Abstract
Xbp1 has been shown to regulate the cell cycle as a transcriptional repressor in budding yeast Saccharomyces cerevisiae. In this study, we demonstrated that Xbp1 regulates DNA double-strand break (DSB) repair in S. cerevisiae. Xbp1 physically and genetically interacts with the histone deacetylase Rpd3 complex. Chromatin immunoprecipitation revealed that Xbp1 is required for efficient deacetylation of histone H4 flanking DSBs by the Rpd3 complex. Deletion of XBP1 leads to the delayed deacetylation of histone H4, which is coupled with increased nucleosome displacement, increased DNA end resection and decreased non-homologous end-joining (NHEJ). In response to DNA damage, Xbp1 is upregulated in a Mec1-Rad9-Rad53 checkpoint pathway-dependent manner and undergoes dephosphorylation. Cdk1, a central regulator of S. cerevisiae cell cycle, is responsible for Xbp1 phosphorylation at residues Ser146, Ser271 and Ser551. Substitution of these serine residues with alanine not only increases the association of Xbp1 with the Rpd3 complex and its recruitment to a DSB, but also promotes DSB repair. Together, our findings reveal a role for Xbp1 in DSB repair via NHEJ through regulation of histone H4 acetylation and nucleosome displacement in a positive feedback manner.
Similar content being viewed by others
Introduction
DNA double-strand breaks (DSBs) can be induced by a variety of extracellular and intracellular insults, such as reactive oxygen species and ionizing radiation. If they remain un-repaired, such DSBs can lead to genome instability 1. In response to DSBs, the DNA damage checkpoint in budding yeast Saccharomyces cerevisiae arrests cells at the G2/M phase 2. The checkpoint is initiated by the recruitment of multiple checkpoint components to the DSBs, including two sensor kinases Mec1 and Tel1 (ATR and ATM in mammals, respectively) 3, 4, 5. Once recruited to the DNA, Mec1 phosphorylates a subset of targets, including Ddc2 (ATRIP) and Rad9 6, 7, 8. Rad9 is an adapter protein mediating the activation of effector kinases Rad53 and Chk1 by Mec1 3, 9. A third sensor, the 9-1-1 clamp, which is made up of Rad17, Mec3 and Ddc1, also promotes Rad9 phosphorylation and Rad53 phosphorylation by Mec1. The DNA damage signaling eventually leads to phosphorylation of histone H2A (H2AX in mammals), recruitment of chromatin remodelers, including the Ino80, Rvb1, NuA4 and Swr1 complexes, to the DSBs, cell cycle arrest and global transcriptional response 3, 10. Eukaryotic cells mainly employ two approaches to repair DSBs: non-homologous end-joining (NHEJ) and homologous recombination (HR). NHEJ entails the direct rejoining of the broken ends of DNA, whereas HR needs a template to repair the break 11.
Histone modification, such as reversible acetylation, phosphorylation, ubiquitination and ADP-ribosylation, plays important roles in various nuclear processes including DNA repair 12, 13, 14. Histone acetylation and deacetylation are catalyzed by histone acetyltransferase (HAT) and histone deacetylase (HDAC), respectively. There are several HAT and HDAC complexes in eukaryotic cells. In S. cerevisiae, Rpd3, one of the major HDACs, exists in the Rpd3L or the Rpd3S complexes, both of which contain the common subunits, Rpd3, Sin3 and Ume1. Pho23, Sap30, Sds3, Cti6, Rxt2, Rxt3, Dep1, Ume6 and Ash1 are exclusively included in the Rpd3L complex, while Rco1 and Eaf3 are specific to Rpd3S 12, 15. Accumulating evidence points to the critical role of both acetylation and deacetylation in DSB repair. Mutation of histone H3 K14 and K23 increases the sensitivity to DSBs induced by HO endonuclease 16, a site-specific endonuclease required for initiating mating-type switching 17, 18, 19, 20. The NuA4 HAT complex is recruited to a DSB concomitantly with H2A S129 phosphorylation 6. Esa1, the catalytic subunit of NuA4, is required for efficient NHEJ 21, and interference of H4K16 deacetylation by Sin3/Rpd3 specifically impairs the NHEJ pathway 22. The HDACs Rpd3 and Hst1 are found to be recruited to the HO lesion during homologous recombination repair and may be responsible for the dynamic changes of histone acetylation 23. Acetylation of free histone H3 on K56 via the HAT Rtt109 has been shown to be critical for chromatin assembly after repair 24. Recently, two human homologs of Rpd3, HDAC1 and HDAC2 have been found to function in DNA damage response by promoting DSB repair 25.
Xbp1, which is homologous to the transcription factors, Swi4 and Mbp1, has been shown to regulate the cell cycle as a transcriptional repressor in S. cerevisiae. Xbp1 expression can be induced by stress and starvation during the mitotic cycle, while overexpression of Xbp1 results in growth retardation, G1 arrest and volume increase 26. Xbp1 has also been shown to function in efficient sporulation and morphology changes of yeast cells during nitrogen-limited growth 27, 28. Consistent with the role of Xbp1 in cell cycle control, a screen searching for direct Cdk1 substrates in S. cerevisiae identified Xbp1 as a potential Cdk1 substrate 29.
Although considerable progress has been made in understanding the role of histone acetylation in DNA repair, some important questions still remain to be answered. For instance, it is not clear how chromatin disassembly and assembly is affected by histone acetylation. The yeast HDACs Hst3 and Hst4 have been suggested to be downregulated by DNA damage in a checkpoint-dependent manner and to be responsible for the cell cycle regulation of histone H3K56 acetylation in the genome 30, but whether and how the cell cycle and checkpoint regulate histone acetylation flanking DSBs remain largely unclear. In this study, we identified Xbp1 as a regulator of DSB repair. We found that Xbp1 promotes deacetylation of histone H4 flanking DSBs, which is dependent on Sin3 and is important for subsequent inhibition of nucleosome displacement, slowdown of DNA end resection and promotion of NHEJ. In addition, Xbp1 can be upregulated in response to DNA damage in a Mec1-Rad9-Rad53 checkpoint pathway-dependent manner. Xbp1 can be phosphorylated by Cdk1 at Ser146, Ser271 and Ser551, and DNA damage-induced dephosphorylation probably facilitates its association with the Rpd3 complex and recruitment to a DSB site, which in turn promotes DSB repair.
Results
Xbp1 is required for cell survival upon methylmethane sulfonate (MMS)-induced DSBs
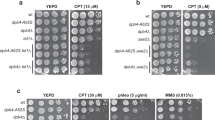
S. cerevisiae DNA repair-related candidate genes identified by bioinformatics approaches were tested for their contribution to cell survival in the presence of DNA damage agents MMS, hydroxyurea (HU) and 4-nitroquinoline-1-oxide (4-NQO) when each of these genes was deleted. Alkylating agent MMS induces DSBs during replication 31, 32. HU, a ribonucleotide reductase inhibitor, inhibits replication and induces formation of DSBs 33. The 4-NQO can mimic UV radiation and generate DNA lesions that are removed mainly by nucleotide excision repair (NER) 34. We found that deletion of XBP1 (xbp1Δ), one of the test candidates, endowed sensitivity to MMS as shown by measuring the growth rate at 5-fold serial dilution (Figure 1A and data not shown), and that re-introduction of Xbp1 with a low-copy vector into xbp1Δ cells fully suppressed this sensitivity. In contrast, deletion of XBP1 had no effect on cell survival in the presence of HU and 4-NQO. Because the 4-NQO-induced damage is mainly repaired by NER, the insensitivity of xbp1Δ cells to 4-NQO suggests no role of Xbp1 in NER. As shown before, deletion of RAD52, which plays important roles in all known pathways of HR as a recombination mediator 35, endowed the cells strong sensitivity to all these agents tested.
