Abstract
A wide variety of nuclear regulators and enzymes are subjected to acetylation of the lysine residue, which regulates different aspects of protein functions. The MYST family histone acetyltransferase, human ortholog of MOF (hMOF), plays critical roles in transcription activation by acetylating nucleosomal H4K16. In this study, we found that hMOF acetylates itself in vitro and in vivo, and the acetylation is restricted to the conserved MYST domain (C2HC zinc finger and HAT), of which the K274 residue is the major autoacetylation site. Furthermore, the class III histone deacetylase SIRT1 was found to interact with the MYST domain of hMOF through the deacetylase catalytic region and deacetylate autoacetylated hMOF. In vitro binding assays showed that non-acetylated hMOF robustly binds to nucleosomes while acetylation decreases the binding ability. In HeLa cells, the recruitment of hMOF to the chromatin increases in response to SIRT1 overexpression and decreases after knockdown of SIRT1. The acetylation mimic mutation K274Q apparently decreases the chromatin recruitment of hMOF as well as the global H4K16Ac level in HeLa cells. Finally, upon SIRT1 knockdown, hMOF recruitment to the gene body region of its target gene HoxA9 decreases, accompanied with decrease of H4K16Ac at the same region and repression of HoxA9 transcription. These results suggest a dynamic interplay between SIRT1 and hMOF in regulating H4K16 acetylation.
Similar content being viewed by others
Introduction
Posttranslational modifications (PTMs) are critical for modulating the functions of a variety of eukaryotic proteins. Among different PTMs, reversible acetylation of the lysine residue can neutralize the positive charge of this amino acid (aa) and regulate protein activities in different ways 1, 2. First identified in histones, lysine acetylation has been extended to play regulatory roles on more than 80 transcriptional factors, and a lot of other nuclear factors and cytoplasmic proteins 3. Acetylation of lysine residues can affect protein stability, enzymatic activity and protein-protein interactions, and these changes then contribute to dynamic control of the cellular signaling under diverse conditions 4.
Lysine acetylation is catalyzed by lysine acetyltransferases (KATs), which mainly include Gcn5-related N-acetyltransferases, E1A-associated protein of 300 kDa (p300)/CREB-binding protein and MYST family proteins 3. KATs transfer acetyl groups to their substrates, and conversely, they can also receive acetyl groups from other KATs or even their own catalytic activities. Both of the two well-known acetyltransferases p300 and PCAF can be acetylated and many sites are autoacetylated. Autoacetylation of PCAF increases its histone acetyltransferase activity 5 and autoacetylation of p300 induces the conformational change, which is important for assembly of different p300-containing complexes 6.
The MYST family is one of the largest but, so far, intensive studies are still needed to fully understand their functions, although some members have been shown to play crucial roles in diverse cellular processes 7, 8. PTMs, specifically acetylation and phosphorylation, are emerging to show regulatory roles on proteins of this family. TIP60, a MYST family member, was previously shown to be phosphorylated, and phosphorylation modulates its histone acetyltransferase activity 9. A recent study on TIP60 also showed that its autoacetylation leads to the dissociation of TIP60 oligomer and strengthens its interaction with the substrates 10.
MOF (also called MYST1 or KAT8) is another well-studied MYST family member, which is the major enzyme acetylating histone H4 lysine 16 and required for dosage compensation in Drosophila 11, 12, 13, 14. The human ortholog of MOF (hMOF) has the same substrate specificity for nucleosomal H4K16 and is involved in regulating the chromatin structure in mammals 15, 16.
Until now, however, PTMs of hMOF and their potential effects still remain elusive. In this study, we found that hMOF is autoacetylated in vitro and in vivo, and SIRT1, the deacetylase for H4K16Ac, is responsible for deacetylating acetylated hMOF. In vitro binding assays showed that autoacetylation of hMOF diminishes its binding activity to nucleosomes. Furthermore, in vivo studies showed that modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment and activities on the chromatin.
Results
hMOF acetylates itself in vitro and in vivo
Many acetyltransferases involved in transcriptional regulation and chromatin modifications, such as p300 and TIP60, were found to be autoacetylated 6, 10. As a member of the MYST family acetyltransferases, it is quite possible that hMOF can also be autoacetylated. To test this, in vitro acetylation assay was carried out using purified GST-hMOF, and acetylation was detected by an acetylated lysine-specific antibody (Figure 1A). GST-hMOF displayed strong acetylation signals when supplemented with acetyl CoA and showed no acetylation signals without the cofactor. However, this is not sufficient to verify hMOF as autoacetylated, as GST could also be responsible for the detected acetylation signals. To exclude this, GST was incubated with GST-hMOF in the presence or absence of acetyl CoA. In both situations, GST showed no acetylation signal. In addition, we used thrombin to cleave the GST tag from GST-hMOF, and the pure hMOF proteins also showed comparable acetylation signals (Supplementary information, Figure S1). These data indicate that hMOF can autoacetylate itself in vitro.
hMOF is self-acetylated in vitro and in vivo. (A) GST-hMOF was incubated in acetylation buffer with or without acetyl CoA at 30 °C for 1 h, and in parallel, GST-hMOF was mixed with GST under the same condition. The reaction product was then resolved in 10% SDS-PAGE followed by Commassie blue staining or immunoblotting with anti-acetyl lysine antibody (Cell signaling Technology). (B) hMOF lacking acetyl CoA-binding site was referred to as hMOF Δ314–333. 293T cells were transiently transfected with FLAG-HA-hMOF FL (full-length) or Δ314-333, and the glycine eluate of FLAG-IP was used to detect acetyl lysine by immunoblotting. The eluate of FLAG-IP from 293T cells transfected with vectors was used as a negative control.
In vivo acetylation analysis of hMOF was also performed to detect autoacetylation. Immunoprecipitated FLAG-HA-hMOF (referred to as FH-hMOF hereafter) showed acetylation signals, and moreover, the hMOF mutant with deletion of the acetyl-CoA binding site showed significantly reduced acetylation signals compared with the wild type (Figure 1B). Disruption of the hMOF catalytic activity reduced its acetylation level, suggesting that hMOF is autoacetylated in vivo.
