Abstract
The mammalian target of rapamycin (mTOR) has drawn much attention recently because of its essential role in cell growth control and its involvement in human tumorigenesis. Great endeavors have been made to elucidate the functions and regulation of mTOR in the past decade. The current prevailing view is that mTOR regulates many fundamental biological processes, such as cell growth and survival, by integrating both intracellular and extracellular signals, including growth factors, nutrients, energy levels, and cellular stress. The significance of mTOR has been highlighted most recently by the identification of mTOR-associated proteins. Amazingly, when bound to different proteins, mTOR forms distinctive complexes with very different physiological functions. These findings not only expand the roles that mTOR plays in cells but also further complicate the regulation network. Thus, it is now even more critical that we precisely understand the underlying molecular mechanisms in order to directly guide the development and usage of anti-cancer drugs targeting the mTOR signaling pathway. In this review, we will discuss different mTOR-associated proteins, the regulation of mTOR complexes, and the consequences of mTOR dysregulation under pathophysiological conditions.
Similar content being viewed by others
Introduction
Easter Island is a small triangular-shaped Chilean island located in the South Pacific Ocean. This island, known as Rapa Nui in the native language, is world famous for its numerous moai or large stone head statues, which are listed as one of the New Seven Wonders of the World. However, most of people do not realize that it is also the humble origin of the wondrous story of TOR (target of rapamycin).
Roughly three decades ago, a bacterial strain, Streptomyces hygroscopicus, was first isolated from this island. These bacteria secrete a potent anti-fungal macrolide that was named rapamycin after Rapa Nui, the location of its discovery. Rapamycin was initially developed as an anti-fungal agent. However, its major application quickly changed after rapamycin was proven to have immunosuppressive and anti-proliferative properties. To date, rapamycin (sirolimus as the trade name) has become an FDA (Food and Drug Administration) approved drug for immunosuppression for organ transplantation, prevention of restenosis post-angioplasty, and chemotherapy for soft-tissue and bone sarcomas 1, 2, 3.
It soon became realized that the anti-proliferative properties of rapamycin were a very powerful tool to study cell growth regulation. In the 1990s, yeast genetic screens identified two rapamycin target genes, mutations of which allowed yeast to escape the cell cycle arrest caused by rapamycin treatment 4, 5. These two genes were named the target of rapamycin 1 and 2 (TOR1 and TOR2). Further studies revealed the molecular mechanism of rapamycin inhibition on TOR 6, 7, 8. Upon entering the cells, rapamycin binds a small protein receptor called FKBP12 (FK506-binding protein 12 kDa). The rapamycin/FKBP12 complex specifically binds to TOR and potently interferes with its function, causing cell growth arrest. Extensive genetic studies in yeast established that TOR plays essential roles in cell growth regulation, particularly in response to nutrients. The identification of TOR genes in yeast led to the subsequent discovery of TOR genes in higher eukaryotes, including mammals. The high degree of conservation among species strongly suggests that TOR is an essential cell growth controller. In addition, the mechanism by which rapamycin inhibits TOR in higher eukaryotes also appears to be conserved 9, 10, 11, 12. Recognition of the importance of TOR and the availability of rapamycin led to studies in yeast, flies, worms, and mammals to elucidate a basic understanding of TOR biology.
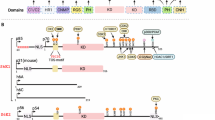
The mammalian TOR, mTOR, is an atypical serine/threonine protein kinase, belonging to the phosphatidylinositol kinase-related kinase (PIKK) family, with a predicted molecular weight of 290 kDa 13, 14. The physiological importance of mTOR is undoubtedly demonstrated by the fact that the knockout of mTOR in mice is primordially embryonic lethal 15, 16, 17. Structurally, mTOR possesses up to 20 tandem HEAT (a protein-protein interaction structure of two tandem anti-parallel α-helices found in huntingtin, elongation factor 3, PR65/A and TOR) repeats at the amino-terminal region, followed by an FAT (FRAP, ATM, and TRRAP, all PIKK family members) domain (Figure 1A) 18. The kinase domain is between the FRB (FKBP12/rapamycin binding) domain, which is C-terminal to the FAT domain, and the FATC (FAT C-terminus) domain, located at the C-terminus of the protein. It is speculated that the HEAT repeats serve to mediate protein-protein interactions, the FRB domain as suggested by its name provides a docking site for the FKBP12/rapamycin complex, and the FAT and FATC domains modulate mTOR kinase activity via unknown mechanisms.
Schematic of mTOR complex components. HEAT: a protein-protein interaction structure of two tandem anti-parallel a-helices found in huntingtin, elongation factor 3, PR65/A and TOR; FAT: a domain structure shared by FRAP, ATM and TRRAP, all of which are PIKK family members; FRB: FKBP12/rapamycin binding domain; FATC: FAT C-terminus; RNC: Raptor N-terminal conserved domain; WD40: about 40 amino acids with conserved W and D forming four anti-parallel beta strands; CRIM: conserved region in the middle; RBD: Ras binding domain.
