Abstract
Magnesium (Mg2+) is abundant in plant cells and plays a critical role in many physiological processes. A 10-member gene family AtMGT (also known as AtMRS2) was identified in Arabidopsis, which belongs to a eukaryote subset of the CorA superfamily, functioning as Mg2+ transporters. Some family members (AtMGT1 and AtMGT10) function as high-affinity Mg2+ transporter and could complement bacterial mutant or yeast mutant lacking Mg2+ transport capability. Here we report an AtMGT family member, AtMGT9, that functions as a low-affinity Mg2+ transporter, and is essential for pollen development. The functional complementation assay in Salmonella mutant strain MM281 showed that AtMGT9 is capable of mediating Mg2+ uptake in the sub-millimolar range of Mg2+. The AtMGT9 gene was expressed most strongly in mature anthers and was also detectable in vascular tissues of the leaves, and in young roots. Disruption of AtMGT9 gene expression resulted in abortion of half of the mature pollen grains in heterozygous mutant +/mgt9, and no homozygous mutant plant was obtained in the progeny of selfed +/mgt9 plants. Transgenic plants expressing AtMGT9 in these heterozygous plants can recover the pollen phenotype to the wild type. In addition, AtMGT9 RNAi transgenic plants also showed similar abortive pollen phenotype to mutant +/mgt9. Together, our results demonstrate that AtMGT9 functions as a low-affinity Mg2+ transporter that plays a crucial role in male gametophyte development and male fertility.
Similar content being viewed by others
Introduction
Magnesium (Mg2+) is an essential mineral nutrient for plants 1, 2, 3 and is the most abundant free divalent cation in plant cells. In animals it has been shown that individual cell types maintain different levels of Mg2+ 4, 5, 6, 7 and this seems to be also true in plants 8, 9. The uptake and efflux of Mg2+ are precisely regulated to maintain homeostasis of this crucial cation. Although the importance of Mg2+ in plant growth and development has been recognized for a long time, the molecular mechanisms of Mg2+ homeostasis and transport remain poorly understood.
The CorA family of Mg2+ transporters is the most extensively studied Mg2+ transport system, which exists widely in bacteria, fungi, animals, and plants, mediating both influx and efflux of Mg2+, and appears to be the primary Mg2+ transport system 10, 11, 12, 13. Topological analysis of CorA protein in bacteria reveals a unique structure that contains a large, acidic N-terminal domain and two transmembrane domains in the C-terminal region. There is an absolutely conserved GMN motif near the end of the first hydrophobic region, and mutagenesis of CorA has shown that these three conserved residues are essential for functional Mg2+ transport 14. Recently, crystal structure of the pentameric cone-shaped CorA Mg2+ transporter from Thermotoga maritima had been elucidated by several research groups 15, 16, 17, 18, demonstrating CorA as a channel-like transport system.
Li et al. 12 and Schock et al. 13 independently identified a 10-member Arabidopsis AtMGT gene family encoding Mg2+ transport proteins that are homologous to the bacterial CorA. Members of the AtMGT family are expressed in a broad range of plant tissues. One family member, AtMGT1, functionally complements an Mg2+ transport mutant of Salmonella typhimurium, whereas a second member, AtMGT10, complements yeast mutant deficient in CorA-like transporters, suggesting that these plant MGT proteins function as Mg2+ transporters. Tracer assays showed that both members mediate the uptake of cations with similar kinetics to that of CorA, a high-affinity Mg2+ transporter in bacteria 19, 20. The AtMGT1 was localized in plant plasma membrane 12 and AtMGT10 (also known as AtMRS2-11) was localized in plant chloroplast envelope membrane system 21, implicating its role in Mg2+ uptake and translocation into chloroplasts. A third member, AtMGT2 (also known as AtMRS2-1), complements the yeast deletion mutant mrs2 and restores the intramitochondrial Mg2+ concentration to nearly wild-type level 13. Very recently, Li et al. 22 identified the fourth member AtMGT5, functioning as a dual-functional Mg2+-transporter that mediates uptake or efflux in a concentration-dependent manner. AtMGT5 is localized in the mitochondria and plays an essential role in pollen development. Mao et al. 23 reported the fifth member AtMGT7, functioning as a low-affinity Mg2+-transporter. Here, we present evidence that another AtMGT member, AtMGT9, functions as a low-affinity Mg2+ transporter and is essential for pollen development and male fertility.
Results
AtMGT9 complements a bacterial strain deficient in Mg2+ uptake
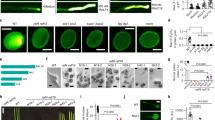
The S. typhimurium mutant strain MM281, which lacks Mg2+ transport systems CorA, MgtA, and MgtB 24 and requires 10 mM Mg2+ in the medium for its normal growth, has served as a good model system to identify proteins that mediate Mg2+ uptake and translocation 12. We transformed this strain with either the vector alone or vector expressing an AtMGT member. Complementation analysis showed that the negative control (MM281 and MM281 transformed with empty pTrc99A vector) did not grow in medium containing less than 10 mM of Mg2+, and the positive control (MM281 transformed with AtMGT1) could grow in medium containing as low as 10 μM Mg2+. MM281 transformed with AtMGT9 could survive in medium containing 500 μM Mg2+, suggesting that AtMGT9 is capable of mediating Mg2+ uptake in the range of sub-millimolar Mg22+ concentrations, but with lower affinity than AtMGT1 (Figure 1A). Furthermore, the results from the liquid growth curve analysis were also consistent with the complementation results observed on agar plates (Figure 1B).