In a recent study using HDACI/II inhibitors SAHA- and MS275-treated human cell lines (Jurkat, MV4-11 and A549), two lysines (K113 and K238) within hMOF were identified as acetylated sites 17. Hence, to test whether these two lysines were the self-acetylation sites, we generated cognate mutants. Single or double mutants of K113R and K238R GST-hMOF proteins were purified to determine their acetyltransferase activities by in vitro acetylation assay using the recombinant histone H4 as substrates. All the three mutant forms demonstrated similar enzymatic activities on recombinant H4 and similar levels of autoacetylation compared with the wild-type proteins (Figure 2A). Point mutations of these two lysines did not change hMOF enzymatic activity, and their inability to be acetylated did not influence hMOF self-acetylation level either. Two possibilities for the observations exist: first, neither K113 nor K238 is the self-acetylation lysine; second, like p300, there are several self-acetylated lysines and mutations of one or two of them do not change the overall acetylation level.
hMOF is self-acetylated in its MYST domain. (A) hMOF point mutations K113R and K238R are marked in hMOF schematic diagram. In vitro acetylation assays using similar amounts of purified GST-hMOF wild type, K113R, K238R and K113R-K238R were carried out as described, and equal amounts of recombinant histone H4 was used as substrates to test their catalytic activities. H4 acetylation was detected by immunoblotting using anti-H4K16 Ac antibody (Millipore). Self-acetylation of these GST-hMOF proteins was measured by immunoblotting. Even loading of recombinant H4 and GST-hMOF proteins were shown by Commassie blue stain. (B) Sequence alignment of MOF MYST domain among human, mouse, Zebrafish, Drosophila and C. elegans was carried out using ClustalX. Lysines conserved among no less than four species are highlighted in black rectangles. (C) Fragments of hMOF were cloned and expressed as GST fusion proteins, and the schematic diagram demonstrates their length and location compared with full-length hMOF. Similar amounts of the three fragments aa 1-130, 157-458 and 231-458 were used as substrates of acetylation assay catalyzed by full-length GST-hMOF. Acetyl lysine was detected by immunoblotting, and CBB stain was used to display even loading of proteins among individual reactions. (D) Pull-down of FLAG-HA-hMOF was performed between GST-hMOF FL or aa 1-130 and 293T cell lysate expressing FLAG-HA-hMOF, and GST was used as pull-down control. GST beads were boiled in 2× sample buffer after several washes, and the supernatant was resolved in 10% SDS-PAGE for immunoblotting and CBB stain. FLAG-HA-hMOF was detected using anti-HA (Roche) antibody.
Although no detailed studies are available, Drosophila MOF has been noticed to be autoacetylated 12, 13; thus, autoacetylation is likely to be conserved in evolution. MYST domain, composed of C2HC zinc finger and HAT domain, is the signature of MYST family proteins and is conserved among even distant species. Sequence comparison of MYST domains among five different species using ClustalX reveals that, within hMOF catalytic domain, there are eight lysines conserved in Caenorhabditis elegans, Drosophila, Zebrafish, mouse and human, and seven conserved in the latter four species (Figure 2B). We speculated that these conserved lysines within MYST domain are more likely to be responsible for hMOF self-acetylation.
To test this, in vitro acetyltransferase assay was performed using recombinant GST-hMOF derivatives aa 1-130, 157-458 and 231-458 as substrates (Figure 2C). Equal amounts of full-length GST-hMOF were used to catalyze the separate reactions. Acetylation signals were observed on aa 157-458 and 231-458, while aa 1-130 showed no acetylated band. The aa 231-458 fragment contains the core HAT domain, while aa 157-458 includes an additional C2HC zinc finger, which is required for substrate recognition and efficient enzyme activity. So it can be inferred that self-acetylation also contributes to the robust acetylation signals of aa 157–458. The reactions were repeated with higher or lower doses of full-length hMOF, and all of them showed similar patterns of acetylation signals as above (data not shown). Therefore, autoacetylation of hMOF occurs mainly on the MYST domain containing several conserved lysine residues.
In a subsequent GST pull-down experiment, full-length GST-hMOF and N terminal GST-hMOF aa 1-130 were used to pull-down hMOF from lysates of 293T cells expressing FH-hMOF (Figure 2D). The results showed that GST-hMOF, but not the N terminal fragment GST-hMOF aa 1-130, can interact with FH-hMOF, indicating that hMOF can form a dimer or oligomer mediated by the MYST domain. In vitro HAT assay also showed that the G327D mutant of GST-hMOF, which bears little enzyme activity 16, can be acetylated by the pure hMOF without GST tag (Supplementary information, Figure S2). These results indicate that the catalytic region could catalyze inter-molecular acetylation of hMOF itself, but an intra-molecular mechanism may act concomitantly.
To further map the autoacetylated sites, we adopted the serial point-mutation strategy. By using the predictor of acetylation at PHOSIDA (http://www.phosida.com), combined with our sequence alignment of MYST domains, we selected lysines 268, 274, 330, 407 and 410 as the most likely sites. We first converted all potential lysines (113, 238, 268, 274, 330, 407 and 410) to arginines to generate a 7KR mutant, and tested its autoacetylation and enzyme activity. The in vitro HAT assays showed that autoacetylation signals (Figure 3A) and enzyme activities on histone H4 (Figure 3B) dramatically decreased. Then we tested the effects of 3KR (113, 238 and 274), 4KR (113, 238, 274 and 268) and 6KR (113, 238, 274, 268, and 330), and all these mutants showed apparently decreased autoacetylation signals compared with the wild-type proteins (Figure 3C). As all the mutants contain the K274R mutation, it is most probable that K274 serves as the key point of autoacetylation. Thus, we generated the single K274R mutation and carried out HAT reactions using the wild-type hMOF without GST tag as the enzyme. The results showed that the single K274R mutation is sufficient to cause apparent decrease of acetylation signals (Figure 3D). To further confirm this, we generated K268R, K330R and K407-410R mutants, and these mutants showed efficient autoacetylation and catalytic activities on H4K16 (Figure 3E). Taken together, we conclude that K274 residue is the major site of hMOF autoacetylation, and K274R mutation impairs the enzyme activity on H4K16 of recombinant histone H4 in vitro.