The binding of rapamycin/FKBP12 to the mTOR FRB domain in vivo clearly blocks some of the physiological functions of mTOR. However, whether rapamycin directly inhibits mTOR's intrinsic kinase activity is not clear. While some scientists believe that the binding mainly prevents mTOR from interacting with its substrates, others have shown that mTOR autophosphorylation (intrinsic mTOR activity) is inhibited by rapamycin 19, 20, 21. Further studies on mTOR phosphorylation are needed to reveal whether rapamycin does, indeed, contribute to the regulation of mTOR intrinsic kinase activity. So far, quite a few phosphorylation sites have been identified in mTOR and many more are expected to come 22, 23, 24, 25, 26. While the phosphorylation of Ser2481 in mTOR has been considered the major indicator of mTOR intrinsic kinase activity, the contributions of other phosphorylation sites towards mTOR activity are not entirely understood.
The earliest identified and best-studied mTOR downstream effectors are S6K1 (p70 ribosomal protein S6 kinase 1) and 4EBP1 (eIF4E binding protein 1) 13. Under basal conditions, S6K1 and 4EBP1 are bound to eIF3 (eukaryotic initiation factor 3) and remain inactive 27. Upon growth stimulations, mTOR binds to eIF3 and phosphorylates S6K1 and 4EBP1. The phosphorylation of S6K1 releases it from eIF3 and activates the kinase. The active S6K1 promotes translation and growth by phosphorylating cellular substrates, such as S6 28, 29. 4EBP1 inhibits cap-dependent mRNA translation via binding to the translation initiator eIF4E (eukaryotic translation initiation factor 4E) 30. The phosphorylation of 4EBP1 by mTOR frees it from eIF4E, relieves its inhibitory effect and stimulates translation initiation. Together, active mTOR enhances cell growth by promoting protein translation and increasing cell mass. Cells with hyperactive mTOR often gain growth advantages and display a larger size 13, 31, 32.
Tuberous sclerosis complex inhibits mTOR activity
Much earlier than the discovery of rapamycin and mTOR, hamartoma syndromes have been documented along the course of human pathological history. These diseases are characterized by multiple benign tumors occurring in a variety of organs. Hamartomas are formed by normally differentiated but structurally disorganized cells, which are often enlarged. Among different hamartoma syndromes, tuberous sclerosis complex (TSC) is an autosomal disorder with a population prevalence of 1/5 000 to 1/10 000 33. TSC tumors can be found in many organs, including brain, heart, kidney, muscles and skin. Like typical hamartoma syndromes, TSC tumors are normally benign, but their presence in these tissues may result in severe clinical manifestations. Although the first documentation of TSC can be traced back to the 19th century, the cause of this disease remained unknown until the recent identification of the TSC1 and TSC2 tumor suppressor genes. Mutation of either one of these genes is sufficient to cause TSC. This was later explained by biochemical evidence demonstrating that TSC1 and TSC2 form a physical and functional complex in vivo. TSC1 stabilizes the complex, while TSC2 exerts GTPase activating protein (GAP) activity towards downstream effectors.
The seemingly parallel TSC syndrome and the mTOR-controlled cell growth were tied together in 2002, when our laboratory together with others 34, 35, 36, 37, inspired by the TSC genetic studies in Drosophila 38, 39, 40, 41, showed that the major function of TSC1/TSC2 is to inhibit mTOR. This finding provided the first piece of evidence for mTOR involvement in human tumorigenesis and opened the door for a plethora of studies on the regulation and functions of the TSC-mTOR signaling network.
The TSC1/TSC2 complex (TSC1/2) has been established as the major upstream inhibitory regulator of mTOR 42, 43. Functioning as a rheostat, TSC1/2 suppresses mTOR's activity to restrain cell growth under stress conditions, and releases its inhibition when conditions are favorable for growth. In TSC syndrome patients, TSC mutations (loss of mTOR inhibition) lead to a hyperactive mTOR, causing cell overgrowth and tumor formation. Interestingly, elevated mTOR activity has been detected in many other hamartoma syndromes. Together, these results implicate a possible common cause underlying different benign tumor syndromes, and place mTOR under the spotlight as an anti-cancer drug target. Naturally, rapamycin immediately became the ideal candidate to treat TSC syndrome due to its exquisitely specific and potent inhibition of mTOR. Indeed, three rapamycin analogs, CCI-779 (Wyeth), RAD001 (Novartis), and AP23573 (Ariad Pharmaceuticals Inc.) are currently in clinical trials for cancer treatment.