Complementation of MM281 mutant by AtMGT9. (A) Growth of different bacterial strains on the N-minimal medium containing 10 μM, 500 μM, 1 mM, 2 mM, 5 mM, and 10 mM MgSO4. Rows 1–4 of each plate showed growth status of MM281 or MM281-transformed cells: row 1, MM281; row 2, MM281 transformed with pTrc99A vector as negative control; row 3, MM281 transformed with AtMGT1 cDNA in pTrc99A vector as positive control; row 4, MM281 transformed with AtMGT9 cDNA in pTrc99A vector. The horizontal spots of each row stand for the bacteria grown from cultures diluted in a 10-fold series from left to right. (B) Bacterial growth curves when cultured in liquid medium. MM281 cells and MM281 overexpressing AtMGT1 and AtMGT9 were grown in N-minimal liquid medium containing increasing concentrations of MgCl2 from a to d: 100 μM (a), 500 μM (b), 1 mM (c), and 10 mM (d). The growth of the cultures was monitored every 2 h by OD600. Data are average values of three independent experiments and are presented as mean ± SE.
AtMGT9 functions as a low-affinity Mg2+ transporter
The transport properties of AtMGT9 were analyzed in uptake experiments using 63Ni2+ as a tracer, based on previously described procedures 12, 22, 23, 24. A number of studies have demonstrated that Mg2+ and Ni2+ can use the same transport system with similar kinetics in the cell 14, 20. Inhibition of Ni2+ tracer uptake by non-radioactive Mg2+ or other cations represents the uptake efficiency of Mg2+ or other cations. Studies have proved that AtMGT1 acts as a high-affinity Mg2+ transporter 12. Figure 2A and 2B show that the uptake kinetics for Ni2+ and Mg2+ displayed distinct patterns between AtMGT1 and AtMGT9. In the MM281 mutant strain, Mg2+ began to inhibit tracer uptake at a concentration of 10 mM, which is consistent with the fact that MM281 required 10 mM MgSO4 to grow. In MM281 expressing AtMGT1, tracer uptake inhibition took place in concentrations of 1–10 μM of Mg2+ or 10–100 μM of Ni2+. In MM281 expressing AtMGT9, tracer uptake inhibition took place in concentrations of 100 μM-1 mM of Mg2+ or 1–10 mM of Ni2+, and the uptake kinetics curve is between AtMGT1 and MM281, suggesting that AtMGT9 functions as a low-affinity Mg2+ transporter.
Ni2+ and Mg2+transport measured by inhibition of 63Ni2+ uptake. (A) Mg2+inhibition of 63Ni2+ uptake. (B) Ni2+ inhibition of 63Ni2+ uptake. Transport assay was performed using three Salmonella strains as described in Materials and Methods. Tracer uptake is presented as the percentage of the maximal uptake by each strain. Data are average values of three independent experiments and are presented as mean ± SE. Uptake was measured for 3 min with triplicates at each cation concentration.
To further address the functional identity of AtMGT9, we performed tracer inhibition assays with a number of other divalent cations to determine the ionic selectivity of AtMGT9. As shown in Figure 3, several divalent cations, including Co2+, Mn2+, Cd2+, Cu2+, and Fe2+, significantly inhibited tracer uptake by AtMGT9. For example, the concentrations required for 50℅ inhibition are in the range of 100 μM-1 mM for Mg2+, Co2+, Mn2+, Fe2+, and Cu2+; more than 10 μM for Cd2+. These results indicate that AtMGT9 is capable of transporting Mg2+ under physiological conditions (cellular Mg2+ levels overlap with its transporting range of 100 μM-1 mM). Although AtMGT9 might be able to transport other divalent cations, the concentrations required for activity went beyond physiological range for those cations that are considered as trace elements for plant nutrition and exist well below the micromolar levels.
Cation selectivity of AtMGT9. Inhibition of AtMGT9-mediated tracer uptake by divalent cations. The data shown are normalized to the percentage of control (63Ni2+ uptake with 100 μM Ni2+ in the solution without other divalent cations.) Data are average values of three independent experiments and are presented as mean ± SE. Uptake was measured for 3 min with triplicates at each cation concentration.
AtMGT9 displays distinct expression patterns in Arabidopsis
The expression pattern of AtMGT9 was analyzed by semi-quantitative reverse transcription (RT)-PCR with different PCR reaction cycles of 20, 24, 28, 30, 32, and 35. The PCR products increased linearly when amplified from 20 to 32 cycles, and they showed similar expression pattern under 24, 28, and 30 cycles of amplification. The result revealed that AtMGT9 transcripts were present in all tissues tested, including roots, stems, young cauline, or rosette leaves, flowers and siliques (Figure 4A). To analyze AtMGT9 gene expression pattern in more detail, we made a reporter gene construct by using the promoter of AtMGT9 to drive the expression of β-glucuronidase (GUS) gene and produced transgenic plants containing this construct. Seven GUS transgenic lines were selected randomly and their GUS activities were identified with similar expression pattern. GUS activity was detected in anther (Figure 4B-a, b), vascular tissues of the cotyledon and young true leaves (Figure 4B-e), and the root tips (Figure 4B-c, d). In addition, GUS activity was also detected in the dry seed, but not in the imbibed seed. GUS activity was not detected in mature rosette or cauline leaves, or young siliques (data not shown).