Autoacetylaiton of hMOF mainly occurs at lysine 274. (A) Wild-type GST-hMOF or 7KR (K113, K238, K274, K268, K330, K407 and K410 to R) proteins were used for in vitro HAT reactions. Acetylation signals were detected by anti-acetyl lysine antibody. (B) Wild-type GST-hMOF or 7KR proteins were incubated with recombinant histone H4 in HAT buffer at 30 °C for 1 h. H4K16Ac was detected by western blot using specific H4K16Ac antibody. (C) Wild-type GST-hMOF, 3KR (113, 238 and 274 to R), 4KR (113, 238, 274 and 268 to R), 6KR (113, 238, 274, 268 and 330 to R) and 7KR proteins were used for in vitro HAT reactions. Acetylation signals were detected by anti-acetyl lysine antibody. (D) Wild-type GST-hMOF or K274R-mutant proteins were used for in vitro HAT reactions catalyzed by pure hMOF proteins without GST tags. Acetylation signals were detected by anti-acetyl lysine antibody and the CBB staining showed equal loading. (E) Wild-type GST-hMOF, K274R, K268R, K330R or K407-410R proteins were incubated with recombinant histone H4 in HAT buffer at 30 °C for 1 h. Acetylation signals of MOF proteins were detected by anti-acetyl lysine antibody and acetylation of H4 was detected by the specific H4K16Ac antibody.
SIRT1 physically interacts with hMOF
Among the MYST family histone acetyltransferases, TIP60 and hMOF are more closely related due to their structural analogy and are divided into the same subgroup of MYST proteins. TIP60 is self-acetylated, and can interact with and be deacetylated by SIRT1 10. TIP60 and SIRT1 also share some common substrates, like p53 18, 19. Similarity between TIP60 and hMOF suggests that hMOF may also be deacetylated by SIRT1 and have mutual functional interactions with SIRT1.
To test this hypothesis, localizations of hMOF and SIRT1 were studied in 293T cells stably expressing FH-hMOF. Immunofluorescence was performed to stain ectopic hMOF using antibody against HA (green) and endogenous SIRT1 with anti-SIRT1 antibody (red; Figure 4A). Co-immunoprecipitation (co-IP) between FH-hMOF and myc-SIRT1 was also performed in 293T cells (Figure 4B). Both experiments verified the interaction between these two proteins.
hMOF interacts with SIRT1 in vivo. (A) The 293T cells stably expressing FLAG-HA-hMOF were fixed in 4% paraformaldehyde, and immunofluorescence was carried out using primary antibodies anti-FLAG (mouse IgG) and anti-SIRT1 (rabbit IgG) followed by secondary antibodies anti-mouse IgG/FITC (hMOF, green) and anti-rabbit IgG/TRITC (SIRT1, red). Nuclei were stained with DAPI. (B) The 293T cells were co-transfected with FLAG-HA-hMOF and myc-SIRT1. Cells were then lysed in IP buffer, and immunoprecipitation was performed using anti-FLAG antibodies with equal amounts of preimmune mouse IgG as the negative control. Immunoprecipitated FLAG-HA-hMOF was shown by using CBB stain, while co-IP product was detected with anti-myc antibody.
Furthermore, the deacetylase-defective mutant SIRT1 (H363Y) showed less interaction signals compared with wild type (Figure 5A), and this observation suggested that the catalytic domain of SIRT1 may mediate the interaction with hMOF. Therefore, we performed co-IP between truncated fragments of SIRT1 and full-length hMOF (Figure 5B). It turned out that the catalytic domain of SIRT1 mediates the interaction with hMOF. Co-IP between full-length SIRT1 and fragments of hMOF showed that both fragments aa 157-458 and 231-458 bind to SIRT1, while aa 1-130 does not (Figure 5C). To further investigate whether their interaction is direct, GST pull-down was performed between purified GST-hMOF of different length and purified MBP-SIRT1 (Figure 5D). Using western blot, we detected binding of SIRT1 in hMOF aa 157-458, 231-458 and full-length groups. This is in agreement with the co-IP experiment. Thus, hMOF interacts directly with SIRT1, and the catalytic domains of these two enzymes mediate the interaction.
The MYST domain of hMOF interacts directly with SIRT1 catalytic domain. (A) The 293T cells stably expressing FH-hMOF were transfected with myc-tagged SIRT1 (wild type), SIRT1 H363Y mutant or empty vectors. IP was performed using anti-FLAG agarose, and eluted with glycine. hMOF was immunoblotted with anti-HA, and co-IP was detected with anti-myc. Overexpression of SIRT1 was detected with anti-SIRT1 antibody to compare with endogenous expression. (B) SIRT1 fragments were expressed with myc tag, as described, and the schematic diagram showed their length and relative location compared with full-length SIRT1. IP was done as described above. Co-IP and overexpression of SIRT1 fragments were immunoblotted with anti-myc, and immunoprecipitated hMOF was detected by HA antibody. (C) Truncated hMOF, as represented in the schematic picture, was expressed with N terminal FLAG-HA tag. Myc-SIRT1 full-length was co-expressed with these truncated hMOF, and IP was performed as above. Co-IP and overexpression of myc-SIRT1 were immunoblotted with anti-myc, while immunoprecipitated hMOF proteins were detected by CBB stain. (D) MBP-SIRT1 was purified from E. coli and was subjected to pull-down by GST-hMOF FL, aa 1-130, 157-458 and 231–458. GST served as the pull-down control. Equal amounts of input MBP-SIRT1 were measured by CBB stain, while the pull-down product was detected using anti-SIRT1 antibody.