The emergence of mTOR complex 1
TOR is a large protein with many domains known to mediate protein-protein interactions. By gel filtration chromatography, TOR elutes in a fraction corresponding to a molecular weight much larger than its predicted size, which prompted many research groups to purify TOR binding partners. In 2002, seminal works from Hall's group first identified multiple TOR-associated proteins in yeast, including KOG1, AVO1, AVO2, AVO3 (AVO1/2/3) and LST8 44. Curiously, either TOR1 or TOR2 can complex with KOG1 and LST8 to form a rapamycin-sensitive complex, termed TOR complex 1 (TORC1), while only TOR2 binds AVO1/2/3 and LST8 to form a rapamycin-insensitive complex, termed TOR complex 2 (TORC2). Almost at the same time, Raptor (regulatory associated protein of mTOR) was also identified as an mTOR-binding protein 45, 46. Amino-acid alignment reveals that Raptor is the mammalian homolog of the yeast KOG1. mTOR, Raptor, and the later identified mammalian LST8 (mLST8) 47 form a complex that is sensitive to rapamycin inhibition, termed mTOR complex 1 (mTORC1).
Raptor is an essential and presumably non-enzymatic subunit of mTORC1 45, 46. It contains a highly conserved amino-terminal region followed by three HEAT repeats and seven WD40 (about 40 amino acids with conserved W and D forming four anti-parallel beta strands) repeats (Figure 1B). Knockout of Raptor, similar to knockout of mTOR, is also early embryonic lethal. It is agreed that Raptor is indispensable for mTOR to phosphorylate S6K1 and 4EBP1, but whether Raptor is a positive or negative regulator regarding mTOR activity remains ambiguous. Raptor has been proposed as a scaffolding protein to recruit substrates for mTOR and thereby demonstrates a positive effect on mTOR activity 45. Alternatively, other studies have shown that Raptor negatively regulates mTOR when tightly bound to the kinase 46. The interaction between mTOR and Raptor is very dynamic. It can be largely sustained in 0.3% CHAPS containing buffer, but not in 1% NP-40 containing buffer. In addition, some experimental stimuli also alter the amount of Raptor associated with mTOR. Amino acid withdrawal (nutrient deprivation) or rapamycin treatment enhances or reduces Raptor's binding to mTOR, respectively 46, 48. It has been pointed out that the Raptor-mTOR interaction requires multiple regions on Raptor and at least the HEAT repeats of mTOR.
mLST8 is another mTORC1 subunit identified after Raptor 47. It consists almost entirely of seven WD40 repeats and possibly binds to the kinase domain of mTOR (Figure 1C). Little has been reported regarding the regulation of the mLST8 and mTOR interaction. mLST8 seems to bind mTOR constitutively, probably due to the “sticky” WD40 structure. Because it binds to mTOR's kinase domain, it was speculated that mLST8 regulates mTOR kinase activity 47. However, there is no substantial evidence to support this hypothesis. When first identified, mLST8 was believed to be important for mTORC1 activity. Knockdown of mLST8 by RNAi (RNA interference) in cells suppressed S6K1 and 4EBP1 phosphorylation, the two well-characterized mTORC1 effectors. However, the most recent murine studies suggest that mLST8 is dispensable for mTORC1 function 49. The phosphorylation of S6K1 or 4EBP1 is not impaired in mLST8−/− MEFs (mouse embryo fibroblasts). Moreover, mTORC1 purified from these cells can still phosphorylate S6K1 in vitro. In addition, the interaction between mTOR and Raptor remains normal in the mLST8−/− MEFs, suggesting that mLST8 is not important for mTORC1 integrity. In line with the observations in mammals, knockdown of dLST8 (Drosophila LST8) by double strand RNA (dsRNA) in cultured S2 cells did not decrease dS6K (Drosophila S6K1) phosphorylation, whereas knockdown of dTOR (Drosophila TOR) or dRaptor (Drosophila Raptor) eliminated dS6K phosphorylation 50. Collectively, these results suggest that mLST8 is not essential for mTORC1 function. The discrepancy between the initial and later conclusions might be explained by the variation in RNAi efficiency in cells versus animals, the compensation for mLST8 in mice developed during early embryonic stages, or the different genetic backgrounds between cultured cancer cell lines and the knockout animals. Clearly, further studies are needed to elucidate the functions of mLST8 in mTORC1.