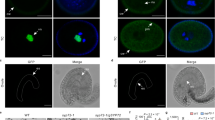
Expression patterns of AtMGT9 by RT-PCR and GUS activity analysis. (A) Transcriptional expression patterns of AtMGT9 by semi-quantitative RT-PCR. The size of the AtMGT9 DNA fragment produced in the RT-PCR experiment is 640 bp. (B) GUS histochemical staining in a floral bud cluster from a plant transformed with AtMGT9 promoter-GUS. GUS activity (indicated by blue color) was observed in two discontinuous developmental stages (a and b). GUS activity was also detectable in the tip of the main root, young lateral root (c and d), and in the vascular tissue of young leaves (e). The scale bars are 1 mm. (C) Longitudinal sectioning of flowers at various stages revealed specific AtMGT9/GUS activity in the anther, with distinctive temporal pattern during pollen development. Pollen developmental stages classification referred to Sanders et al. 25. (a) Flower buds at stages 1–5. Stage 5 is the earliest time AtMGT9 promoter activity was detectable. (b and c) GUS activity was detectable in anther during pollen developmental stages 5–10. (d and e) GUS activity was disappeared at stages 11–12. (f) GUS activity was detected in anther again from stage 13. The scale bars are 200 μm. (D) Cross sectioning of flowers at various stages revealed specific AtMGT9/GUS activity in the anther, with distinctive spatial and temporal pattern during pollen development. (a) Flower buds at 1–5, no GUS activity was detected earlier than stage 5. (b and b′) Stage 5 is the earliest time AtMGT9 promoter activity was detectable. (b′) is the enlarged image of one anther sectioning in (b). (c and c′) GUS was evenly distributed throughout the anther at stage 9. (d and d′) GUS activity was shifted to tapetum at stage 10. (e and e′) No GUS activity was detected in anther at stages 11. (f and f′) GUS was evenly distributed throughout the anther and the mature pollen grains during stages 13–14. The scale bars are 50 μm.
Perhaps the most striking finding in the histochemical analysis is that GUS activity appeared to fluctuate significantly during different stages of the anther development. The GUS activity was strong at some stages of anther development, but weak or not detectable at others. Strong GUS activity was detected from stages 5 to 10 and from stages 13 to 14 as defined by Sanders et al. 25 (Figure 4B-a, b). From the longitudinal and cross-sections of flower buds of the transgenic plants, a rather unique temporal and spatial pattern of GUS activity was observed (Figure 4C and 4D). The onset of AtMGT9 promoter activity began as early as stages 5–6 of anther development (Figure 4D-b, b′). The GUS accumulation at stages 7–9 is evenly distributed throughout the anther (Figure 4D-c, c′). As the anther develops to stages 9–10, the onset of pollen maturation, the spatial pattern of AtMGT9 expression is shifted to tapetum, the “nursing tissue” of pollen development (Figure 4D-d, d′). When the tapetum tissue is degraded, no GUS activity was detected in the anther at stages 11 and 12 (Figure 4D-e, e′). The GUS activity then reappeared in the entire anther including the mature pollen grains (Figure 4D-f, f′).
Taken the above results together, the expression patterns of AtMGT9 varied temporally and spatially in tissues of anther, roots, and leaves.
Isolation and analysis of T-DNA insertion mutant of AtMGT9
One T-DNA insertion mutant (mgt9, salk_030174) of AtMGT9 gene was identified from ABRC collections of T-DNA transformed Arabidopsis lines. The insertional site was mapped to the +91 nucleotide in relation to the first nucleotide (+1) of the translation start codon (ATG) (Figure 5A). Genotyping of T2 plants obtained from the seed pool led to the identification of several heterozygous +/mgt9 mutant plants. However, attempts to identify homozygous mgt9/mgt9 mutant plants were unsuccessful. Moreover, no homozygous mutant seedlings were obtained in the progeny of selfed +/mgt9 plants, suggesting that gametophyte and/or embryo development may have been defective when AtMGT9 function is disrupted.
Phenotypic analysis of pollen grains from control and +/mgt9 mutant plant. (A) Scheme of AtMGT9 gene structure and position of the T-DNA insertion. The ATG start codon and TGA stop codon are indicated. The T-DNA insertion is shown as the triangle above the gene diagram. (B) Bright-field microscopy images of mature pollen grains from wild type (a), and from T-DNA insertion line +/mgt9 (b). Approximately 50% of the mature pollen grains from +/mgt9 heterozygous plant were aborted (b and c). Arrows showed abnormal pollen grains. (C) Vigor detection of pollen grains. Bright-field microscopy image of mature pollen grains from +/mgt9 showed the bigger normal pollen grains and the smaller abortive pollen grains (a). Fluorescent microscopic images of the corresponding pollen grains showed that the bigger pollen grains can emit fluorescence under UV-excitation after DAPI staining, but the small abortive pollen grains that have undergone cell death and DNA degradation cannot emit any fluorescence (b). Arrows showed abnormal pollen grains. (D) SEM of the anthers and pollen grains. Whole anther structure at the dehiscence/senescence transition stage of Col.0 (a) and +/mgt9 heterozygous mutant (b), scale bars are 50 μm; higher magnification images reveal healthy wild type (c) and about 50% aborted pollen grains in the +/mgt9 mutant (d), scale bars are 10 μm; the image of the spalling pollen grains of +/mgt9 mutant (e and f).
The T-DNA insertion in the +/mgt9 mutant plants confers kanamycin resistance (Kanr). Segregation of Kanr was analyzed in the progeny of eight selfed heterozygous +/mgt9 plants (Table 1). From a total of 1 106 offspring, 536 seedlings were kanamycin resistant. This segregation ratio of 1:1 is consistent with the expected ratio for a fully penetrant mutation in a gene required for either the male or female gametophyte. Reciprocal cross-tests between the heterozygous +/mgt9 mutant and wild-type plants were carried out. The F1 progenies were genotyped by PCR and analyzed for Kanr or kanamycin sensitivity (Kans). When the heterozygous mutant was used as the male parent, the progeny population did not have kanamycin resistant seedlings or seedlings bearing the mutated mgt9 allele. When the heterozygous mutant was used as the female parent, 154 seedlings from 323 offspring were kanamycin resistant, and bore the mutated allele. These results showed that all the pollen grains, but not ovules, bearing the mutated mgt9 allele were unable to transmit this allele. In other words, the pollen grains carrying the mutant allele are not fertile.