SIRT1 deacetylates hMOF
Interaction between SIRT1 and hMOF brought up the possibility that SIRT1 deacetylates acetylated hMOF. In vitro deacetylation assay was performed, and self-acetylated hMOF was used as the substrate (Figure 6A). SIRT1 erased the lysine acetylation signals from hMOF only when NAD+ was supplemented. Treatment of the FH-hMOF-stable cell line with nicotinamide (NAM), a specific sirtuins inhibitor, led to elevated acetylation signals. SIRT1 overexpression in FH-hMOF 293T cells reduced the acetylation level of immunoprecipitated FH-hMOF, and on the contrary, knockdown of SIRT1 increased hMOF acetylation level slightly (Figure 6B).
Ac-hMOF is deacetylated by SIRT1 in vitro and in vivo. (A) GST-hMOF was first acetylated in vitro, and then coupled to GST beads followed by four washes to remove traces of acetyl CoA. GST beads were then incubated with purified SIRT1 in the presence of NAD+ or not at 30 °C for 1 h, and then boiled in 2× sample buffer. Acetylation level was determined by anti-acetyl lysine antibody. (B) 293T cells stably expressing FLAG-HA-hMOF were treated with 2 mM nicotinamide for 16 h. 293T cells expressing FLAG-HA-hMOF were infected with SIRT1 RNAi and luciferase RNAi retroviruses separately. Myc-SIRT1 was transfected transiently into 293T cells expressing FLAG-HA-hMOF. FLAG-HA-hMOF was immunoprecipitated with anti-FLAG agarose and eluted with glycine for detection of acetylated lysines.
Binding ability of hMOF to nucleosomes is regulated by its reversible acetylation
Although hMOF manifests its acetyltransferase activity upon all four histones in vitro, immunoprecipitated hMOF catalyzes only H4K16 acetylation on nucleosomes. Depletion of hMOF in HeLa cells leads to specific decrease in H4K16Ac 16. Therefore, regulation of binding of hMOF to nucleosomal substrates is critical for its proper physiological functions.
As acetylation changes surface charges of proteins, we proposed that acetylation of hMOF may influence its conformation and its interaction with substrates. To test this, we measured the in vitro binding of HeLa nucleosomes, reconstituted mononucleosomes and recombinant histones by acetylated GST-hMOF in comparison with non-acetylated GST-hMOF and included GST as a negative control (Figure 7A, 7B and 7D). HeLa nucleosomes were measured for their length, and > 80% were mononucleosomes. Western blot shows that ac-GST-hMOF obviously binds less to HeLa nucleosomes or reconstituted mononucleosomes. In a reverse pull-down experiment, reconstituted mononucleosomes immobilized on streptavidin beads also bind less to ac-GST-hMOF (Figure 7C). However, western blot analysis showed that ac-GST-hMOF and GST-hMOF have similar binding ability to recombinant histone octamers (Figure 7D).
Acetylation of hMOF reduces its affinity with nucleosomes. Equal amounts of acetylated or non-acetylated GST-hMOF were employed to pull-down HeLa nucleosomes (A), reconstituted nucleosomes (B) or recombinant histone octamers (D), and GST served as the negative control. Acetylation of GST-hMOF was shown by western blot in panel A. The pull-down product was detected with mixed anti-H3 and H4 antibodies. Biotin-labeled DNA was reconstituted into mononucleosomes and further coupled to streptavidin beads (C). After several washes, streptavidin beads were used to pull-down GST-hMOF (acetylated or not, shown by immunoblotting), and the pull-down product was detected by CBB stain. GST served as the negative control.
Theoretically, the isoelectric point of full-length hMOF is 8.48, while that of the MYST domain is 9.2 (calculated using Expasy pI computation tool). Under physiological conditions, hMOF and its MYST domain are both positively charged. Positive charges on surface of hMOF (especially the catalytic domain) likely facilitate its binding to the negatively charged nucleosomes and hence are of great importance to hMOF function in chromatin modifications. Acetylation of the hMOF catalytic domain neutralizes its positive charges, which would disturb hMOF binding to nucleosomes. Also, acetyl groups in hMOF catalytic domain may lead to insufficient space in the catalytic pocket for substrate binding.
Nucleosomes and recombinant histone octamers differ in some ways, such as surface charges and histone accessibility, and this may explain the different performance of acetylated hMOF in binding nucleosomes and recombinant histone octamers compared with non-acetylated hMOF.
Modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment on the chromatin
To understand how modulations of hMOF autoacetylation by SIRT1 affect hMOF affinity to the nucleosomes in cells, we performed chromatin retention assays to detect hMOF recruitment to the chromatin in response to changes of SIRT1 level. After overexpression of SIRT1 in HeLa cells, the cells were crosslinked with formaldehyde followed by fractionation into whole-cell soluble part and chromatin part. The results showed that upon SIRT1 overexpression, recruitment of hMOF to chromatin increases, while its expression levels in the whole-cell lysate fractions showed no apparent difference (Figure 8A). In SIRT1 knockdown HeLa cells, the recruitment of hMOF to chromatin decreased compared with the control group (Figure 8B), while the expression level of hMOF in the whole-cell lysate remained unchanged. It should be noted that the level of p53, which is suppressed by SIRT1, apparently increased upon SIRT1 knockdown as expected, but the global H4K16Ac level did not increase. Two previous studies observed similar results for the global H4K16Ac level upon depletion of SIRT1 20, 21, suggesting that deacetylation of H4K16Ac by SIRT1 is restricted to specific sites on the chromatin. Our results suggested a possibility that reduced hMOF recruitment upon knockdown of SIRT1 may also be responsible for this observation.