Akt joins TSC-mTORC1 regulation
After the discoveries of Raptor and mLST8, using “mTORC1” instead of “mTOR” to describe mTOR's function in cell growth control appears to be more accurate. While new mTOR binding proteins are still being identified, a considerable amount of work has also been devoted to elucidating the upstream regulation of mTORC1. One important function of mTORC1 is to sense growth factor signals to regulate cell growth. Many growth factors such as insulin initiate their intracellular signaling cascades by activating phosphatidylinositol-3-kinase (PI3K) through cell surface receptors 51. A major effector of PI3K is Akt, also termed PKB (serine/threonine protein kinase B). Akt is one of the most important survival kinases, involved in regulating a wide array of cellular processes, including metabolism, growth, proliferation and apoptosis 52, 53, 54. At the cell membrane, active PI3K leads to the generation of the lipid second messengers PIP3 (phosphatidylinositol-3,4,5-trisphosphate or PtdIns(3,4,5)P3) and PIP2 (PtdIns(3,4)P2) 55, 56. One important contribution of PIP3 is to recruit Akt via its PH (pleckstrin homology) domain. After translocation to the plasma membrane, Akt is phosphorylated at the activation loop site T308 (Threonine 308) and the hydrophobic motif site S473 (Serine 473) by PDK1 (3-phosphoinositide-dependent protein kinase-1) and PDK2, respectively 57, 58. PDK1 is also recruited to the membrane through the binding of its PH domain to PIP3 59. The identity of PDK2 remained unknown until recently. Akt is fully activated by dual phosphorylation by PDK1 and PDK2. Although the mechanism of Akt activation was fairly clear, how Akt contributes to cell growth control at the molecular level remained unclear. Genetic epistasis analysis in Drosophila placed TSC1/2 downstream of PI3K and Akt but upstream of S6K1. Complementing the studies in fly, several labs discovered that Akt directly phosphorylates TSC2 on multiple sites, directly linking PI3K-Akt to TSC-mTORC1 35, 37, 60, 61. These phosphorylations inhibit TSC1/2's function, thus upregulating mTORC1 activity. This important finding filled the gap between extracellular growth factor signals and intracellular TSC-mTORC1 regulation, leading to the establishment of the growth factor (insulin)-PI3K-Akt-TSC-mTORC1-S6K1/4EBP1 signaling pathway that largely explains how mTOR promotes cell growth under growth factor stimulation.
Rheb: a positive mTORC1 regulator
Another great example of mammalian biochemical studies complementing Drosophila genetic studies is the identification of Rheb as the mTORC1 regulator. Genetic screens in Drosophila identified Rheb (Ras-homolog enriched in brain), a small GTPase, as a positive cell growth regulator 62, 63. Simultaneously, mammalian TSC2 was biochemically proven to accelerate the intrinsic rate of GTP hydrolysis of Rheb, converting Rheb from the GTP-bound (active) to the GDP-bound (inactive) form 64, 65, 66. This suggests that Rheb is a direct target of TSC2 GAP activity, and that TSC2 inhibits Rheb function. Consistently, substantial evidence demonstrates that Rheb positively regulates mTORC1. In particular, Rheb over-expression stimulates S6K1 and 4EBP1 phosphorylation, which are indications of mTORC1 activity. This effect can be blocked by rapamycin and dominant-negative mTOR, suggesting that Rheb functions through mTORC1. Recently, Long and colleagues reported that Rheb binds to mTORC1 directly. Although the interaction does not require Rheb GTP loading, the GTP-bound rather than the GDP-bound Rheb stimulates mTOR kinase activity in vitro 67. The identification of Rheb as the mTORC1 stimulator advanced our understanding of TSC-mTORC1 regulation at the molecular level 68. The establishment of TSC2 as the Rheb GAP triggered the search for the putative Rheb GEF (guanine/guanyl nucleotide exchange factor) or GDI (guanyl nucleotide dissociation inhibitor). However, these efforts yielded little or no result for years. The difficulties of this task cast doubts on the existence of the Rheb GEF, owing to Rheb's low basal GTPase activity. It has also been speculated that multiple Rheb GEFs might exist with regulatory redundancy. Curiously, TCTP (translationally controlled tumor protein) in Drosophila has been recently identified as a candidate for dRheb GEF 69. Expression of the human TCTP rescued the phenotype of dTCTP mutant in Drosophila. The genetic data in Drosophila strongly indicate that TCTP is a Rheb GEF, but the biochemical studies show that the GEF activity of TCTP towards Rheb is very low. Furthermore, TCTP is an abundant protein expressed at a level higher than actin. Therefore, it is unclear how such an abundant protein controls Rheb activity in a regulatory fashion. Further studies are needed to clarify whether TCTP indeed functions as a Rheb GEF in mammalian cells.
PRAS40 negatively affects mTORC1 activity
While TSC1/2 relays its inhibition on mTORC1 through the small GTPase Rheb, PRAS40 (proline-rich Akt substrate 40 kDa or AKT1S1 (Akt1 substrate 1)) has been identified to more directly hinder mTORC1 function (Figure 1D) 70, 71. Initially, PRAS40 was proposed to be a new mTORC1 subunit that inhibits mTORC1 activity. Vander Haar et al. demonstrated that PRAS40 binds directly to the mTOR kinase domain, whereas Sancak and colleagues showed that it associates with mTOR via Raptor. Curiously, high salt concentrations weaken the attachment of PRAS40 to mTORC1 and thereby increase mTORC1 kinase activity in vitro. PRAS40 mediates growth factor signals to mTORC1, but whether PRAS40 senses nutrient availability is still being debated. It has been known that Akt can phosphorylate PRAS40 near its C-terminus on T246 (Thr246) 72. This phosphorylation appears to release its inhibitory effect on mTORC1. Thus, PRAS40 provides mTORC1 with a direct sensor for PI3K-Akt signaling, bypassing TSC-Rheb. Recent studies by Oshiro et al. 73 and Wang et al. 74 have identified a TOR signaling motif (TOS motif) in PRAS40 . The TOS motif is known to be present in mTORC1 substrates and is important for substrate binding to the kinase complex. Furthermore, the binding between over-expressed Raptor and PRAS40 appeared to be very stable. They proposed PRAS40 as a novel mTORC1 substrate. In support of their hypothesis, the authors discovered that mTORC1 phosphorylates PRAS40 on Ser183, and that this phosphorylation is sensitive to rapamycin. In addition, over-expression of PRAS40 suppressed the phosphorylation of S6K1 and 4EBP1, and likewise over-expression of S6K1 and 4EBP1 suppressed PRAS40 phosphorylation. This can be explained by a competition between substrates for the same kinase, mTOR. Regardless whether PRAS40 is an mTORC1 subunit or substrate, it is clear that PRAS40 exhibits negative effect on phosphorylation of other TORC1 substrates. Further studies are required to clarify the role of PRAS40 on mTORC1 function. Nevertheless, the identification of PRAS40 in mTORC1 has potential therapeutic implications. In TSC tumors, mutations of TSC typically activate Rheb, leading to constitutive activation of mTORC1. It has been shown in vitro that PRAS40 antagonizes Rheb effects in a dose-dependent manner 71. Correspondingly, an increase in PRAS40 by over-expression in cells retards cell growth. Therefore, drugs mimicking PRAS40 behavior may be beneficial for TSC patients.