AtMGT9 heterozygous mutant +/mgt9 shows abortive pollen phenotype
We examined the pollen grains in the heterozygous mutant (+/mgt9) flowers and found that about 50% of the pollen grains collapsed at stages close to maturation (Figure 5B-b, c). Pollen grains were also observed under scanning electron microscopy (SEM). Anthers and pollen grains from control plants appeared normal (Figure 5D-a, c), whereas a number of abnormal and collapsed pollen grains were obvious in the anthers from heterozygous mutant plants (Figure 5D-b, d, e, f). The percentage of aborted pollen grains was consistent with that observed by light microscopy (Figure 5B). The death ratio of pollen grains was also calculated by UV-fluorescent microscopy after staining with DAPI (Figure 5C-a, b).
Anther development in the +/mgt9 mutant was also analyzed by paraffin sectioning (Figure 6). The aborted pollen grains were clearly distinguishable at stage 11 of anther development in the heterozygous mutant (Figure 6B′). Except for the pollen phenotype, no other phenotype in non-pollen tissues is observed.
Cross sectioning of anthers from wild type and +/mgt9 mutant plants. Anthers were stained with gentian violet. Abortive pollen grains were discerned by size and color discrepancy of the pollen grains with wild type. Pollen grains in each anther of wild type were evenly stained (A–C). In heterozygous +/mgt9 mutant, pollen grains at stage 9 or earlier stages in each anther were colored evenly (A′); however, pollen grains color discrepancy appeared at stage 10 and obviously from stages 11–12 (B′ and C′). The scale bars are 50 μm.
Since there is only one T-DNA insertion mutant, we performed two more experiments to verify that the observed abnormal development of microspores was caused by the loss-of-function in the AtMGT9 gene (Figure 7A). First we performed a genetic complementation experiment by introducing pCAMBIA1300-PMGT9-cDNA-TMGT9 construct into +/mgt9 mutant plant. In T2 populations, obtained from self-pollinated T1 plants, four of the 25 tested independent lines were homozygous for both the mgt9 allele (assessed by PCR) and the AtMGT9 transgene (assessed by hygromycin resistance). In all tested T3 progenies of these four lines, pollen development was normal (Figure 7B-c), demonstrating that the AtMGT9 transgene fully complemented the male sterile phenotype.
AtMGT9 mutant and other transgenic plants. (A) AtMGT9 mutant and other transgenic plants. From left to right, they are wild-type plant (a), heterozygous T-DNA insertion mutant plant (b), complementation transgenic plant (c), RNAi transgenic plant (d). (B) Pollen grains from wild-type plant (a), heterozygous T-DNA insertion mutant plant (b), complementation transgenic plant (c), and RNAi transgenic plant (d). Arrows showed abnormal pollen grains. (C) The expression levels of AtMGT9 transcripts in wild-type plant (wt), heterozygous T-DNA insertion mutant plant (+/mgt9), complementation transgenic plant (pMGT9PMGT9 in mgt9/mgt9), and RNAi transgenic plant (MGT9-RNAi).
Secondly, we used the RNA interference approach to connect the function of AtMGT9 to pollen development. We obtained 12 independent transgenic T1 plants harboring the RNAi construct (Figure 8). Among these transgenic plants, nine independent plants have defects in the gametophytic development, similar to that of +/mgt9 (Figures 7B-d, 8C and 8D). Other three independent transgenic plants are indistinguishable from wild type (Figures 7B-a and 8D). Taking ACTIN2 gene as a control, we analyzed the expression levels of AtMGT9 in anther tissues of the transgenic plants by semi-quantitative RT-PCR method. The results showed that the mRNA levels of AtMGT9 were decreased in the transgenic plants with abortive pollen phenotype, whereas the mRNA levels were normal in the three transgenic plants that exhibited normal phenotype (Figures 7C and 8B).
Downregulation of AtMGT9 by RNAi. (A) Schematic diagram of AtMGT9 RNAi construct pRNAi-AtMGT9. Part of the AtMGT9 sequence, the RNAi cassette, which contains insertion of AtMGT9 specific fragment in opposite direction, assembled in pKANNIBAL, and transferred into the binary vector pART27. (B) The expression pattern analysis of anther tissues of the MGT9-RNAi transgenic plants with semi-quantitative RT-PCR. Compared with wild-type Col.0, AtMGT9 transcripts were greatly repressed in MGT9-RNAi transgenic plants with abortive pollen phenotype (lines 1, 2, 3, 4, 6, 8, 9, 10, and 12), but remained normal in MGT9-RNAi transgenic plants without obvious phenotype (lines 5, 7, and 11). (C) Twelve MGT9-RNAi transgenic lines were analyzed. (D) Pollen grains of the MGT9-RNAi transgenic plants. Nine of the twelve transgenic lines showed partial pollen grain abortive phenotype, in consistence with the repression of AtMGT9 transcripts in plants (B).