Modulations of hMOF autoacetylation by SIRT1 regulates hMOF recruitment to the chromatin. (A) HeLa cells were transfected with pcDNA3.1 or SIRT1 expression plasmids and 48 h later, the cells were harvested for chromatin retention assays. The cells were crosslinked as in the ChIP assay and lysed in lysis buffer for 20 min followed by centrifugation. The pellet was washed with lysis buffer for another 5 min and the two parts of supernatant were combined to obtain the whole-cell lysate fraction. The pellet was then sonicated in ChIP sonication buffer as the chromatin fraction. The hMOF recruitment to the chromatin was detected by western blot. (B) The SIRT1 knockdown HeLa cells and control cells were harvested as in A, and hMOF recruitment to the chromatin was detected by western blot. (C) HeLa cells were infected with retroviruses expressing wild-type hMOF, K274A, K274R or K274Q mutants, and 72 h after infection, the cells were harvested as in A and B for chromatin retention assay. (D-E) HeLa cells were infected with either shLuc or shMOF viruses, and knockdown efficiency of hMOF was detected by western blot using MOF antibody and H4K16Ac antibody. The mRNA level of hMOF was measured by qRT-PCR. (F) Transcription of HoxA9 in hMOF knockdown cells or control cells was detected by qRT-PCR. (G) HeLa cells were infected with shLuc or shSIRT1 viruses, and the knockdown efficiency was confirmed by western blot. (H) Transcription of HoxA9 was detected in SIRT1 knockdown cells and control cells by qRT-PCR. (I, J) MOF ChIP and H4K16Ac ChIP were performed in SIRT1 knockdown and control cells. Primers occupying the promoter region (A) and gene body region (B) of HoxA9 locus were used for qPCR.
To further characterize the effects of hMOF autoacetylation, we generated HeLa cell lines expressing FH-hMOF-wt, FH-hMOF-K274A, FH-hMOF-K274R and FH-hMOF-K274Q, and measured hMOF recruitment to the chromatin and levels of H4K16Ac in these cells. Compared with the wild-type hMOF, chromatin recruitment and modifying activities of K274R, which mimics the deacetylated status, did not change apparently. However, the K274Q form, which mimics the acetylated status, cannot be efficiently recruited to chromatin, and the H4K16Ac levels greatly decreased (Figure 8C). K274A mutation also caused reduction of H4K16Ac level, as the K to A mutation dramatically changed the polarity and structure of this residue, which appears critical for the enzyme activities of hMOF (Figure 8C). It should be noted that the recombinant GST-hMOF-K274R protein showed no activity on histone H4 in the in vitro assay (Figure 3E). This suggests that for the K274R mutant, the formation of MOF complex with other subunits is required for its activity. Therefore, we purified the mammalian hMOF-K274R complexes from the stable cell line by FLAG-IP and performed in vitro HAT assay. The results showed that the hMOF-K274R complexes can acetylate histone H4 in vitro (Supplementary information, Figure S3). To further characterize the effects of these hMOF mutants, the H4K16Ac levels of the stable cell lines were compared with that of the blank HeLa cells. The results showed that ectopic expression of hMOF leads to elevated H4K16Ac levels, and the different mutants have no dominant-negative effects (Supplementary information, Figure S4).
Next, we examined whether the modulations of hMOF autoacetylation by SIRT1 affect transcription of specific genes. To this end, we selected a previously characterized hMOF target gene HoxA9 as the model. To confirm HoxA9 as a target gene of hMOF, we generated hMOF knockdown HeLa cell lines, of which the RNAi efficiency was determined by western blot (Figure 8D) and Quantitative RT-PCR (qRT-PCR) (Figure 8E). Meanwhile, depletion of hMOF resulted in decreased global H4K16Ac level (Figure 8D). HoxA9 transcription was detected by qRT-PCR and it was reduced as previously reported (Figure 8F). Then we detected HoxA9 transcription in SIRT1 knockdown cells (Figure 8G). The qRT-PCR data showed that HoxA9 mRNA level decreased upon depletion of SIRT1 (Figure 8H). ChIP experiment showed that the level of H4K16Ac at the promoter region of HoxA9 locus decreased slightly, and it decreased more apparently at the gene body region (Figure 8I). Meanwhile, the ChIP experiments showed that recruitment of hMOF at the promoter region of HoxA9 remained unchanged in SIRT1 knockdown cells, but apparently decreased at the gene body region (Figure 8J). This is consistent with the fact that the MOF-containing MSL complex binds to the gene body regions and potentially facilitates transcription elongation 22, 23.
Discussion
Recent studies demonstrated that hMOF participates in many biological processes, including chromatin decondensation, formation of chromatin boundaries and DNA repair 13, 24, 25, 26. Effects of PTMs on hMOF, so far, have remained poorly studied. In this study, we have revealed that hMOF undergoes reversible autoacetylation, and the acetylation status regulates its binding activity toward nucleosomal substrates. It was also shown that SIRT1 is responsible for deacetylating acetylated hMOF, and the modulations of hMOF autoacetylation by SIRT1 contribute to regulation of hMOF recruitment and modifying activities on the chromatin. Thus, our study provides novel insight into regulatory roles of hMOF acetylation, which is a new substrate of SIRT1.
Accumulating evidence shows that posttranslational automodification of chromatin-modifying factors represents a general regulatory mechanism. PCAF, p300 and TIP60 are autoacetylated, G9a can be automethylated 27 and PARP-1 can be auto-ADP-ribosylated 28. Here, we found that hMOF is autoacetylated both in vitro and in vivo. Although a previous study using the high-throughput mass spectrometry method identified two acetylated lysines on hMOF, our data indicate that they are not required for autoacetylation. As autoacetylation of Drosophila MOF was noticed by Akhtar and coworkers 12, 13, autoacetylation of MOF may be conserved among species. This is further supported by our data that the conserved MYST domain is heavily acetylated, while the N terminal region showed no acetylation. Furthermore, the multiple sequence alignment of MYST domain from five species clearly showed that K238 is not shared by hMOF and Drosophila MOF. Therefore, K113 and K238 receive acetyl groups from other KATs and their functions may be mammal specific. Sequence alignment also showed that there are multiple lysines within the MYST domain that are conserved in no less than four species, thus autoacetylation may occur on these lysines. By using a serial point-mutation strategy, we determined that the autoacetylation of hMOF mainly occurs on K274. Mutation of K274 to Q mimics the acetylated form, which impairs the efficient recruitment of hMOF to chromatin and its modifying activities on chromosomal H4K16. Interestingly, in addition to MOF proteins of different species, the K274 site is also conserved in Tip60, of which the K327 site (equivalent to K274 of hMOF) is among several lysines undergoing acetylation 10. These data indicate that autoacetylation of MOF may play conserved regulatory roles for this acetyltransferase.