Xbp1 is required for the survival of MMS-induced DSBs. (A) DNA damage sensitivity analysis of xbp1Δ mutant. WT and xbp1Δ yeast strains expressing full-length Xbp1-FLAG under control of the ADH promoter in low-copy plasmid pRS315 or empty vector, or rad52Δ strain were spotted at 5-fold serial dilutions on SD-Leu plates containing MMS, HU or 4-NQO. A typical experiment was shown from at least three experiments. (B) Xbp1 is not required for checkpoint activation in response to MMS. Cells were treated with 0.1% MMS for the indicated time. Lysates were analyzed by anti-Rad53 immunoblotting.
To investigate whether the sensitivity to MMS of xbp1Δ cells is a result of its role in MMS-induced checkpoint signaling, we examined the Rad53 phosphorylation level, an indicator of checkpoint activation, and found that XBP1 deletion does not affect MMS-induced Rad53 phosphorylation, while deletion of RAD17, a component in 9-1-1 clamp 3, resulted in reduced Rad53 phosphorylation (Figure 1B). Taken together, these results suggested that Xbp1 is required for MMS-induced DSB repair, but does not function in MMS-induced checkpoint signaling.
Xbp1 physically and functionally interacts with the Rpd3 HDAC complex
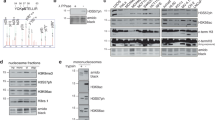
To gain insight into the molecular mechanism of Xbp1 in DSB repair, we tried to identify interacting proteins of Xbp1 by employing tandem affinity purification (TAP) with epitope-tagged Xbp1 and mass spectrometry. Ume1 and Sin3, components of the Rpd3 HDAC complex, were among the potential interacting proteins. Two components of this complex, Sin3 and Rpd3, were previously implicated in DSB repair mediated by both NHEJ and HR via regulating the acetylation of histone N-terminal tails 22, 23. Therefore, we wanted to determine whether Xbp1 interacts with the Rpd3 complex and then contributes to DSB repair by regulating histone acetylation via the Rpd3 complex. To this purpose, we tagged the HA epitope to the C-terminus of Ume1, Rpd3 and Sin3, three common subunits shared by both Rpd3L and Rpd3S and the epitope FLAG to the C-terminus of Xbp1 in WT, Ume1-HA, Rpd3-HA and Sin3-HA cells. Cell lysates of untreated or MMS-treated cells expressing Ume1-HA, Rpd3-HA, Sin3-HA and/or Xbp1-FLAG were subjected to anti-HA immunoprecipitation followed by anti-FLAG immunoblotting. As shown in Figure 2A, Xbp1 could interact with Ume1, Rpd3 and Sin3. Moreover, MMS treatment seemed to enhance this interaction. The relative weak interaction between Xbp1 and Sin3 in the presence of MMS might be owing to the decreased expression of Sin3. These interactions were insensitive to ethidium bromide, which interferes with DNA-protein interactions 36, 37, indicating that these interactions are not bridged by DNA (Figure 2B).
Xbp1 physically and functionally interacts with the Rpd3 complex that plays an important role in DSB repair. (A) Xbp1 interacts with Ume1, Rpd3 and Sin3. Cell extracts of untreated or MMS-treated cells expressing Ume1-HA, Rpd3-HA, Sin3-HA and/or Xbp1-FLAG were subjected to anti-HA immunoprecipitation and then anti-FLAG immunoblotting to detect protein interaction (upper panel), or to direct immunoblotting to verify protein expression (middle and lower panels). (B) The interaction between Xbp1 and the Rpd3 complex is insensitive to ethidium bromide. Ethidium bromide (100 μg/ml; final concentration) was added as indicated to interfere with DNA protein interactions. (C) The C-terminus of Xbp1 is important for the Xbp1-Ume1 interaction. Full-length (FL) and C-terminus deletion (ΔC) Xbp1 proteins are shown on the top. Gray rectangle indicates DNA-binding domain and black vertical bar stands for the CDK consensus phosphorylation site. (D) Deletion of UME1 abolishes the interaction between Xbp1 and Rpd3 or Sin3. Immunoprecipitations in B-D were performed similarly as in A. (E) The DSB repair function of Xbp1 depends on Rpd3 and Sin3. Indicated yeast cells were spotted at 5-fold serial dilutions on YPD plates containing MMS. A typical experiment was shown from at least three experiments.
The fact that Xbp1 has a strong interaction with Ume1 raised the possibility that Xbp1 may interact with the Rpd3 complex through Ume1. To test this idea, we examined protein interactions in the UME1 deletion strain and found that deletion of UME1 abolished both Xbp1-Rpd3 and Xbp1-Sin3 interaction (Figure 2D), indicating that the interaction between Xbp1 and the Rpd3 complex is probably mediated by Ume1. Furthermore, deletion of the C-terminal 200 amino acids of Xbp1 eliminated the Xbp1-Ume1 interaction (Figure 2C), suggesting that the C-terminus of Xbp1 is important for the Xbp1-Ume1 interaction.
We then investigated whether the DSB repair function of Xbp1 depends on the Rpd3 complex. XBP1 was deleted in rpd3Δ or sin3Δ background, yielding xbp1Δrpd3Δ and xbp1Δsin3Δ cells. As shown in Figure 2E, xbp1Δ cells were sensitive to MMS compared with wild-type (WT) cells. rpd3Δ and sin3Δ cells showed similar sensitivity to xbp1Δ cells. Interestingly, xbp1Δrpd3Δ and xbp1Δsin3Δ cells showed no more sensitivity than xbp1Δ, rpd3Δ or sin3Δ cells, suggesting that the function of Xbp1 in MMS-induced DSB repair is dependent on the Rpd3 complex.
Xbp1 promotes deacetylation of histone H4 flanking DSBs through the Rpd3 complex
Considering that Xbp1 can physically and functionally interact with the Rpd3 complex, we wanted to test whether Xbp1 has a role in regulating histone acetylation through the Rpd3 complex, which may contribute to the DSB repair function of Xbp1. The Rpd3 complex is one of the major HDAC complexes in S. cerevisiae and deletion of RPD3 results in histone H3 and H4 hyperacetylation 15, 38. To examine whether Xbp1 influences histone acetylation, we explored global acetylation level of histone H3 or H4 in WT and xbp1Δ cells with or without MMS treatment with polyclonal antibodies, which recognizes acetylated histone H3 or H4 on multiple lysine residues. We found that deletion of XBP1 affected neither histone H3 acetylation nor H4 acetylation regardless MMS treatment (Supplementary information, Figure S1A).