S6K-dependent negative feedback inhibition
Although the newly identified components and regulations largely explain mTORC1 functions, some observations did not fit in the proposed model at the time. In TSC1−/− or TSC2−/− MEFs with hyperactive mTORC1 and S6K1, PI3K-Akt signaling cannot be stimulated by insulin 35, 50, 75, 76. However, this can be rescued by prolonged rapamycin treatment. Similarly, Drosophila TSC1−/− larvae display attenuated Akt activity, which can be restored by knockout of dS6K 41. These observations suggest that active mTOR-S6K1 impinges on Akt signaling. Interestingly, in cells with an elevated S6K1 activity, the expression of IRS1 (insulin receptor substrate 1) was reduced 77. IRS1 is the intermediary molecule bridging insulin receptors and PI3K via direct binding 78, 79. Additional studies revealed that active S6K1 phosphorylates IRS1 on multiple inhibitory sites and promotes its degradation 80, 81, 82. Taken together, the newly identified consequence of mTOR-S6K1 activation is to attenuate Akt signaling 83. This auto-regulatory pathway is defined as S6K1-dependent negative feedback inhibition 84. Surprisingly, this theory may help explain why TSC tumors are normally benign, since over-activation of AKT, such as in the case of PTEN mutations, often leads to malignancy. Consequently, when TSC mutations enhance mTOR and S6K1 activity, leading to tissue overgrowth, they simultaneously restrain other pathways responsible for decreased apoptosis and increased proliferation by inhibiting Akt signaling. In addition to modulating PI3K-Akt signaling, S6K1 was also recently found to directly phosphorylate mTOR on a C-terminal site 26. The consequence of this phosphorylation is not explicit. It may possibly increase mTOR intrinsic kinase activity, providing an additional mechanism for mTORC1 to regulate its function via S6K1.
Additional upstream regulations of mTORC1
It has been known for several years that mTORC1 can sense cellular energy levels, owing to the observations that energy depletion by low glucose culture or 2-DG (2-deoxyglucose, a non-hydrolysable glucose analog) treatment severely decreases S6K1 phosphorylation 85. On the other hand, ribosomal biogenesis and protein translation are major consumers of cellular energy. Thus, mTORC1 must be able to sense cellular ATP levels and shut down protein translation via S6K1 and 4EBP1, when necessary. The study of mTOR's energy-sensing mechanism has recently achieved remarkable progress. The AMP-activated protein kinase (AMPK) has been identified as the cellular energy sensor for mTORC1 86. AMPK monitors the cellular energy reservoir by measuring the ratio of AMP to ATP. A large amount of AMP or a high AMP/ATP ratio reflects low-energy status. Under such energy-stress conditions, AMPK is activated by direct AMP binding. Subsequently, it phosphorylates TSC2 and stimulates TSC2 GAP activity toward Rheb. mTORC1 activity is rapidly downregulated, leading to dephosphorylation of S6K1 and 4EBP1 and inhibition of protein translation. This finding not only answers the long-standing question of how mTORC1 integrates cellular energy signals to control cell growth but also linked TSC and Peutz-Jeghers syndrome (PJS) 87. PJS is a benign tumor syndrome caused by the mutation of tumor suppressor gene LKB1 (also called STK11). LKB1 can phosphorylate AMPK on T172 and accordingly activate AMPK to regulate cellular energy homeostasis. In PJS patients, mutation of LKB1 impairs the ability of AMPK to suppress TSC-mTORC1-S6K1 signaling and therefore promotes tumor formation. In support of this hypothesis, PJS and TSC tumors share striking histological similarities. Furthermore, LKB1 can indeed inhibit mTORC1 via AMPK and TSC 88. In summary, PJS tumors are likely caused by upregulation of the mTORC1 pathway; therefore, inhibition by rapamycin or rapamycin-derived drugs may be beneficial for PJS patients.