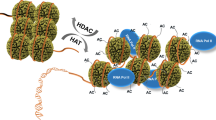
SIRT1 is a class III histone deacetylase that is able to deacetylate H3K9Ac and H4K16Ac and plays a role in chromatin regulation 29. Besides the histone substrates and many other nuclear factors, SIRT1 also deacetylates chromatin-modifying enzymes, including PCAF, p300, TIP60 and SUV39H1 10, 30, 31, 32. In this study, we identified hMOF as another substrate of SIRT1. It is of note that hMOF and SIRT1 share common substrates like H4K16Ac and p53 19, suggesting their joint participations in different cellular processes. A previous study showed that depletion of SIRT1 in cancer cells reactivated some silenced tumor suppressor genes accompanied by increased H4K16 and H3K9 acetylation at their promoter regions 33. Recruitment of SIRT1 to promoters of target genes and its deacetylation activities toward H4K16Ac and H3K9Ac contribute one means of transcriptional regulation (Figure 9). In this study, we show that modulations of hMOF autoacetylaiton by SIRT1 regulate hMOF recruitment and modifying activities on chromatin. This adds another means of regulation at the global level and illustrates the cooperative functions between the two nuclear enzymes (Figure 9).
Recruitment of hMOF to the chromatin is regulated by the switch of hMOF status. SIRT1 has been shown to be recruited to specific gene promoters and directly deacetylate H4K16Ac, which then suppresses gene transcription. Here, we found that autoacetylation prevents hMOF from efficient binding to nucleosomes, probably by neutralizing positive charges on hMOF surface. The class III histone deacetylase SIRT1 can decrease hMOF acetylation and enhance its recruitment to the chromatin and further affect gene transcription by modifying H4K16 at the gene body regions. Modulations of hMOF autoacetylation by SIRT1 provide another means for concerted regulation of gene expression.
So far, hMOF is known to be contained in two different complexes, MOF-MSL and MOF-MSL1v1 15, 34. Both the complexes have acetyltransferase activity toward nucleosomal H4, but free hMOF barely showed any activity toward HeLa nucleosomes 35, 36, even though MOF has been shown to be able to bind to the chromatin 37. Our results showed that free hMOF acetylates itself in the MYST domain, and autoacetylation greatly decreased its binding to reconstituted nucleosomes or native HeLa nucleosomes. This provides at least one explanation for little activity of free hMOF toward nucleosomal H4, as the nucleosomal substrate cannot efficiently bind to the catalytic site of autoacetylated hMOF. When hMOF is complexed with other factors, however, the HAT activity can be regulated in different ways. First, certain deacetylases (for example SIRT1) may remove acetylation of hMOF and increase its interaction with the nucleosomal substrate (Figure 9). Second, the well-studied proteins in different hMOF complexes, like MSL1/2/3 and MSL1v1 35, 36, may interfere with the autoacetylation of hMOF so as to facilitate targeting of hMOF to specific substrates, or meanwhile, stabilize its interaction with the substrates.
The mutation analyses showed that K274Q and K274A mutations greatly reduced hMOF recruitment and global H4K16Ac level, while the K274R mutant behaves similarly to the wild type. These data suggest that the structure and charge of aa 274 is important for assembly of the MOF complex, and autoacetylaiton may impede the process. A recent study focusing on the two MOF complexes showed that they have different substrate preferences, and proposed a model that the MSL1v1 complex is recruited to the promoters of target genes through interactions with MLL and p53, while the recruitment of the MSL complex by H3K36me3 is important for acetylating H4K16 downstream of the genes 36. Another study in Drosophila also showed that the NSL complex (equivalent to MSL1v1 complex) binds to the promoters of target genes to facilitate transcription initiation, while the MSL complex is important for elongation 38. Our ChIP analyses showed that upon SIRT1 knockdown, decreased hMOF recruitment and H4K16Ac level took place at the gene body region, indicating that the autoacetylation mainly affects the MSL complex.
During the submission of our manuscript, another study also identified K274 residue as being acetylated 39. Their data showed that the K274A MOF was less stable in a thermal denaturation assay and not active in acetylating histone H4 N terminal peptides. Their data on the K274A mutant support our study, although they did not explore further the functional relevance of K274 acetylation because of their main focus on the effects of MSL1 mutations on the complex structure.
In addition to acetylation, other modifications of hMOF, for example, phosphorylation, also have potential regulatory roles, and may even crosstalk with acetylation. Therefore, it is important to explore further the post-translational modifications of MOF, which will lead to a better understanding of this important acetyltransferase.
Materials and Methods
DNA constructs
Full-length hMOF cloned from HeLa cDNA by RT-PCR was ligated into pGEX-KG in fusion with GST tag for prokaryotic expression and into MSCV-puro in fusion with FLAG-HA tag for eukaryotic expression. The hMOF-derived fragments, including hMOF aa 1-130, 157-458 and 231-458 16, were PCR amplified and inserted into the same vectors. Single or multiple-point mutations of GST-hMOF, as well as Δ314-333-aa K274A, K274R and K274Q mutants of FLAG-HA-hMOF were generated using site-directed mutagenesis facilitated by DpnI (NEB) selection. SIRT1 expression vector pcDNA-SIRT1 was a gift from Prof Ishikawa 40, pcDNA-myc-SIRT1 was from Professor Shengqi Wang 41 and myc-tagged SIRT1 fragments have been described earlier 42. MBP-SIRT1 was expressed by cloning SIRT1 cDNA into pMAL-p4X vector. Oligos of shRNAs against hMOF, SIRT1 and luciferase were cloned into retroviral vector pSIREN-RetroQ at the BamHI / EcoRI site, and their targeting sequences are 5′-GCAAGATCACTCGCAACCAAA-3′, 5′-GATGAAGTTGACCTCCTCA-3′ and 5′-CGCCTGAAGTCTCTGATTA-3′, respectively.