The finding that Sin3 acts to deacetylate lysine 16 of histone H4 near a DSB site 22 prompted us to investigate whether Xbp1 also modulates histone acetylation near the DSB lesions. We employed a HO-induced single chromosomal DSB system in JKM179, which has deletion of the HML and HMR loci and expresses HO endonuclease under the control of the galactose inducible promoter 39. As XBP1 harbors several mutations in this strain (our sequencing results, data not shown) and these mutations may compromise Xbp1 function, we thus re-introduced an empty vector or WT XBP1 containing two FLAG tags at its C-terminus under the control of its own promoter to this strain after removing the original XBP1. To observe the kinetics of histone H4 acetylation, we collected samples at 0, 40, 80 and 150 min after the HO endonuclease was induced, chromatin immunoprecipitation (ChIP) assay was then performed with anti-acetyl-histone H4 or anti-histone H4 antibodies, followed by quantitative PCR analysis. The primers that specifically amplify the region 0.4 or 1.6 kb upstream of the HO recognition site on chromosome III were used to examine acetylation near the HO site. Histone acetylation was measured as the ratio of the HO signals (derived from HO primers PCR) to the control signals (derived from control primers PCR) after normalized to the abundance of histone H4. In agreement with previous studies 6, 40, we observed transient hyperacetylation of histone H4 followed by deacetylation in the region 0.4 kb upstream of the break (Figure 3A). We also noticed that the deacetylation was weaker than that observed in previous studies. That could be due to the usage of different anti-acetyl-histone H4 antibody. In this study, we used an antibody that recognizes acetylated histone H4 on 5, 8, 12 and 16 lysine residues, so the acetyl histone H4 level observed could represent the comprehensive effect of these residues. Interestingly, in both the 0.4 and 1.6 kb region, the acetylation level of histone H4 appeared higher in XBP1 deletion cells than in xbp1Δ cells with re-introduction of XBP1. Quantitative PCR analysis revealed no significant difference in the extent of DSB induction in these strains, as shown by the percentage of intact MAT locus (Supplementary information, Figure S1B). These results suggest a role of Xbp1 in promoting deacetylation of local histone H4 flanking DSB sites.
Xbp1 mediates deacetylation of histone H4 flanking DSBs. (A) Xbp1-dependent deacetylation of histone H4. The xbp1Δ JKM179 cells expressing Xbp1-FLAG under control of its native promoter in pRS315 or empty vector were cultured to exponential phase in raffinose-containing SD-Leu medium. Samples were collected at 0, 40, 80 and 150 min after HO endonuclease was induced by adding 2% galactose. ChIP with antibodies, which can recognize histone H4 or acetylated histone H4 on lysines 5, 8, 12 and 16, was carried out followed by quantitative PCR. Data were quantitated as described in Materials and Methods. (B) Xbp1-dependent deacetylation of histone H4 relies on Sin3. ChIP was performed with xbp1Δ and xbp1Δsin3Δ strains containing Xbp1-FLAG under control of its native promoter in pRS315, Sin3 under control of its native promoter in pRS314 or empty vector. Samples were collected after HO endonuclease was induced for 150 min. ChIP was performed as in A, and data were quantitated as described in Materials and Methods. DSB induction efficiency determined by quantitative PCR was indicated. The asterisks indicate a statistically significant difference, which was analyzed by Student's t-test, and data are presented as mean ± SEM (n = 3, *P < 0.05; **P < 0.01). A typical experiment was shown from at least three experiments.
To test whether Xbp1-mediated deacetylation of histone H4 is dependent on the Rpd3 complex, we made double knockout of the XBP1 and SIN3 genes, and then introduced empty vector, the XBP1 and/or the SIN3 construct to the xbp1Δ or xbp1Δsin3Δ cells. As the increase in histone H4 acetylation level is more significant and persistent in the 1.6 kb region than the 0.4 kb region in xbp1Δ cells (Figure 3A), the acetylation level of histone H4 in the 1.6 kb region was then determined by ChIP assay after inducing the HO endonuclease for 150 min. In accordance with the data in Figure 3A, re-introduction of XBP1 accelerated deacetylation of histone H4 in xbp1Δ cells. XBP1 re-introduction had no effect in xbp1Δsin3Δ cells, however, expression of both XBP1 and SIN3 accelerated deacetylation of histone H4 in xbp1Δsin3Δ cells, indicating that the Xbp1 function depends on Sin3 (Figure 3B). Based on all these observations, we conclude that Xbp1 mediates deacetylation of histone H4 near DSBs and the deacetylation is dependent on the Rpd3 complex.
Xbp1 is required for efficient NHEJ and can be recruited to the vicinity of DSBs
Sin3 and Rpd3 have been reported to affect NHEJ, as their deletion led to a defect in religation of linearized plasmids 22. To test whether Xbp1 is also involved in NHEJ, we performed plasmid religation assay as described previously 41, 42. Briefly, the yeast centromere plasmid pRS313 was linearized by EcoRI in a region with no significant homology to the yeast genome. Yeast cells were then transformed in parallel with limiting amounts of cut or uncut plasmid DNA. Because plasmid maintenance requires religation of the linear plasmids, the relative transformation efficiency obtained with linear versus circular plasmid DNA can reflect the NHEJ activity of the yeast cells tested. The results in Figure 4A showed that RPD3 and SIN3 deletion impaired the religation efficiency, and YKU70 deletion almost completely blocked religation. xbp1Δ cells exhibited similar religation efficiency as rpd3Δ and sin3Δ cells, indicating that they may function together. We noticed that the decrease of religation efficiency in rpd3Δ and sin3Δ cells was mild compared with previous report 22, which might be because of the different religation system used in this study. However, blockage of religation by YKU70 deletion clearly indicates that our system worked well. We also examined the kinetics of NHEJ in JKM179 cells with or without Xbp1 and found that re-introduction of Xbp1 accelerated NHEJ repair (Figure 4B). These lines of evidence support a role of Xbp1 in NHEJ repair.
Xbp1 is required for efficient NHEJ and can be recruited to a region near a DSB. (A) Xbp1 is required for efficient DNA religation. WT, rpd3Δ, sin3Δ, xbp1Δ and yku70Δ cells were transformed in parallel with limiting amounts of EcoRI-linearized pRS313 plasmids or mock-digested circular pRS313 plasmid. Religation efficiency was calculated as described in Materials and Methods. (B) Re-introduction of Xbp1 accelerates NHEJ repair. The xbp1Δ cells in the JKM179 background expressing Xbp1-FLAG under control of its native promoter in pRS314 or empty vector were cultured to the exponential phase in raffinose-containing SD-Trp medium. HO endonuclease was induced by addition of galactose for 2 h. Glucose was then added to turn off the expression of HO endonuclease, and cells were allowed to grow for another 4 h. Samples were collected at 0, 2, 4 and 6 h after HO endonuclease was induced. Data were quantitated as described in Materials and Methods. (C) Xbp1 and Rpd3 are similarly recruited to a region near a DSB. The xbp1Δ cells in the JKM179 background expressing Xbp1-FLAG under control of its native promoter in pRS315 or Rpd3-FLAG cells were cultured to exponential phase in raffinose-containing SD-Leu medium (Xbp1) or YEP-lactate (Rpd3). Samples were collected at 0, 40, 80 and 150 min after HO endonuclease was induced. ChIP with anti-FLAG antibodies was carried out followed by quantitative PCR. (D, E) The Rpd3 complex promotes the recruitment of Xbp1 to DSB. The xbp1Δ or rpd3Δxbp1Δ cells expressing Xbp1-FLAG under control of its native promoter in pRS314 or RPD3-FLAGxbp1Δ cells expressing Xbp1 under control of its native promoter in pRS314 or empty vector were cultured to the exponential phase in raffinose-containing SD-Trp medium. Samples were collected at 0, 80 and 150 min after HO endonuclease was induced. ChIP was performed as in C, and data were quantitated as described in Materials and Methods. The asterisks indicate a statistically significant difference, which was analyzed by Student's t-test, and data are presented as mean ± SEM (n = 3, *P < 0.05; **P < 0.01). A typical experiment was shown from at least three experiments.