Surprisingly, AMPK is not only involved in energy sensing but also bridges Wnt signals to TSC-mTORC1 regulation 89. The Wnt (wingless and int) family binds to the cell-surface receptors of the Frizzled family and thus initiates intracellular signaling cascades. Activation of the Wnt pathway inhibits glycogen synthase kinase 3 (GSK3) and thereby stabilizes β-catenin, which translocates into the nucleus and activates the transcription of a wide array of growth-promoting genes 90, 91, 92, 93, 94. Therefore, Wnt signaling plays a pivotal role in cell growth, differentiation, and development. Although β-catenin-dependent transcription regulations have been studied extensively, little is known about the role of Wnt signaling in the regulation of protein translation. Inoki and colleagues recently discovered that Wnt activates the mTOR pathway in both cultured cell and murine model systems. Interestingly, activation of the mTOR pathway by Wnt is GSK3 dependent, but has little to do with β-catenin-related regulation. Furthermore, Inoki et al. revealed that, under low cellular energy conditions, activated AMPK phosphorylates TSC2 on Ser1345, which serves as a priming phosphorylation for subsequent TSC2 phosphorylations by GSK3β (GSK3 β isoform) on Ser1341 and Ser1337. The hyperphosphorylated TSC2 has increased activity, which leads to mTORC1 inhibition. Therefore, Wnt stimulation releases GSK3β inhibition on mTORC1 and promotes cell growth. Consistently, rapamycin, the specific mTOR inhibitor, potently retards the growth of Wnt-1-expressing tumor cells in nude mice. These findings are significant because the Wnt-mTOR-translation regulation is novel and independent of the well-established canonical Wnt pathway that activates β-catenin-dependent transcription. In summary, the study demonstrates that Wnt can stimulate cell growth by increasing translation via the TSC-mTORC1 pathway.
So far we have gained a basic understanding of how mTORC1 senses growth factors, cellular energy levels, and Wnt signals to regulate cell growth. One important question left unanswered is how mTORC1 responds to nutrient levels or amino acid availability. It is well documented that amino acids, particularly leucine, can dramatically boost S6K1 and 4EBP1 phosphorylation in an mTOR-dependent manner 95. Consistently, cells cultured under low nutrient conditions show strong reduction of mTORC1 activity. Many research groups have been keen to unravel this mystery, but consensus on a unifying model remains to be established. The major debate focuses on whether TSC1/2 is engaged in the process, as some believe that amino acids activate mTORC1 by inhibiting TSC1/2 34 or stimulating Rheb 62, 65, 66, 96. Contrary to this belief, it has been observed that TSC−/− cells can still respond to nutrients 97, and both TSC and Rheb homologs are missing in fission yeast, which still promptly senses amino acid availability. Recently hVPS34, a class III PI3K, has been reported to signal amino acid availability to mTORC1 independent of the TSC-Rheb axis 98, 99. hVPS34 is activated by amino acid stimulation and is inhibited by AMPK. Solving the complexity of this puzzle will require considerable efforts to test these different hypotheses.
Emergence of mTOR complex 2
As mentioned earlier, two TOR complexes have been identified in yeast 44, 100, 101. However, much of our current knowledge of mTOR is limited to mTORC1, because of the availability and wide use of rapamycin, the potent and specific mTORC1 inhibitor. In recent years, exciting progress has been made in identifying mTOR associated proteins, leading to the discovery of mTOR complex 2 (mTORC2), which has a distinctive physical structure and physiological functions compared to mTORC1 14, 102. This finding added breadth and depth to our knowledge regarding mTOR biology.
Contrary to mTORC1, mTORC2 is not sensitive to rapamycin inhibition. One postulation is that some mTORC2 components may block the binding of rapamycin/FKBP12 complex to the FRB domain of mTOR. Under this assumption, prolonged rapamycin treatment may decrease mTORC2 by competing for newly synthesized mTOR. The rapamycin/FKBP12 complex could compete against mTORC2 components to bind the newly synthesized mTOR and prevent further assembly of mature mTORC2. Supporting this model, reduction of mTORC2 has been observed in only certain types of cells undergoing prolonged rapamycin treatment at higher concentrations than was necessary to inhibit mTORC1 103. However, further structural studies are needed to prove this mechanism.
So far four components, mTOR, Rictor (rapamycin-insensitive companion of mTOR), Sin1 (also called Mip1), and mLST8 have been identified to assemble mTORC2 21, 104, 105, 106. Both Rictor and Sin1 are unique subunits of mTORC2, whereas mTOR and mLST8 are also present in mTORC1.