Generation of stable cell line
The 293T cells or HeLa cells were infected with retroviruses expressing FLAG-HA-tagged hMOF or mutant forms, and selected with puromycin (2 μg/ml) for 2 weeks.
Immunoprecipitation and western blot
The 293T cells were transfected with combinations of different myc-tagged SIRT1 and FLAG-HA-tagged hMOF constructs or control vectors, and were lysed in IP buffer (20 mM Tris-Cl (pH 8.0), 150 mM NaCl and 1% Triton X-100), supplemented with protease inhibitor cocktail (Sigma) for co-IP and additional 2 mM NAM and 10 μM sodium butyrate for acetyl lysine detection. After continuous gentle agitation at 4 °C for 30 min, the lysate was cleared by centrifugation, and anti-FLAG-M2 agarose was added to capture FLAG-tagged proteins. Immunoprecipitated proteins were eluted by 0.1 M glycine (pH 3.0) and detected by western blot using anti-myc antibody (Santa Cruz, sc-40) for co-IP and anti-acetyl lysine antibody (Cell Signaling Technology, 9441) for detection of acetylation.
In vitro acetylation and deacetylation assays
GST-tagged full-length hMOF and three hMOF fragments were purified from Escherichia coli according to the GE purification protocol. For each reaction, 5 μg of substrate protein was incubated with 3 μg of full-length hMOF in reaction buffer with or without acetyl-CoA (20 μM) at 30 °C for 1 h, as described previously 43. GST-hMOF was coupled to glutathione-sepharose 4B beads after in vitro autoacetylation, and the beads were washed four times with PBS and once with deacetylation buffer to remove traces of acetyl-CoA before subsequent deacetylation assays. The immobilized GST-hMOF was then incubated with 1 μg purified human His-SIRT1 (Sigma, S8446) with or without NAD+ (Sigma, N1663) in the deacetylation buffer (Sigma, A6480) at 30 °C for 1 h with occasional inversions. The products of the reactions were analyzed by western blot using anti-acetyl lysine antibody (Cell Signaling Technology, 9441).
GST pull-down assay
GST-hMOF proteins and MBP-SIRT1 were purified from E. coli. Different GST-hMOF proteins (full-length or fragments) and GST were coupled to GST beads, and incubated with MBP-SIRT1 in IP buffer with agitation at 30 °C for 3 h. GST beads were then washed four times with high-salt buffer (20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 1% Triton X-100), and boiled with 2× sample buffer for western blot using anti-SIRT1 antibody (Santa Cruz, sc-15404).
Mononucleosome-binding assay
HeLa mononucleosomes were prepared as described 44. For reconstituted mononucleosomes, recombinant HeLa histones and 194-bp satellite DNA (5′ biotin labeled) were employed, and reconstitution of nucleosome core particles (NCPs) followed the salt jump-down protocol 45. The reconstituted NCPs were further captured by streptavidin agarose. Equal amounts of Ac-GST-hMOF or GST-hMOF coupled to GST-beads were used for binding of HeLa mononucleosomes or reconstituted NCPs. Conversely, reconstituted NCPs that were immobilized on streptavidin agarose were used to bind products from parallel GST-hMOF acetylation assays in the presence of acetyl-CoA or not. All nucleosome-binding experiments were performed as described 37.
Immunofluorescence
The 293T cells expressing FLAG-HA-tagged hMOF were fixed in 4% paraformaldehyde at room temperature for 10 min. Post-fixative permeabilization was carried out in PBS containing 0.1% Triton X-100 for 10 min. Expression of hMOF was stained with mouse anti-FLAG-M2 (Sigma, F3165) in a dilution ratio of 1:2 000, and SIRT1 was stained with rabbit anti-SIRT1 (Santa Cruz, sc-15404) in a dilution ratio of 1:200, followed by anti-rabbit IgG/TRITC (Zhongshan Goldenbridge Biotechnology) and anti-mouse IgG/FITC (Zhongshan Goldenbridge Biotechnology) staining. The nuclei were stained with DAPI.
Quantitative RT-PCR
Total RNA from HeLa cells was isolated by using Trizol (Invitrogen) reagent and reverse transcription was carried out per the manufacturer's manual (New England Biolabs). Real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa) on RotorGene Q cycler (Qiagen). The primers used are listed in Supplementary information, Table S1.
Chromatin immunoprecipitation assay
ChIP assays were performed as described by the Upstate protocol with some modifications. HeLa cells were crosslinked by adding formaldehyde to a final concentration of 1% at room temperature for 10 min. After washing four times with 20 ml PBS in 50 ml conical tubes, cells were scraped and swelled in hypotonic swelling buffer (25 mM HEPES (pH 7.8), 1.5 mM MgCl2, 10 mM KCl, 0.1% NP-40, protease inhibitor cocktail from Sigma) and incubated on ice for 10 min. Following centrifugation at 2 000 rpm. for 5 min, the nuclei were lysed in SDS lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris (pH 8.1)) and sonicated with Branson 150 sonicator. Antibodies against H4K16Ac (Millipore, 07-329) and MOF (Bethyl, A300-992A) were used for IP. PCR data were analyzed by using the 2−ΔΔCt method and normalized to input samples. The results were expressed as fold changes over control groups. The primers used are listed in Supplementary information, Table S1.
Chromatin retention assay
HeLa cells were crosslinked with formaldehyde as in the ChIP assay, and glycine was added to the final concentration of 0.125 M to quench the crosslinking. After three washes with cold PBS, cells were extracted in the lysis buffer (20 mM Tris-Cl (pH 8.0), 150 mM NaCl, 0.2 mM EDTA, 10% Glycerol, 1% Triton X-100, protease inhibitor cocktail from Sigma) by end-to-end rotation for 30 min. After centrifugation at 3 000 rpm. and transfer of the supernatant, the pellet was again resuspended in the same lysis buffer and kept on ice for 5 min. After centrifugation, the two parts of supernatant were combined as the whole-cell lysate. The nuclei were then resuspended in ChIP-SDS buffer and sonicated as in the ChIP assay. The soluble chromatin fraction was used to detect retention of hMOF by western blot.
References
Yang XJ . Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 2004; 26:1076–1087.