Xbp1 has been identified as a DNA-binding protein 26. In agreement with its role in regulating histone H4 acetylation, we also detected more than 2-fold enrichment of Xbp1 protein in the 0.4 kb region upstream of the HO-induced DSBs (Figure 4C). Interestingly, the kinetics of recruitment of Rpd3 is similar to that of Xbp1, further suggesting that they may function together. Next, we asked whether Xbp1 recruits the Rpd3 complex to DSBs or vice versa. ChIP assays showed that the recruitment of Xbp1 to a DSB site was decreased in the absence of RPD3, while XBP1 deletion had no effect on the recruitment of Rpd3 (Figure 4D and 4E and Supplementary information, Figure S2A, S2B), suggesting that the Rpd3 complex can promote the recruitment of Xbp1 to DSBs. Quantitative PCR analysis revealed no significant difference in the extent of DSB induction in these experiments (Supplementary information, Figure S2C and S2D). These data indicate that the Rpd3 complex facilitates the DSB binding of Xbp1, which in turn promotes histone H4 deacetylation by the Rpd3 complex and subsequently increases NHEJ repair.
Xbp1 slows down nucleosome displacement concomitantly with decreased DNA end resection
Histone acetylation controls chromatin assembly and disassembly processes 6, 24, 43. To test whether Xbp1-mediated deacetylation of histone H4 affects nucleosome displacement, we employed ChIP to measure the abundance of histone H4 flanking the DSBs. As shown in Figure 5A and 5B, in response to DSBs, histone H4 was quickly removed from both the 0.4 and 1.6 kb regions upstream of the HO-induced breaks. XBP1 deletion accelerated histone H4 removal. As histone removal usually accompanies with single-stranded DNA (ssDNA) formation 44, 45, we then examined whether Xbp1 also affects ssDNA formation by quantitative amplification of ssDNA (QAOS) analysis. As shown in Figure 5C, XBP1 deletion enhanced ssDNA formation. These data revealed that XBP1 deletion led to increased histone H4 acetylation, elevated histone removal and increased DNA end resection, suggesting that Xbp1-mediated deacetylation of histone H4 may protect the DNA ends by slowing down nucleosome displacement and DNA end resection and then promote NHEJ repair.
Xbp1 slows down nucleosome displacement and ssDNA formation. (A, B) Xbp1 delays nucleosome displacement. The xbp1Δ cells in the JKM179 background expressing Xbp1-FLAG under control of its native promoter in pRS315 or empty vector were cultured to exponential phase in raffinose-containing SD-Leu medium. Samples were collected at 0, 40, 80 and 150 min after HO endonuclease was induced by adding 2% galactose. ChIP with antibodies to histone H4 was carried out followed by quantitative PCR. (C) Xbp1 slows down ssDNA formation. Input samples from A and B were used to determine ssDNA levels by QAOS assay as described in Materials and Methods. The asterisks indicate a statistically significant difference, which was analyzed by Student's t-test, and data are presented as mean ± SEM (n = 3, *P < 0.05; **P < 0.01). A typical experiment was shown from at least three experiments.
DNA damage induces Xbp1 dephosphorylation
Xbp1 was recognized as a potential Cdk1 substrate in a screen searching for direct Cdk1 substrates in budding yeast 29. When XBP1 was expressed under the control of the ADH promoter instead of its own promoter to prevent the induction of Xbp1 by these DNA damage agents (see below), we noticed that the band of Xbp1 protein appeared as a smear on SDS-PAGE gels, and the smear was reduced under the treatment of MMS and HU but not phleomycin (PHL; Figure 6A). These findings together suggest that Xbp1 protein probably undergoes post-translational modification, most likely phosphorylation. Indeed, calf intestinal alkaline phosphatase (CIAP) significantly reduced the upper part of the Xbp1 band, confirming that the smear is due to phosphorylation (Figure 6B), which suggests that DNA damage provokes dephosphorylation of Xbp1.
The DSB binding and DSB repair activities of Xbp1 are regulated by Cdk1-mediated phosphorylation and DNA damage-induced dephosphorylation. (A) DNA damage agents induce Xbp1 dephosphorylation. The exponentially growing cells expressing Xbp1-FLAG under control of ADH promoter in pRS315 or empty vector (vec) were untreated (−) or treated with either 0.1% MMS, 200 mM HU or 40 μg/ml phleomycin for 1 h. Immunoblotting was performed with total cell lysates. (B) Xbp1 is a phosphoprotein. The exponentially growing cells expressing Xbp1-FLAG under control of its native promoter in pRS315 were untreated (−) or treated with 0.1% MMS for 1 h, protein extracts were then prepared. Half of each extract was treated with CIAP; and the remaining half was mock treated. Samples were analyzed by immunoblotting. (C) Mutation of S146, S271 and S551 to Ala (3SA) abolishes Xbp1 phosphorylation. Exponentially growing cells expressing Xbp1-FLAG or Xbp1-3SA-FLAG under control of its native promoter in pRS315 or empty vector were treated with either 0.1% MMS, 200 mM HU or 40 μg/ml phleomycin for 1 h. Immunoblotting was performed with total cell lysates. (D) Inhibition of Cdk1 activity by Sic1 overexpression decreases Xbp1 phosphorylation. RJD1026 (expressing inducible nondegradable Sic1-Δ3P), W303-1A and RJD1025 (expressing inducible WT Sic1) cells carrying Xbp1-FLAG or Xbp1-3SA-FLAG under control of the ADH promoter in pRS315 or empty vector were cultured to the exponential growing phase in raffinose-containing SD-Leu medium. Sic1 expression was transiently induced by adding 2% galactose. Aliquots were withdrawn every 30 min and processed for immunoblotting. Tubulin served as a loading control. (E) Xbp1-3SA increases the survival efficiency of both WT and xbp1Δ cells in response to MMS. Survival efficiency of WT and xbp1Δ strains expressing WT Xbp1-FLAG or Xbp1-3SA-FLAG under control of the ADH promoter in pRS315 or empty vector was determined as described in Materials and Methods. (F) The 3SA mutation strengthens the Xbp1-Ume1 interaction. Cell extracts of WT or Ume1-HA cells expressing Xbp1-FLAG, Xbp1-3SA-FLAG under control of its native promoter in pRS313 or empty vector were subjected to anti-HA immunoprecipitation and then anti-FLAG immunoblotting (upper panel), or to direct immunoblotting to verify protein expression (middle and lower panels). The numbers are the relative intensities of the immunoprecipitated protein against the total. (G) The 3SA mutation enhances the binding of Xbp1 to DSBs. The xbp1Δ JKM179 cells expressing Xbp1-FLAG or Xbp1-3SA-FLAG under control of its native promoter in pRS314 were cultured to the exponential phase in raffinose-containing SD-Trp medium. Samples were collected at 0, 80 and 150 min after HO endonuclease induction. Anti-FLAG ChIP was performed similarly as in Figure 4C. The asterisks indicate a statistically significant difference, which was analyzed by Student's t-test, and data are presented as mean ± SEM (n = 3, *P < 0.05; **P < 0.01). A typical experiment was shown from at least three experiments.