Rictor was the first identified subunit that is unique to mTORC2 21, 104. It is the homolog of AVO3 in yeast, which was identified independently, and is the defining member of this rapamycin-insensitive complex. Therefore, mTORC2 is also called the Rictor complex. Rictor is a large protein with a predicted molecular weight of 190 kDa. It has some domain structures in the amino terminal region that are relatively conserved among species, but the functions of these domains are not known (Figure 1E). It is speculated that these domains may mediate substrate binding and are important for mTORC2 assembly. The interaction between Rictor and mTOR is not blocked by rapamycin nor affected by nutrient levels, which are conditions known to regulate mTORC1. Thus, it is not surprising that knockdown of Rictor by RNAi in cultured cells does not change the phosphorylation status of S6K1 and 4EBP1. This suggests that mTORC2 has different physiological functions from mTORC1. The overall physiological importance of Rictor is emphasized by the fact that the Rictor knockout mice die around E10.5, possibly due to defects in vascular development 49, 106, 107.
The functions of mTORC2
Based on the remarkable conservation of TOR among species, mTORC2 may have similar functions as yeast TORC2, which mainly regulates the actin cytoskeleton, possibly through the Rho small GTPase family and protein kinase C (PKC). Indeed, knockdown of Rictor in HeLa cells increased F-actin accumulation, indicating that mTORC2 is important for cytoskeleton dynamics 21, 104. Moreover, decreased PKCα phosphorylation as well as protein levels were detected in both Rictor knockdown cells and Rictor−/− MEFs, but the mechanism by which mTORC2 regulates PKCα is unknown (21, 49, 104 and unpublished observations).
Probably the most important discovery regarding mTORC2 function is the identification of mTORC2 as the long-sought PDK2, which phosphorylates Akt on the hydrophobic motif (HM) site S473 108, 109. Sabatini's research group showed that knockdown of TORC2 by RNAi dramatically decreased Akt HM phosphorylation in both cultured mammalian cell lines and Drosophila S2 cells. In addition, purified mTORC2 can phosphorylate recombinant Akt on S473 in vitro, and this effect is blocked by knockdown of either mTOR or Rictor. Together, these studies establish mTORC2 as the long-sought PDK2 that is required for Akt HM phosphorylation both in vivo and in vitro. Originally, mTOR was not considered a PDK2 candidate because the phosphorylation of S473 in Akt is not sensitive to rapamycin; however, the later discovery that mTORC2 was insensitive to rapamycin helped correct this initial misconception. The role of mTORC2 as the PDK2 is further supported by the recent studies of Rictor knockout mice 49, 106, 107. In the Rictor−/− MEFs, Akt phosphorylation on S473 is diminished. This fact strongly implicates mTORC2 as the major PDK2 in vivo. These discoveries accelerated further studies of mTORC2, as the S473 phosphorylation of Akt could be used as an indicator of mTORC2 activity.
Other mTORC2 subunits
Recently Sin1 was identified as a new integral subunit of mTORC2, which is important for the complex assembly as well as function 70, 105, 106. Actually AVO1, the Sin1 homolog in yeast, was first discovered by Hall's group as one of the yeast TORC2 components 44. Sin1 is conserved among all eukaryotic species with a tissue expression pattern similar to that of mTOR 110. It has a conserved region in the middle of the sequence, but the amino acid homology among the species is very limited (Figure 1F) 111. A Ras-binding domain and a C-terminal PH domain have been identified recently 112. Sin1 was proven to be a bona fide mTORC2 component by various experimental approaches, including mass spectrometry, gel-filtration chromatography, and immunoprecipitation. The interaction between Sin1 and Rictor appears to be relatively stable. 1% NP-40 containing buffer disrupts mTORC1, but only modestly decreases the Sin1/Rictor association. It is postulated that hSin1 and Rictor stabilize each other through binding, building the structural foundation for mTORC2. This hypothesis is supported by the fact that knockdown of either Rictor or Sin1 in cells leads to a decrease of other mTORC2 component protein levels 106. Particularly, knockdown of Rictor in either HeLa or HEK293 cells reduces Sin1 protein levels by more than 90%. In addition, knockdown of Sin1 decreases the interaction between mTOR and Rictor, suggesting that Sin1 is important for mTORC2 assembly. More importantly, knockdown of Sin1 in both mammalian and Drosophila cells dramatically decreases the Akt phosphorylation on the HM site. Purified mTORC2 phosphorylates Akt on S473 in vitro, which is blocked by Sin1 RNAi. Moreover, Akt S473 phosphorylation is diminished in Sin1−/− MEFs 105. Taken together, these data demonstrate that Sin1 is an essential component of mTORC2 and important for mTORC2 functions both in vivo and in vitro.
mLST8 is another mTORC2 subunit 44. It was first identified in mTORC1 and later also found in mTORC2 47. Knockout of mLST8 in mice is embryonic lethal 49. As mentioned earlier, murine studies of mLST8 suggest that it is important for mTORC2 function. First, the phenotypes of mLST8 knockout mice resemble that of Rictor knockout mice. The mLST8 knockout mice also die around E10.5, also possibly due to defects in vascular development. Second, knockout of mLST8 only disrupts mTORC2 assembly though it exists in both of the mTOR complexes. In addition, Akt phosphorylation on S473 is dramatically decreased in mLST8−/− MEFs, whereas S6K1 and 4EBP1 phosphorylation is not affected, indicating an impaired mTORC2 and a functional mTORC1, respectively. These facts demonstrate that mLST8 is functionally important for mTORC2, but may not be essential for mTORC1 function.