Kouzarides T . Acetylation: a regulatory modification to rival phosphorylation? EMBO J 2000; 19:1176–1179.
Yang XJ, Seto E . Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 2008; 31:449–461.
Zhao S, Xu W, Jiang W, et al. Regulation of cellular metabolism by protein lysine acetylation. Science 2010; 327:1000–1004.
Santos-Rosa H, Valls E, Kouzarides T, Martinez-Balbas M . Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res 2003; 31:4285–4292.
Black JC, Choi JE, Lombardo SR, Carey M . A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell 2006; 23:809–818.
Thomas T, Voss AK . The diverse biological roles of MYST histone acetyltransferase family proteins. Cell Cycle 2007; 6:696–704.
Voss AK, Thomas T . MYST family histone acetyltransferases take center stage in stem cells and development. Bioessays 2009; 31:1050–1061.
Lemercier C, Legube G, Caron C, et al. Tip60 acetyltransferase activity is controlled by phosphorylation. J Biol Chem 2003; 278:4713–4718.
Wang J, Chen J . SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J Biol Chem 2010; 285:11458–11464.
Gu W, Szauter P, Lucchesi JC . Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev Genet 1998; 22:56–64.
Akhtar A, Becker PB . Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell 2000; 5:367–375.
Morales V, Straub T, Neumann MF, Mengus G, Akhtar A, Becker PB . Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J 2004; 23:2258–2268.
Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC . A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol 2005; 25:9175–9188.
Dou Y, Milne TA, Tackett AJ, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 2005; 121:873–885.
Taipale M, Rea S, Richter K, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol 2005; 25:6798–6810.
Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009; 325:834–840.
Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001; 107:149–159.
Sykes SM, Mellert HS, Holbert MA, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 2006; 24:841–851.
Vaquero A, Scher MB, Lee DH, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev 2006; 20:1256–1261.
Miller KM, Tjeertes JV, Coates J, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 2010; 17:1144–1151.
Larschan E, Alekseyenko AA, Gortchakov AA, et al. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 2007; 28:121–133.
Wang Z, Zang C, Cui K, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009; 138:1019–1031.
Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL . Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006; 311:844–847.
Gupta A, Sharma GG, Young CS, et al. Involvement of human MOF in ATM function. Mol Cell Biol 2005; 25:5292–5305.
Sharma GG, So S, Gupta A, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and DSB repair. Mol Cell Biol 2010; 30:3582–95.
Chin HG, Esteve PO, Pradhan M, et al. Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res 2007; 35:7313–7323.
Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL . NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 2004; 119:803–814.
Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D . Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell 2004; 16:93–105.
Bouras T, Fu M, Sauve AA, et al. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem 2005; 280:10264–10276.
Fulco M, Schiltz RL, Iezzi S, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell 2003; 12:51–62.
Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D . SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 2007; 450:440–444.
Pruitt K, Zinn RL, Ohm JE, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet 2006; 2:e40.
Mendjan S, Taipale M, Kind J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 2006; 21:811–823.
Cai Y, Jin J, Swanson SK, et al. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem 2010; 285:4268–4272.
Li X, Wu L, Corsa CA, Kunkel S, Dou Y . Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell 2009; 36:290–301.
Akhtar A, Becker PB . The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO Rep 2001; 2:113–118.
Raja SJ, Charapitsa I, Conrad T, et al. The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol Cell 2010; 38:827–841.
Kadlec J, Hallacli E, Lipp M, et al. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat Struct Mol Biol 2011; 18:142–149.
Takata T, Ishikawa F . Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun 2003; 301:250–257.
Dai JM, Wang ZY, Sun DC, Lin RX, Wang SQ . SIRT1 interacts with p73 and suppresses p73-dependent transcriptional activity. J Cell Physiol 2007; 210:161–166.
Zhang R, Chen HZ, Liu JJ, et al. SIRT1 suppresses activator protein-1 transcriptional activity and cyclooxygenase-2 expression in macrophages. J Biol Chem 2010; 285:7097–7110.
Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004; 303:2011–2015.
Xu M, Long C, Chen X, Huang C, Chen S, Zhu B . Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 2010; 328:94–98.
Godde JS, Nakatani Y, Wolffe AP . The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res 1995; 23:4557–4564.
Acknowledgements
This work was supported by the National Basic Research Program Grants 2011CB965203, 2011CB964803 and 2011CB503902, Hi-Tech Research and Development Program of China Grant 2007AA021206 and the National Natural Science Foundation of China Grants 31030026 and 31021091.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
In vitro autoacetylation of hMOF. (PDF 987 kb)
Supplementary information, Figure S2
In vitro autoacetylation of GST-hMOF constructs. (PDF 920 kb)
Supplementary information, Figure S3
In vitro HAT assay was carried out using FLAG-IP purified mammalian hMOF-K274R complex. (PDF 1163 kb)
Supplementary information, Figure S4
Measurement of H4K16Ac levels from blank HeLa cells and HeLa cells with ectopic hMOF expression. (PDF 988 kb)
Supplementary information, Table S1
Primers used in this study. (PDF 24 kb)
Rights and permissions
About this article
Cite this article
Lu, L., Li, L., Lv, X. et al. Modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment and activities on the chromatin. Cell Res 21, 1182–1195 (2011). https://doi.org/10.1038/cr.2011.71
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2011.71
Keywords
This article is cited by
-
Revealing the role of epigenetic and post-translational modulations of autophagy proteins in the regulation of autophagy and cancer: a therapeutic approach
Molecular Biology Reports (2024)
-
KAT8 acetylation-controlled lipolysis affects the invasive and migratory potential of colorectal cancer cells
Cell Death & Disease (2023)
-
Acetylation of PAX7 controls muscle stem cell self-renewal and differentiation potential in mice
Nature Communications (2021)
-
NAD+ metabolism: pathophysiologic mechanisms and therapeutic potential
Signal Transduction and Targeted Therapy (2020)
-
Chidamide epigenetically represses autophagy and exerts cooperative antimyeloma activity with bortezomib
Cell Death & Disease (2020)