Cdk1-mediated phosphorylation of Xbp1 regulates its DSB repair activity
Xbp1 protein contains three Cdk consensus phosphorylation sites (S/T*-P-x-K/R) 29. To determine whether the serine residues in these consensus sites contribute to the phosphorylation of Xbp1, we substituted these serine residues (Ser146, Ser271 and Ser551) with alanine (3SA mutant), and examined its smear status by SDS-PAGE. As shown in Figure 6C, 3SA mutation severely diminished the smear of Xbp1, indicating that these serine residues make a major contribution to the smear status, i.e., the phosphorylation of Xbp1. Of note, both MMS and HU could further sharpen the band of 3SA, suggesting that there may be other phosphorylation sites. To further confirm that Cdk1 is the kinase responsible for Xbp1 phosphorylation, we overexpressed the Cdk1 inhibitor Sic1 46. Sic1 protein has a short half-life and mutations on its phosphorylation sites (Sic1-Δ3P, T2A/T5G, T33A and S76A) extends its half-life 46. Overexpression of the long half-life Sic1-Δ3P resulted in dephosphorylation of Xbp1 to the level of Xbp1 3SA mutant. In contrast, Xbp1 remained phosphorylated in WT cells or in cells overexpressing WT Sic1 (Figure 6D). These data strongly support the role of Cdk1 in direct phosphorylation of Xbp1.
To examine whether the Cdk1-mediated phosphorylation serves as a mechanism to regulate Xbp1 function, we examined the survival of WT and xbp1Δ cells expressing empty vector, WT Xbp1 or 3SA mutant in the presence of MMS. As shown in Figure 6E, Xbp1 3SA mutant promoted the survival rate, even higher than those of the cells expressing WT Xbp1 in both WT and xbp1Δ cells, strongly indicating an important role of phosphorylation in regulating Xbp1 activity in DSB repair.
Dephosphorylation of Xbp1 promotes its association with the Rpd3 complex and subsequent recruitment to DSBs
To investigate the function of Xbp1 phosphorylation, we first examined whether mutation of the Xbp1 phosphorylation sites affects its binding with the Rpd3 complex. As shown in Figure 6F, 3SA mutation enhanced the Xbp1-Ume1 interaction. Since the Rpd3 complex promotes the recruitment of Xbp1 to the HO-induced DSBs, we therefore studied whether 3SA mutation would also lead to increased recruitment of Xbp1. Indeed, ChIP assay revealed that the 3SA mutation enhanced the binding of Xbp1 to DSBs (Figure 6G and Supplementary information, Figure S3A), without affecting the DSB induction rate (Supplementary information, Figure S3B). These results suggest that DNA damage-induced dephosphorylation of Xbp1 may promote its interaction with the Rpd3 complex and recruitment to DSBs, then facilitating deacetylation of histone H4 by the Rpd3 complex and NHEJ repair.
Xbp1 is upregulated by DNA damage via the Mec1-Rad9-Rad53 checkpoint pathway
The XBP1 promoter contains several stress-regulated elements, and XBP1 mRNA has been shown to be induced by several stress conditions including MMS 26. To determine whether the Xbp1 protein level is regulated by DNA damage via the checkpoint pathway, we engineered the XBP1 genomic locus in WT and checkpoint mutant cells to encode Xbp1 with the C-terminal FLAG tag and assessed the Xbp1 protein level in the presence of different DNA damage agents. As shown in Figure 7A, Xbp1 protein was upregulated by MMS, HU or PHL, all of which can cause DSBs 31, 32, 33, 47. In order to investigate whether this induction depends on the checkpoint pathway, we examined the Xbp1 protein levels in cells defective in the checkpoint pathway. MEC1 and RAD53 deletion mutants can survive in sml1 mutation background 48. Deletion of SML1 alone had no effect on Xbp1 expression, but the Xbp1 level was very low in untreated sml1Δmec1Δ, sml1Δrad53Δ and rad9Δ cells. Furthermore, these checkpoint mutants curbed the ability of MMS to induce Xbp1 expression, in sharp contrast to the sml1Δ or WT cells (Figure 7B). These results demonstrate that the Xbp1 protein level is regulated by the Mec1-Rad9-Rad53 DNA damage checkpoint pathway.
Xbp1 is induced by DNA damage, and the induction is dependent on the checkpoint pathway. (A) MMS, HU and phleomycin increase the Xbp1 protein level. Exponentially growing cells expressing Xbp1-FLAG or Xbp1 in the endogenous loci were untreated (−) or treated with either 0.1% MMS, 200 mM HU or 40 μg/ml phleomycin for 1 h. Immunoblotting was performed with total cell lysates. (B) The induction of Xbp1 expression by MMS is abrogated in sml1Δmec1Δ, sml1Δrad53Δ and rad9Δ mutants. Exponentially growing WT and various checkpoint mutants cells expressing Xbp1-FLAG in the endogenous loci were untreated or treated with 0.1% MMS for 1 h. Immunoblotting was performed with total cell lysates. Tubulin served as a loading control.
Discussion
In this study, we identified XBP1 as an important gene that facilitates DNA DSB repair via NHEJ in the budding yeast S. cerevisiae. Xbp1 can physically and functionally interact with the Rpd3 complex, one of the major HDAC complexes in the budding yeast. Xbp1 promotes deacetylation of histone H4 in the vicinity of DSBs, and this effect is dependent on the Rpd3 complex. Delayed deacetylation of histone H4 occurs concomitantly with increased nucleosome displacement, increased ssDNA formation and reduced NHEJ in XBP1 deletion cells. In addition, phosphorylation of Xbp1, which is controlled by Cdk1 and DNA damage, is important for regulating its association with the Rpd3 complex, DSB binding and DSB repair activity. Finally, Xbp1 is upregulated by DNA damage in a checkpoint-dependent manner.
The function of Xbp1 in DNA repair
Xbp1 has been identified as a stress-induced transcriptional repressor of the Swi4/Mbp1 family 26. Xbp1 is also induced late in the meiotic cycle, and contributes to sporulation through inhibition of CLN1 expression 27. Furthermore, the Xbp1-mediated repression of CLB2 expression is important for morphology changes of yeast cells during nitrogen-limited growth 28. All these functions of Xbp1 rely on its transcriptional repressor activity. In this study, we uncovered a new function of Xbp1 in DSB repair. Although Xbp1 may indirectly affect DSB repair through regulating the expression of repair genes, our findings indicate that Xbp1 probably contribute to DSB repair directly: (1) it is recruited near a DSB, and its recruitment is probably facilitated by DNA damage-induced dephosphorylation; (2) it regulates histone H4 acetylation, nucleosome displacement and ssDNA formation flanking DSBs; and (3) its expression is regulated by the DNA damage checkpoint pathway.
Chromatin assembly and disassembly have been demonstrated to play an important role in DSB repair 43, 45, 49, 50. In response to DSBs, nucleosomes near the breaks are removed probably by the cooperation of ATP-dependent chromatin remodelers and histone chaperones 45. In addition, DNA end resection may also play a role in nucleosome removal 44, 45. In this study, we observed that XBP1 deletion leads to increased nucleosome displacement, enhanced ssDNA formation and increased histone H4 acetylation. Xbp1-promoted deacetylation of histone H4 slows down nucleosome displacement, probably either by inhibiting the function of some ATP-dependent chromatin remodelers or histone chaperones or by reducing DNA end resection. It has been shown that mutations in HAT Esa1 reduced recruitment of Rvb1 to DSBs 6, 43, which is a subunit of the INO80 chromatin remodeling complex required for efficient nucleosome displacement at a DSB 44. Thus, it is interesting to test whether Xbp1-mediated deacetylation of histone H4 inhibits nucleosome displacement through regulating the recruitment of INO80 complex to DSBs.