Discussion
Though Mg2+ plays very important roles in many biochemical and physiological processes, our understanding of Mg2+ acquisition, translocation, and cellular homeostasis is still in its infancy. Recent studies have demonstrated that CorA-like Mg2+ transporters function as a major transporting system in a broad range of organisms including bacteria, fungi, plants, and some animals. In higher plants, CorA-like Mg2+ transporter family (AtMGT) has been identified from Arabidopsis thaliana and consists of at least 10 members. Two of the family members, AtMGT1 and AtMGT10, were experimentally confirmed to function as high-affinity Mg2+ transporters 12, 13. AtMGT5 was identified to function as a dual-functional Mg2+-transporter mediating both influx and efflux 22. Very recently, by using both functional complementation and tracer uptake assays, AtMGT7 was identified to function as a low-affinity Mg2+ transporter 23. Here, we identified another AtMGT family member, AtMGT9, that serves as an Mg2+ transporter in the range of sub-millimolar Mg2+ concentration in the medium, placing this transporter in the low-affinity category as compared with the high-affinity transporters such as AtMGT1 and AtMGT10. Because this Mg2+ concentration may exist in some of the soil types, AtMGT9 may function in Mg2+ uptake from the soil, consistent with the expression of this gene in the root tip region (Figure 4B-c, d). In addition, AtMGT9 may also function in intraorganelle Mg2+ translocation if it is located in the endomembrane systems.
Concerning the function of AtMGT9, it is relevant to compare this transporter with AtMGT5, another member in the AtMGT family. Both T-DNA knockout mutants of AtMGT9 and AtMGT5 showed abortive pollen phenotype. Genetic analysis also revealed that the mutated allele cannot be transmitted through male gametophyte. These findings suggest a connection between Mg2+ transport and homeostasis in pollen grains and pollen development. How may these two transporters be involved in pollen grain Mg2+ homeostasis? It is interesting to note that AtMGT5 was localized to mitochondria. As Mg2+ is a critical cofactor of ATP synthases and other enzymes in the respiration processes, Mg2+ level should serve as an essential regulatory element of mitochondrial activity. As mitochondrial defects have long been implicated in male sterility, it is not fortuitous that AtMGT5 plays an essential role in pollen development 26, 27, 28, 29. Although due to technical difficulty the sub-cellular localization of AtMGT9 protein has not been determined, it stands as a possibility that this transporter may also be localized in the mitochondria and regulate Mg2+ homeostasis in this organelle. Due to its distinct transport property as compared to that of AtMGT5, it is reasonable to speculate that AtMGT9 may function differently from AtMGT5; while AtMGT5 mediates both influx and efflux, AtMGT9 mediates Mg2+ influx only when the Mg2+ concentration in the external medium reaches submillimolar levels.
Both AtMGT9 and AtMGT5 are essential for pollen development, suggesting that these transporters are not functionally redundant. Several possible scenarios may be proposed regarding the possible mechanism of action. First, as discussed above, they may be both located in the mitochondria but function independently in different processes, each of which is essential for mitochondrial Mg2+ homeostasis. Second, the two transporters may interact with each other and form hetero-oligomer complexes in mitochondria. Third, they may be each located in a different membrane system working coordinately in pollen grain Mg2+ transport. Each of these possibilities is supported by some evidence. The GUS activity analyses showed that AtMGT5 is an anther-specific gene and, like AtMGT9, follows a distinct temporal and spatial pattern along the stages of anther development. However, the expression patterns of these two MGT genes, although with some overlap, are not the same. For example, AtMGT5 is expressed from stages 5 to 13, but not in later stages when pollen grains are mature. For AtMGT9, GUS activity is also detected from stages 5 to 10, but undetectable at stages 11–12, and detected again from stage 13. Sectioning observation revealed that the expression of AtMGT5 gene appears to 'shift' from the filament-anther junction to the tapetum, and eventually to pollen grains along the developmental progression of microspores. AtMGT9 gene seems to express in more cell types in anther. Besides in anther, AtMGT9 is also expressed in vascular tissues of young leaves and root tips. By comparing AtMGT9 and AtMGT5 expression patterns, it is not likely for the two AtMGTs to assemble together into a Mg2+-transport channel (see below), though we can not completely exclude the possibility because expression of the two genes do overlap at some pollen development stages (stages 5–10).
AtMGT family members share significant structural features with the bacterial Mg2+ transporter CorA. The CorA transport system is constitutively expressed, mediates both influx and efflux of Mg2+, and appears to be the primary Mg2+ transport system 11. Recent work shows that a bacterial CorA Mg2+ transporter from T. maritima is assembled into a homo-pentameric structure 15, 16, 17, 18. It is therefore likely that plant MGT members may either function as homopentamers or as heteropentamers. In the context of expression patterns of AtMGT members, some members may indeed work together as heteropentamers. For example, four AtMGT members are shown to express in anther at some stages of flower development. Apart from AtMGT9 and AtMGT5, AtMGT4 and AtMGT6 are also expressed in pollen development (data not shown). AtMGT4 is anther-specific and showed almost identical expression pattern to AtMGT5, while AtMGT6 showed similar expression pattern with AtMGT9. These observations support the possibility that in planta, hetero-pentamer protein(s) may be formed by combinations of AtMGTs.
In an earlier work by our group, one AtMGT member, AtMGT7, was found encoding two mRNAs that have resulted from alternative splicing, AtMGT7a and AtMGT7b. AtMGT7a was identified expressing in all organs of the plant by RT-PCR and the AtMGT7a variant was identified functioning as a low-affinity Mg2+ transporter 23. AtMGT9 is also expressed in all organs and the AtMGT9 protein also functions as a low-affinity Mg2+ transporter. Although unlike AtMGT9, AtMGT7 is not an essential gene, and no phenotype was observed in AtMGT7 T-DNA insertion lines, the similar expression pattern and similar affinity to Mg2+ raise the possibility that they may work together in Mg2+-transport. Our ongoing investigation will test this hypothesis.
