 137:835–848.
Sharma SV, Settleman J . Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev 2007; 21:3214–3231.
Sarthy AV, Morgan-Lappe SE, Zakula D, et al. Survivin depletion preferentially reduces the survival of activated K-Ras-transformed cells. Mol Cancer Ther 2007; 6:269–276.
Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009; 462:108–112.
Chien Y, Kim S, Bumeister R, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 2006; 127:157–170.
Meylan E, Dooley AL, Feldser DM, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature 2009; 462:104–107.
Xue W, Meylan E, Oliver TG, et al. Response and resistance to NF-kappaB inhibitors in mouse models of lung adenocarcinoma. Cancer Discov 2011; 1:236–247.
Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS . Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 2000; 290:134–138.
Tsai FY, Keller G, Kuo FC, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 1994; 371:221–226.
Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF . An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 1996; 379:180–182.
Boronat S, Campbell JL . Linking mitosis with S-phase: Cdc6 at play. Cell Cycle 2008; 7:597–601.
Yoshida K, Sugimoto N, Iwahori S, et al. CDC6 interaction with ATR regulates activation of a replication checkpoint in higher eukaryotic cells. J Cell Sci 2010; 123:225–235.
Gilad O, Nabet BY, Ragland RL, et al. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res 2010; 70:9693–9702.
Liu P, Leong T, Quam L, et al. Activating mutations of N- and K-ras in multiple myeloma show different clinical associations: analysis of the Eastern Cooperative Oncology Group Phase III Trial. Blood 1996; 88:2699–2706.
Dolma S, Lessnick SL, Hahn WC, Stockwell BR . Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell 2003; 3:285–296.
Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA . Creation of human tumour cells with defined genetic elements. Nature 1999; 400:464–468.
Davies AM, Chansky K, Lara PN Jr., et al. Bortezomib plus gemcitabine/carboplatin as first-line treatment of advanced non-small cell lung cancer: a phase II Southwest Oncology Group Study (S0339). J Thorac Oncol 2009; 4:87–92.
Boutros M, Bras LP, Huber W . Analysis of cell-based RNAi screens. Genome Biol 2006; 7:R66.
Eisen MB, Spellman PT, Brown PO, Botstein D . Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998; 95:14863–14868.
Acknowledgements
We would like to thank members of the LRI Signal Transduction Laboratory for helpful discussions and advice. We also thank Graham Clark and the LRI Equipment Park, and Derek Davies and the LRI FACS lab. This work was funded by Cancer Research UK.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Extended characterisation of cell death induction in response to KRAS siRNA treatment of KRAS mutant and wild-type cells. (PDF 673 kb)
Supplementary information, Figure S2
Set-up of a large-scale differential apoptosis siRNA screen and validation of primary hits. (PDF 1093 kb)
Supplementary information, Figure S3
Further validation of top hits from large-scale differential apoptosis siRNA screen. (PDF 1428 kb)
Supplementary information, Figure S4
Secondary screen siRNA controls and analysis. (PDF 508 kb)
Supplementary information, Figure S5
Chemotherapeutic drugs can elicit differential killing of mutant RAS cells. (PDF 1038 kb)
Supplementary information, Figure S6
Drug combinations enhance killing of mutant RAS cells. (PDF 2288 kb)
Supplementary information, Data S1
Experimental procedures (PDF 96 kb)
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0
About this article
Cite this article
Steckel, M., Molina-Arcas, M., Weigelt, B. et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res 22, 1227–1245 (2012). https://doi.org/10.1038/cr.2012.82
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2012.82
Keywords
This article is cited by
-
Genetic screening for anticancer genes highlights FBLN5 as a synthetic lethal partner of MYC
Cell Communication and Signaling (2023)
-
CDC6, a key replication licensing factor, is overexpressed and confers poor prognosis in diffuse large B-cell lymphoma
BMC Cancer (2023)
-
Cytotoxicity of combinations of the pan-KRAS SOS1 inhibitor BAY-293 against pancreatic cancer cell lines
Discover Oncology (2022)
-
Generation and identification of a conditional knockout allele for the PSMD11 gene in mice
BMC Developmental Biology (2021)
-
Cell death in pancreatic cancer: from pathogenesis to therapy
Nature Reviews Gastroenterology & Hepatology (2021)