We found that both MMS and HU could induce the expression and dephosphorylation of Xbp1, while the XBP1 deletion mutant was sensitive only to MMS. This indicates that Xbp1 may have a different role in the HU-induced DSB repair process. Indeed, we observed that deletion of XBP1 increased Rad53 phosphorylation under HU treatment (data not shown), suggesting that Xbp1 may be involved in HU-induced checkpoint signaling.
The Rpd3 complex promotes the recruitment of Xbp1 to the vicinity of DSBs to facilitate deacetylation of histone H4
Although deregulation in global acetylation of both histone H3 and H4 has been linked to DSB repair defect 16, 21, the DSB repair processes may only require changes in the local histone acetylation flanking the lesions to construct a microenvironment that can facilitate repair. In addition, regulation of histone acetylation in the region near the damaged sites should be more specific without affecting the undamaged regions. Consistent with this, the NuA4 HAT complex was found to be recruited to a region proximal to a DSB via phosphorylation of H2A 6, and the HATs Gcn5 and Esa1 and the HDACs Rpd3 and Hst1 are recruited to the HO lesion during HR repair 23. In this study, we showed that Xbp1 can physically and functionally interact with the Rpd3 complex (Figure 2) and act to promote deacetylation of histone H4 flanking a DSB during NHEJ process in a Sin3-dependent manner (Figure 3). In addition, Xbp1 can be recruited to the vicinity of DSBs, which is facilitated by the Rpd3 complex (Figure 4C and 4D and Supplementary information, Figure S2A), and mutation of the Xbp1 phosphorylation sites enhances its association with the Rpd3 complex and then recruitment to DSBs (Figure 6F and 6G and Supplementary information, Figure S3). These observations support the role of Xbp1 as a binding factor of the Rpd3 complex to facilitate deacetylation of histone H4 in the region near DSBs.
Xbp1 can directly bind to DNA 26. Although there is a potential Xbp1-binding site at about 3.5 kb upstream of the HO cut site in the S. cerevisiae genome, our data showed that Xbp1 cannot bind to regions 0.4 or 1.6 kb away upstream of the HO cut site at the MAT locus without HO induction (Supplementary information, Figure S4A). This rules out the possibility of pre-binding of Xbp1 near the HO cut site. Furthermore, HO-induced DSB has no effect on Xbp1 expression (Supplementary information, Figure S4B). These data demonstrate that the DNA binding of Xbp1 to the DSB sites is specifically induced by HO.
Xbp1 acts in a positive feedback mechanism to facilitate DSB repair
It has been demonstrated that Xbp1 is upregulated late in meiosis 27. Our data showed that the Xbp1 protein level is upregulated in response to DNA damage and this regulation depends on the DNA damage checkpoint pathway (Figure 7). We also found that Xbp1 is phosphorylated by Cdk1 and dephosphorylated on DNA damage, and the unphosphorylated form of Xbp1 (Xbp1-3SA) is more active in repairing MMS-induced DSBs (Figure 6). These data indicate that Xbp1 promotes DNA repair in a positive feedback manner.
Together, our findings suggest the following working model. In response to DSBs, Xbp1 is upregulated and undergoes dephosphorylation. Then, Xbp1 binds to a region near DSBs and promotes the Rpd3 complex-mediated deacetylation of histone H4. Deacetylation of histone H4 may protect the DNA end by slowing down nucleosome displacement and ssDNA formation, and then promote direct religation of the two broken DNA ends by NHEJ.
Materials and Methods
Plasmid and strain construction
pRS315-ADH-FLAG or pRS315-FLAG were created by introducing the ADH promoter and/or FLAG tag to the multicloning sites of pRS315. pRS315-ADH-XBP1-FLAG was created by introducing full-length XBP1 gene to pRS315-ADH-FLAG. Mutation of Ser146, Ser271 and Ser551 of Xbp1 by alanine (Xbp1-3SA) was accomplished by PCR. pRS315-XBP1-FLAG was generated by introducing full-length XBP1 gene and its native promoter upstream of FLAG. pRS314-XBP1, pRS314-XBP1-FLAG or pRS314-XBP1-3SA-FLAG were generated by introducing XBP1, XBP1-FLAG or XBP1-3SA-FLAG and its native promoter to pRS314. pRS314-SIN3 was created by introducing the full-length SIN3 gene and its native promoter to pRS314. pRS313-XBP1-FLAG or pRS313-XBP1-3SA-FLAG were generated by introducing XBP1-FLAG or XBP1-3SA-FLAG and its native promoter to pRS313. Two vectors containing FLAG-TADH-URA or HA-TADH-LEU cassettes were generated for C-terminal tagging of proteins.
Yeast strains with complete deletion of the coding sequence of XBP1, RPD3, SIN3, YKU70 or RAD52 genes were constructed based on PCR-mediated gene disruption strategy 51. Construction of double mutant strains was performed by sequential gene disruption. C-terminal tags of Xbp1, Ume1, Rpd3 or Sin3 were constructed by PCR-based gene tagging methods 52. Strains used in these studies are listed in Supplementary information, Table S1.
TAP and mass spectrometry analysis
Yeast cells were first cultured in YPD overnight to a stationary phase. Then the cells were diluted and allowed to grow at 30 °C for about 4 h until the OD660 was between 0.6 and 0.8. Cells were harvested and lysed mechanically with glass beads. TAP purifications were done as described 53, 54.
The eluted proteins from TAP purification were reduced and alkylated with DTT and iodoacetamide, respectively. Digestion was performed with Promega sequencing grade modified trypsin overnight at 37 °C. Peptides were extracted with 10% formic acid and subjected to liquid chromatography tandem MS (LC-MS/MS) analysis.
DNA damage sensitivity assay and survival efficiency assay
Sensitivity assay
Yeast cells were first cultured in appropriate medium overnight to a stationary phase. Then the cells were diluted and allowed to grow at 30 °C for about 4 h until the OD660 was between 0.4 and 0.6. Five-fold dilution series were spotted on YPD or SD-Leu plates containing DNA damage agents as indicated and grown at 30 °C for 2-5 days.
MMS survival efficiency assay
It was performed similarly as in sensitivity assay except that the strains were diluted and plated onto YPD plates containing 0.02% MMS or no drug. Survival efficiency was calculated by counting colonies plated in triplicate in MMS plates after 4-day growth and normalized to that from no drug plates, respectively. P-value was determined using Student's t-test.
Plasmid religation assay
EcoRI linearized pRS313 plasmids (500 ng) were used for yeast transformation with lithium acetate methods. As a transformation-efficiency control, an equal amount of mock-digested circular pRS313 plasmid was also transformed. Religation efficiency was calculated by counting the number of transformants generated from the linearized plasmids and normalized to that from the uncut plasmids after 3-day growth in SD-His plates. P-value was determined using Student's t-test.
Immunoblotting and immunoprecipitation
Exponentially growing cells were untreated or treated with 0.1% MMS, 200 mM HU, 40 μg/ml PHL or 2% galactose for the indicated time, trichloroacetic acid (TCA) extracts were prepared as described 55 and proteins were resolved by 7.5% SDS-PAGE. Immunoblotting was performed with anti-FLAG-M2 monoclonal (Sigma), anti-tubulin monoclonal (Abcam), anti-Rad53 polyclonal (Santa Cruz Biotechnology), anti-acetyl-Histone H4 (Upstate) and anti-acetyl-Histone H3 (Upstate) antibodies.