Materials and Methods
Functional complementation of Mg2+-transport by S. typhimurium mutant MM281 system
The S. typhimurium mutant MM281, which lacks the Mg2+-transport genes – CorA, MgtA, and MgtB, is often used as a system for functional complementation analysis of candidate Mg2+-transporters 24. AtMGT9 cDNA was subcloned into pTrc99A vector as previously described 1. MM281 cells were transformed with pTrc99A vector or pTrc99A-AtMGT9 plasmid by electroporation. Cells were plated onto LB medium supplemented with 20 mM Mg2+ and appropriate antibiotics (100 μg/ml ampicillin, 34 μg/ml chloramphenicol, and 50 μg/ml kanamycin), and incubated at 37 °C overnight. The transformants were confirmed by PCR amplification of both the vector and AtMGT9 coding sequences. Individual positive transformants were grown in LB liquid medium containing 20 mM Mg2+, same antibiotics, and 0.05 mM IPTG for the induction of AtMGT9 expression. MM281-AtMGT1 reported earlier 12 was used as a positive control. The cultures were adjusted to 1.0 OD600, diluted in a 10-fold series, and spotted (2 μl) onto N-minimal medium supplemented with different concentrations of MgSO4 and the antibiotics 30. Growth of different strains was scored after incubation at 37 °C for 24 h. The growth rate of the four strains in liquid medium was also monitored. After growing in LB liquid medium to log phase (OD600 0.6–0.8), the cells were harvested by centrifugation, washed twice with distilled water to remove traces of Mg2+, and resuspended in distilled water. N-minimal liquid medium was prepared containing increasing concentrations of MgSO4 (10 μM, 500 μM, 1 mM, and 10 mM). Cells were then added to a final OD600 of 0.001–0.002. The growth of the cultures was monitored over 26 h and the OD600 was plotted as a function of growth time.
Tracer uptake assays
This assay utilized the fact that Mg2+ and Ni2+ can be transported by the same CorA-like transporters with similar kinetics 14. The percent of inhibition of Ni2+ tracer uptake by non-radioactive Mg2+ represents the uptake efficiency of Mg2+. Uptake of 63Ni2+ was performed as described previously 11, 12, 20, 22. Bacterial mutant MM281 strains containing the pTrc99A empty vector, pTrc99A-AtMGT1, or pTrc99A-AtMGT9, were used to perform the 63Ni2+ uptake assays as follows. Bacterial cultures were grown overnight in LB medium containing 20 mM Mg2+ and the appropriate antibiotics. When the concentration of the cells reached OD600 = 1, 0.05 mM IPTG was added to the culture medium and the culture was grown for additional 5 h. Cells were washed with N-minimal medium, and diluted 1:5 with the same medium containing appropriate antibiotics. After a 3-h subculture in the minimal medium, cells were collected by centrifugation at 1 000×g for 15 min and washed twice with ice-cold Mg2+-free N-minimal medium. The washed cells were resuspended in the same medium and cell density was adjusted to 1.0 OD600. For a standard assay, uptake was initiated by adding 0.2 ml cells to 0.8 ml of N-minimal medium containing 100 μM NiCl2 and 0.5 μCi of 63Ni2+. For the ionic selectivity assay, various concentrations of divalent cations were included in the tracer uptake buffer. Typically, uptake was stopped after 5 min by adding 1 ml of ice-cold washing buffer. Cells were washed four times with 1.5 ml of ice-cold washing buffer and the 63Ni2+ content of bacterial cells was determined by a scintillation counter (model A210001 Packard Instruments). The relative tracer uptake was standardized against the maximal value and presented as % of maximal uptake.
Plant material and growth conditions
A. thaliana plants (ecotype Columbia 0) were used in this study unless otherwise indicated. Plant growth condition and Agrobacterium-mediated transformation were as described previously 31. The transgenic lines were selected on 0.5× Murashige and Skoog medium (Sigma-Aldrich) containing 50 μg/ml kanamycin (for pBI121-, pBI101-based vectors) or 30 μg/ml hygromycin B (for pCAMBIA 1300-based vectors).
AtMGT9 cDNA cloning and expression pattern analysis
Total RNA of 7-week-old Arabidopsis plants was extracted with RNAeasy Kit (Qiagen). Total RNA (2.5 μg) was heated at 65 °C for 10 min and then subjected to RT reaction using SuperTranscriptase II (Invitrogen) and oligo dT18 for 60 min at 42 °C. 1 μl of RT reaction was used as template to amplify AtMGT9 cDNA with ExTaq DNA polymerase (TaKaRa). The primers were as follows: MGT9F (5′-TGT TGA TGC TGG TGC TCT GAA-3′); MGT9R (5′-TCG AAA ACA AGA TCT GCC CAA-3′). The PCR amplifications were performed with initial denaturation at 94 °C for 3 min, followed by 30 standard PCR reaction cycles each consisting of a 30 s denaturing step at 94 °C, an annealing step at 56 °C for 40 s and an elongation step at 72 °C for 1 min 30 s, and a final extension of 10 min at 72 °C. Aliquots of PCR samples were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining under UV light. To conduct AtMGT9 cDNA cloning, target cDNA band was recovered from the agarose gel, and ligated into pMD18-T vector to obtain the AtMGT9 cDNA clone. Several cDNA clones were sequenced to acquire the clones whose sequences matched the published sequence in The Arabidopsis Information Resource.
To analyze AtMGT9 gene expression pattern, total RNAs from root, stem, leaf, flower, and silique tissues were extracted and used in semi-quantitative RT-PCRs as described above except for different PCR reaction cycles of 20, 24, 28, 30, 32, and 35. ACTIN2 gene was used as a quantitative control (with primers ACTIN2-F: 5′-GCC ATC CAA GCT GTT CTC TC-3′; ACTIN2-R: 5′-GCT CGT AGT CAA CAG CAA CAA-3′).