Exponentially growing cell cultures (50 ml) were pelleted, washed two times with TBS (20 mM Tris-HCl, pH 7.6, 150 mM NaCl), and resuspended in 600 μl of lysis buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 1 mM PMSF) plus protease inhibitors (Roche). Proper amount of glass beads were added, and the cells were lysed by vortexing for 1 h at 4 °C. After centrifugation, the supernatant was pre-cleared with protein A sepharose 4B beads (Invitrogen) for 1 h to reduce the background. Then, 2 μl anti-HA antibody and 30 μl of protein A sepharose 4B beads were added and incubated at 4 °C overnight. Immunoprecipitates were washed three times with lysis buffer and then subjected to anti-FLAG or anti-HA immunoblotting. For MMS treatment, cell cultures were treated with 0.1% MMS for 1 h. To interfere with DNA-protein interactions, 100 μg/ml (final concentration) ethidium bromide was added to the lysates together with anti-HA antibody and protein A sepharose 4B beads, and then incubated at 4 °C overnight. Band intensity was quantified using Bandscan software.
ChIP assay
ChIP assay was performed based on the protocol from the Haber Lab (http://www.bio.brandeis.edu/haberlab/jehsite/protocol.html) with the following modifications: yeast cells were precultured in glucose-containing SD-Leu, SD-Trp or YPD medium (for detection of Rpd3 enrichment) overnight. Next day, the cells were washed and diluted in raffinose-containing SD-Leu, SD-Trp, SD-Leu/Trp or YEP-lactate medium (for detection of Rpd3 enrichment; OD660 = 0.4–0.5) and allowed to grow at 30 °C for about 5 h (OD660 = 0.6-07). Then 2% galactose was added to induce HO endonuclease (HO+). At each time point, 45 ml cells were harvested and crosslinked for 15 min at room temperature. After lysis, sonication and centrifugation, the samples were pre-cleared with 30 μl of protein A sepharose 4B beads at 4 °C for 1 h. Then proper amount of anti-acetyl-Histone H4 (Millipore), anti-Histone H4 (Abcam) or anti-FLAG-M2 monoclonal (Sigma) antibodies were added and the samples were incubated at 4 °C overnight. The next day, 40 μl of protein A sepharose 4B beads were added to each sample and incubated at 4 °C for 2 h. Other procedures, including ChIP washes, elution, reversing crosslink, etc. are exactly the same as that described in the protocol from the Haber Lab.
NHEJ kinetics analysis
HO endonuclease was induced in exponentially growing cells by addition of galactose (2% final) for 2 h. Then, glucose (2% final) was added to turn off the expression of HO endonuclease, and cells were allowed to grow for another 4 h. Samples were collected at the indicated time points and the genomic DNA was extracted and subjected to quantitative PCR of the percentage of uncut MAT locus. P-value was determined using Student's t-test.
Quantitative PCR
Immunoprecipitated DNA was analyzed by Mx3000P detection system (Stratagene) using EvaGreen dye (Biotium). HO primers to the regions which are 0.4 kb (SG563: 5′-TCAACCATATATAATAACTTAATAGACGACATTC-3′ and SG564: 5′-CTAGACGTTTTTCTTTCAGCTTTTTTG-3′) or 1.6 kb (SG573: 5′-GTTCTCATGCTGTCGAGGATTTT-3′ and SG574: 5′-AGACGTCCTTCTACAACAATTCATAAGT-3′) upstream of the HO recognition site on chromosome III, and control primers (SG525: 5′-AATTGGATTTGGCTAAGCGTAATC-3′ and SG526: 5′-CTCCAATGTCCCTCAAAATTTCTT-3′) to a region in SMC2 on chromosome VI were used 56. DSB primers (5′-CAGGATAGCGTCTGGAAGTCAAAA-3′ and 5′-GAGCAAGACGATGGGGAGTTTCAA-3′) amplifying the DNA that spans the HO cut site were used to determine the efficiency of DSB induction as described 57.
For analysis of histone acetylation, the ratio of the HO signals (derived from HO primers PCR) to the control signals (derived from control primers PCR) from anti-acetyl-Histone H4 immunoprecipitated samples was first calculated, and then normalized to the same ratio derived from anti-Histone H4 immunoprecipitated samples. For detection of histone H4, Xbp1 or Rpd3 levels near the break, the ratio of the HO signals to the control signals in the immunoprecipitated samples was calculated. For DSB induction efficiency analysis, DSB signals (derived from DSB primers PCR) were normalized to the control signals (derived from control primers PCR) in the input samples from the ChIP experiment. In these experiments, values obtained from the samples before HO endonuclease induction were assigned as one, and then the change after HO endonuclease induction for each group was calculated. For NHEJ kinetics analysis, DSB signals were normalized to the control signals as above. Values obtained from the samples before HO endonuclease induction were assigned as 100%, and then the change after HO endonuclease induction for each group was calculated. P-value was determined using Student's t-test.
QAOS assay
Input samples from ChIP assays were used to determine ssDNA level at 1.6 kb away from the DSB by QAOS assay as described previously 56, 58. Briefly, non-boiled or boiled input DNA samples were added to quantitative PCR reaction mixture containing primers HO1-ss (5′-ATCTCGAGCGTCATATCGGATCACACAATTCATAAGTC-3′), HO1-f (SG573) and Tag (5′-ATCTCGAGCGTCATATCGGATCAC-3′). PCR was carried out as follows: step1: 40 °C for 5 min, ramp to 72 °C, at 2 °C/min; step 2: 72 °C for 10 min; step3: 94 °C for 5 min; step4: 94 °C for 30 s, 56 °C for 30 s and 72 °C for 30 s (50 cycles). The ratio of the signals from non-boiled samples (amplification of ssDNA) to the signals from boiled samples (amplification of total DNA) was calculated to determine the percentage of ssDNA. P-value was determined using Student's t-test.
CIAP dephosphorylation assay
Exponentially growing cells treated with or without 0.1% MMS for 1 h were washed with 1 ml of 20% TCA and lysed in 100 μl of 20% TCA with equal volume of glass beads. Lysates were collected and combined with the washing from the glass beads (100 μl of 5% TCA, 2×). TCA was removed by centrifugation and precipitates were re-solubilized by adding 30 μl of 2 M Tris-HCl, pH 7.5. Then samples were treated with 60 U of CIAP (TaKaRa) or mock for 30 min at 37 °C. Finally, the samples were subjected to immunoblotting analysis.
References
Su TT . Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet 2006; 40:187–208.
Weinert TA, Hartwell LH . The Rad9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 1988; 241:317–322.
Harrison JC, Haber JE . Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 2006; 40:209–235.
Lisby M, Barlow JH, Burgess RC, Rothstein R . Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 2004; 118:699–713.
Melo J, Toczyski D . A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol 2002; 14:237–245.
Downs JA, Allard S, Jobin-Robitaille O, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell 2004; 16:979–990.
Emili A . MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell 1998; 2:183–189.
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM . DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273:5858–5868.
Gilbert CS, Green CM, Lowndes NF . Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell 2001; 8:129–136.
Shroff R, Arbel-Eden A, Pilch D, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 2004; 14:1703–1711.
Daley JM, Palmbos PL, Wu DL, Wilson TE . Nonhomologous end joining in yeast. Annu Rev Genet 2005; 39:431–451.