Isolation of AtMGT9 knock-out allele and phenotypic analysis
Seeds for the T-DNA insertion line of the AtMGT9 gene were purchased from the ABRC (Arabidopsis Biological Resource Center, OH, USA) and identified using a primer specific for the T-DNA left border and AtMGT9 specific primers (allele MGT9 (in salk_030174) was from ABRC). The T-DNA insertion site in the allele is described in the Salk Institute website (http://signal.salk.edu) and was confirmed by PCR-based genotyping (Figure 5A). Pollen grains from the plants harboring the heterozygous T-DNA allele were collected and analyzed using bright field, fluorescent microscopy (Olympus BX51) and SEM as described previously 32.
Transgenic plants
For analyzing AtMGT9 promoter activity, a 2.5-kb fragment upstream of ATG starting codon was PCR-amplified using the primers MGT9-PF (5′-GCTCTAGAA TGT AAA GTT AAT ATT TAC GTG ACT AT-3′), and MGT9-PR (5′-CGGGATCCC ATT GAC TTA CCT GCA GCT GGT CAG AGA-3′). The amplified promoter sequence was digested with XbaI and BamHI, and inserted into the multicloning site in the plant binary vector pBI101.1 to drive the expression of the GUS reporter 33, 34, 35. Transformed Arabidopsis plants carrying the promoter-GUS fusion were selected based on kanamycin resistance. The promoter activity was visualized in the plant tissues by GUS activity stain using 5-bromo-4-chloro-3-indolyl-β-D-glucuronide as a substrate according to published protocols 35. For further analysis of the GUS staining pattern, GUS-stained flower clusters were embedded in paraffin, sectioned, and examined under microscope according to Ruzin 36.
For genetic complementation, the AtMGT9 promoter (PMGT9) and terminator (TMGT9) fragments were amplified from wild-type genomic DNA using the primers (PMGT9-F: 5′-CGGGTACCT CTT GCT CGT CAT TAT G-3′, PMGT9-R: 5′-GCGGATCCA AAA CTA CCT CAC CGT C-3′; TMGT9-F: 5′-GCCTGCAGC CTT ATT AGC TAG AC-3′, TMGT9-R: 5′-CGAAGCTTA TGT GGA AAC ACG A-3′). The AtMGT9 cDNA fragment was amplified from cloned construct pGEM-AtMGT9 (MGT9cds-F: 5′-GCCTGCAGA TGG CGC AAA ACG GG-3′, MGT9cds-R: 5′-CGCTGCAGT CAA GAT CCC ACA AG-3′). Restriction enzyme pairs KpnI and BamHI, PstI and HindIII, and PstI and PstI were used to excise the promoter, terminator, and cDNA fragment respectively. They were then subcloned into binary vector pCAMBIA 1300 (pCAMBIA, Canberra, Australia), resulting in the transcriptional fusion construct PMGT9-cDNA-TMGT9. This construct was introduced into Agrobacterium tumefaciens strain GV3101 (mp90) by electroporation and was selected on kanamycin. AtMGT9 complementation construct was transformed into the +/mgt9 mutant plant with the floral dipping method 37, and the transformants were selected on 30 μg/ml hygromycin.
For preparing the RNAi construct, a 560-bp fragment in the coding region of AtMGT9 was amplified and subcloned into the pKANNIBAL vector as described by Wesley et al. 38. This fragment was chosen because it exhibited low nucleotide sequence identity (less than 40%) to the corresponding regions in other AtMGT genes. The RNAi cassette assembled in pKANNIBAL was transferred into the binary vector pART27 as a NotI fragment, yielding the final T-DNA construct pRNAi-AtMGT9 to produce transgenic plants used in this study.
References
Leroy J . Necessité du magnesium pour la croissance de la souris. Comput Rend Soc Biol 1926; 94: 431–433.
Lusk JE, Williams RJP, Kennedy EP . Magnesium and the growth of Escherichia coli. J Biol Chem 1968; 243:2618–2624.
Marschner H, (eds). Mineral Nutrition in Higher Plants. 2nd edn, Academic Press: San Diego, 1995.
Valberg LS, Holt JM, Paulson E, Szivek J . Spectrochemical analysis of sodium, potassium, calcium, magnesium, copper, and zinc in normal human erythrocytes. J Clin Invest 1965; 44:379–389.
Seiler RH, Ramirez O, Brest AN, Moyer JH . Serum and erythrocytic magnesium levels in congestive heart failure: effect of hydrochlorothiazide. Am J Cardiol 1966; 17:786–791.
Walser M . Magnesium metabolism. Ergeb Physiol 1967; 59:185–296.
Iyengar GV, Kollmer WE, Bowen HJM (eds). The Elemental Composition of Human Tissues and Body Fluids. Verlag Chemie, Weinheim: New York, 1978.
Stelzer R, Lehmann H, Krammer D, Luttge U . X-Ray microprobe analysis of vacuoles of spruce needle mesophyll, endodermis and transfusion parenchyma cells at different seasons of the year. Botanica Acta 1990; 103:415–423.
Shaul O, Hilgemann DW, de-Almeida-Engler J, Van MM, Inze D, Galili G . Cloning and characterization of a novel Mg2+/H+ exchanger. EMBO J 1999; 18:3973–3980.
Lusk JE, Kennedy EP . Magnesium transport in Escherichia coli. J Biol Chem 1969; 244:1653–1655.
Snavely MD, Florer JB, Miller CG, Maguire ME . Magnesium transport in Salmonella typhimurium: expression of cloned genes for three distinct Mg2+ transport systems. J Bacteriol 1989; 171:4752–4760.
Li L, Tutone AF, Drummond RS, Gardner RC, Luan S . A novel family of magnesium transport genes in Arabidopsis. Plant Cell 2001; 13:2761–2775.
Schock I, Gregan J, Steinhauser S, Schweyen R, Brennicke A, Knoop V . A member of a novel Arabidopsis thaliana gene family of candidate Mg2+ ion transporters complements a yeast mitochondrial group II intron-splicing mutant. Plant J 2000; 24:489–501.
Szegedy MA, Maguire ME . The CorA Mg2+ transport protein of Salmonella typhimurium: mutagenesis of conserved residues in the second membrane domain. J Biol Chem 1999; 274:36973–36979.
Lunin VV, Dobrovetsky E, Khutoreskaya G . Crystal structure of the CorA Mg2+ transporter. Nature 2006; 440:833–837.
Eshaghi S, Niegowski D, Kohl A, Martinez Molina D, Lesley SA, Nordlund P . Crystal structure of a divalent metal ion transporter CorA at 2.9 angstrom resolution. Science 2006; 313:354–357.
Payandeh J, Pai EF . A structural basis for Mg2+ homeostasis and the CorA translocation cycle. EMBO J 2006; 25:3762–3773.
Maguire ME . The structure of CorA: a Mg2+-selective channel. Curr Opin Struct Biol 2006; 16:432–438.
Smith RL, Banks JL, Snavely MD, Maguire ME . Sequence and topology of the CorA magnesium transport systems of Salmonella typhimurium and Escherichia coli: identification of a new class of transport protein. J Biol Chem 1993; 268:14071–14080.
Smith RL, Szegedy MA, Kucharski LM, et al. The CorA Mg2+ transport protein of Salmonella typhimurium: mutagenesis of conserved residues in the third membrane domain identifies a Mg2+ pore. J Biol Chem 1998; 273:28663–28669.
Drummond RSM, Tutone A, Li YC, Gardner RC . A putative magnesium transporter AtMRS2-11 is localized to the plant chloroplast envelope membrane system. Plant Sci 2006; 170:78–89.
Li L, Sokolov LN, Yang Y, et al. A mitochondrial magnesium transporter functions in Arabidopsis pollen development. Mol Plant 2008; 1:675–685.
Mao DD, Tian LF, Li LG, et al. AtMGT7: an Arabidopsis gene encoding a low affinity magnesium transporter. J Integr Plant Biol; 50:1530–1538.
Snavely MD, Gravina SA, Cheung TT, Miller CG, Maguire ME . Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. J Biol Chem 1991; 266:824–829.
Sanders PM, Bui AQ, Weterings K, et al. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 1999; 11:297–322.
Cui X, Wise RP, Schnable PS . The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 1996; 272:1334–1336.
Sabar M, Gagliardi D, Balk J, Leaver CJ . ORFB is a subunit of F1F(O)-ATP synthase: insight into the basis of cytoplasmic male sterility in sunflower. EMBO Rep 2003; 4:381–386.
Heazlewood JL, Whelan J, Millar AH . The products of the mitochondrial orf25 and orfB genes are FO components in the plant F1FO ATP synthase. FEBS Lett 2003; 540:201–205.
Hanson MR, Bentolila S . Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 2004; 16:S154–S169.
Nelson DL, Kennedy EP . Magnesium transport in Escherichia coli. Inhibition by cobaltous ion. J Biol Chem 1971; 246:3042–3049.
Zhang S, Raina S, Li H, et al. Resources for targeted insertional and deletional mutagenesis in Arabidopsis thaliana. Plant Mol Biol 2003; 53:133–150.
Gupta A, Ludäscher B, Martone ME . Registering scientific information sources for semantic mediation. 21st International Conference on Conceptual Modeling (ER), 2002:182–198.
Datla RS, Hammerlindl JK, Panchuk B, Pelcher LE, Keller W . Modified binary plant transformation vectors with the wild-type gene encoding NPTII. Gene 1992; 211:383–384.
Jefferson RA, Burgess SM, Hirsh D . Beta-glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA 1986; 83:8447–8451.
Jefferson RA, Kavanagh TA, Bevan MW . GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 1987; 6:3901–3907.
Ruzin SE . Plant Microtechnique and Microscopy. Oxford: Oxford University Press, 1999:322.
Clough SJ, Bent AF . Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998; 16:735–743.
Wesley SV, Helliwell CA, Smith NA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 2001; 27:581–590.
Acknowledgements
We are grateful to Dr Michael E Maguire (Case Western Reserve University, USA) for providing the Salmonella mutant strain MM281 and advice on tracer uptake. This work was supported by grants from National Natural Science Foundation of China (30370794), Hunan Young Scientist Program (03JJY1003), and Hunan Provincial NSFC (08JJ3088).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Chen, J., Li, Lg., Liu, Zh. et al. Magnesium transporter AtMGT9 is essential for pollen development in Arabidopsis. Cell Res 19, 887–898 (2009). https://doi.org/10.1038/cr.2009.58
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2009.58
Keywords
This article is cited by
-
New insights into comprehensive analysis of magnesium transporter (MGT) gene family in rice (Oryza sativa L.)
3 Biotech (2023)
-
Discovery and characterization of magnesium transporter (MGT) gene family in Citrus sinensis and their role in magnesium deficiency stress
Plant Growth Regulation (2023)
-
Magnesium fertilization reduces high-temperature damages during anthesis in spring wheat (Triticum aestivum L.) by affecting pollen viability and seed weight
Environmental Science and Pollution Research (2023)
-
Genome-wide identification and expression analysis of magnesium transporter gene family in grape (Vitis vinifera)
BMC Plant Biology (2022)
-
24-Epibrassinolide induces protection against nickel excess in soybean plants: anatomical evidences
Brazilian Journal of Botany (2021)