Kurdistani SK, Grunstein M . Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Bio 2003; 4:276–284.
Peterson CL, Laniel MA . Histones and histone modifications. Curr Biol 2004; 14:R546–R551.
Roth SY, Denu JM, Allis CD . Histone acetyltransferases. Annu Rev Biochem 2001; 70:81–120.
Yang XJ, Seto E . The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Bio 2008; 9:206–218.
Qin S, Parthun MR . Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol Cell Biol 2002; 22:8353–8365.
Haber JE . Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair 2006; 5:998–1009.
Jensen R, Sprague GF, Herskowitz I . Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci USA 1983; 80:3035–3039.
Kostriken R, Strathern JN, Klar AJ, Hicks JB, Heffron F . A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell 1983; 35:167–174.
Strathern JN, Klar AJ, Hicks JB, et al. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 1982; 31:183–192.
Bird AW, Yu DY, Pray-Grant MG, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 2002; 419:411–415.
Jazayeri A, McAinsh AD, Jackson SP . Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc Natl Acad Sci USA 2004; 101:1644–1649.
Tamburini BA, Tyler JK . Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol 2005; 25:4903–4913.
Chen CC, Carson JJ, Feser J, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 2008; 134:231–243.
Miller KM, Tjeertes JV, Coates J, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 2010; 17:1144–1151.
Mai B, Breeden L . Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol Cell Biol 1997; 17:6491–6501.
Mai B, Breeden L . CLN1 and its repression by Xbp1 are important for efficient sporulation in budding yeast. Mol Cell Biol 2000; 20:478–487.
Miled C, Mann C, Faye G . Xbp1-mediated repression of CLB gene expression contributes to the modifications of yeast cell morphology and cell cycle seen during nitrogen-limited growth. Mol Cell Biol 2001; 21:3714–3724.
Ubersax JA, Woodbury EL, Quang PN, et al. Targets of the cyclin-dependent kinase Cdk1. Nature 2003; 425:859–864.
Maas NL, Miller KM, DeFazio LG, Toczyski DP . Cell cycle and checkpoint regulation of histone H3K56 acetylation by Hst3 and Hst4. Mol Cell 2006; 23:109–119.
Kupiec M, Simchen G . Regulation of the RAD6 gene of Saccharomyces cerevisiae in the mitotic cell cycle and in meiosis. Mol Gen Genet 1986; 203:538–543.
Schwartz JL . Monofunctional alkylating agent-induced S-phase-dependent DNA damage. Mutat Res 1989; 216:111–118.
Merrill BJ, Holm C . A requirement for recombinational repair in Saccharomyces cerevisiae is caused by DNA replication defects of mec1 mutants. Genetics 1999; 153:595–605.
Friedberg EC, Walker GC, Siede W . DNA Repair and Mutagenesis. Washington DC: ASM Press, 1995.
San Filippo J, Sung P, Klein H . Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 2008; 77:229–257.
Yano KI, Morotomi-Yano K, Wang SY, et al. Ku recruits XLF to DNA double-strand breaks. EMBO Rep 2008; 9:91–96.
Lai JS, Herr W . Ethidium-bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA 1992; 89:6958–6962.
Rundlett SE, Carmen AA, Kobayashi R, et al. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA 1996; 93:14503–14508.
Moore JK, Haber JE . Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 1996; 16:2164–2173.
Lin YY, Qi Y, Lu JY, et al. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev 2008; 22:2062–2074.
Boulton SJ, Jackson SP . Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res 1996; 24:4639–4648.
Schar P, Herrmann G, Daly G, Lindahl T . A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev 1997; 11:1912–1924.
van Attikum H, Gasser SM . ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle 2005; 4:1011–1014.
Tsukuda T, Fleming AB, Nickoloff JA, Osley MA . Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 2005; 438:379–383.
Linger JG, Tyler JK . Chromatin disassembly and reassembly during DNA repair. Mutat Res 2007; 618:52–64.
Verma R, Annan RS, Huddleston MJ, et al. Phosphorylation of Sic1p by G(1) Cdk required for its degradation and entry into S phase. Science 1997; 278:455–460.
Moore CW . Internucleosomal cleavage and chromosomal degradation by bleomycin and phleomycin in yeast. Cancer Res 1988; 48:6837–6843.
Zhao XL, Muller EGD, Rothstein R . A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 1998; 2:329–340.
Groth A, Rocha W, Verreault A, Almouzni G . Chromatin challenges during DNA replication and repair. Cell 2007; 128:721–733.
Osley MA, Tsukuda T, Nickoloff JA . ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res 2007; 618:65–80.
Burke D, Dawson D, Stearns T . Methods in Yeast genetics. New York: Cold Spring Harbor Laboratory Press, 2000.
Bahler J, Wu JQ, Longtine MS, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998; 14:943–951.
Puig O, Caspary F, Rigaut G, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 2001; 24:218–229.
Rigaut G, Shevchenko A, Rutz B, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 1999; 17:1030–1032.
Pellicioli A, Lucca C, Liberi G, et al. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J 1999; 18:6561–6572.
van Attikum H, Fritsch O, Hohn B, Gasser SM . Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 2004; 119:777–788.
Morrison AJ, Highland J, Krogan NJ, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 2004; 119:767–775.
Booth C, Griffith E, Brady G, Lydall D . Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res 2001; 29:4414–4422.
Acknowledgements
We are grateful to Dr James E Haber (Brandeis University, USA) for yeast strains, insightful suggestions and critical comments on the manuscript, Drs Aaron D Gitler (University of Pennsylvania School of Medicine, USA), Raymond J Deshaies (California Institute of Technology, USA) and Stephen P Jackson (University of Cambridge, UK) for yeast strains, Dr Jürg Bähler (Wellcome Trust Sanger Institute, UK) for plasmids, Drs Jin-Qiu Zhou (Institute of Biochemistry and Cell Biology, SIBS, CAS, China) and Jijie Chai (National Institute of Biological Sciences, China) for technical support, and Dr Haiying Hang (Institute of Biophysics, CAS, China) for technical advice. We also thank Ting Martin Ma for critical reading and Junjie Luo for statistical analysis. This work was supported by grants from the National Natural Science Foundation of China (30930050, 30921004) and 973 Program (2006CB943401, 2006CB910102, 2010CB833703, 2010CB833706).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Xbp1 could not affect global acetylation level of histone H3 or H4. (PDF 136 kb)
Supplementary information, Figure S2
The Rpd3 complex promotes the recruitment of Xbp1 to DSB. (PDF 155 kb)
Supplementary information, Figure S3
3SA mutation enhances the binding of Xbp1 to DSBs. (PDF 116 kb)
Supplementary information, Figure S4
Xbp1 cannot bind to regions 0.4 kb or 1.6 kb away upstream of the HO cut site at the MAT locus without HO induction. (PDF 113 kb)
Supplementary information, Table S1
Yeast strains used in this study. (PDF 89 kb)
Rights and permissions
About this article
Cite this article
Tao, R., Chen, H., Gao, C. et al. Xbp1-mediated histone H4 deacetylation contributes to DNA double-strand break repair in yeast. Cell Res 21, 1619–1633 (2011). https://doi.org/10.1038/cr.2011.58
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2011.58
Keywords
This article is cited by
-
The down-regulation of XBP1, an unfolded protein response effector, promotes acute kidney injury to chronic kidney disease transition
Journal of Biomedical Science (2022)
-
A common strategy for initiating the transition from proliferation to quiescence
Current Genetics (2017)