Akt regulation by mTORC2
As a growing body of evidence demonstrates that mTORC2 phosphorylates S473 in Akt, opinions on the effect of this phosphorylation in regards to the phosphorylation of T308, the PDK1 site, appear to be divisive. In cells whose mTORC2 was transiently knocked down by RNAi, the phosphorylation of both S473 and T308 was decreased 106, 108. However, in the MEFs obtained from either Rictor or Sin1 knockout animals, the T308 phosphorylation remained normal though S473 phosphorylation was diminished 49, 105. The observation in transient knockdown cells fits the model that the phosphorylated S473 serves as a docking site for PDK1 to phosphorylate T308 113, 114. Without the prior phosphorylation of S473 by mTORC2, the phosphorylation of T308 by PDK1 was inhibited. The results from the MEFs suggest that these two phosphorylation events are independent of each other. PDK1 can phosphorylate Akt on T308 without the priming phosphorylation of S473 109, 115, 116. The discrepancy may be explained by how long the cells were deprived of Akt signaling. Akt is such an important survival kinase that lack of Akt activity is catastrophic for cells. In the Rictor or Sin1 knockout animals, the cells suffer from low Akt activity from the primordial developmental stage. The chronic pressure may force the cells to develop some as-of-yet-unidentified compensatory mechanisms that would restore Akt T308 phosphorylation. Therefore, the MEFs derived from these animals show a normal Akt T308 phosphorylation. RNAi knockdown in established cell lines is relatively transient. The well-differentiated cells may not be able to develop compensation in such a short time. For this reason, knockdown of mTORC2 leads to a decrease in T308 phosphorylation.
Although whether S473 and T308 phosphorylations are independent of each other remains an important question to answer, a consensus has been reached that Akt is most active when both S473 and T308 are phosphorylated 114, 117, 118, 119. Therefore, it has been proposed that mTORC2 is essential for maintaining high levels of Akt signaling 120. The functions and regulations of TOR complexes are highly conserved between mammalian species and Drosophila. Sometimes the lack of genetic redundancy among TOR pathway components makes it easier to dissect their roles in the Drosophila system 120. Recent studies from Cohen's group elegantly proved that dTORC2 (Drosophila TORC2) plays a key role in sustaining high levels of Akt signaling. They fine-tuned Akt signaling in Drosophila by either knockout of dTORC2 or knockin of Akt mutants, and evaluated the consequences by using both biochemical and physiological readouts. Clearly, knockout of dTORC2 by disrupting either dRictor (Drosophila Rictor) or dSin1 (Drosophila Sin1) led to the loss of HM phosphorylation in dAkt and a decreased Akt activity accordingly. Curiously, this residual dAkt activity is enough to support Drosophila development under normal or low PI3K signaling conditions. However, the HM phosphorylation is indispensable for permitting a high-level of PI3K-Akt signaling. Over-expression of the PI3K catalytic subunit elevated PI3K signaling, causing tissue over-growth in Drosophila 121. This abnormality was efficiently suppressed by the loss of dTORC2 activity in the dRictor mutant flies. Similarly, dPTEN mutation led to increase of PIP3 and hyperplasia in developing Drosophila eye 122. The hyperplasia was blocked by the diminished Akt HM phosphorylation in the dTORC2 mutant flies. Moreover, replacing the endogenous dAkt with an HM mutant dAkt inhibited the PI3K-induced hyperplasia. Together, these results suggest that HM phosphorylation of Akt is vital to sustain high-levels of insulin/PI3K signaling. It is worth noting that knockout of Rictor, Sin1, or mLST8 in mice leads to embryonic lethality, while knockout of dTORC2 in Drosophila only causes a modest developmental delay 120. It is possible that a broader spectrum of regulation that depends on a high level of Akt activity exists in mice than in Drosophila. Alternatively, additional downstream effectors of mTORC2 in mammals may play pivotal roles in early developmental stages.
The studies of mTORC2 function place Akt downstream of mTORC2. However, overwhelming evidence also suggests that Akt is upstream of mTORC1. It phosphorylates TSC2 and subsequently promotes mTORC1 activity and cell growth. This can be reconciled by addressing whether it is the same Akt that carries out these distinctive functions. If the same Akt responded to both mTORC1 and mTORC2, a plausible model would be that it is first activated by mTORC2 and PDK1 at the cell membrane and then phosphorylates TSC2 to activate mTORC1. Activated mTORC1 stimulates S6K1 activity that in turn shuts down Akt signaling by accelerating IRS1 degradation (the S6K1-mediated negative feedback inhibition). Consistent with this speculation, insulin stimulates Akt phosphorylation within minutes, while S6K1 activation takes a longer period of time (about 30 min) 55. On the other hand, it is also possible that there are multiple pools of Akt in the cells, reacting to different stimulations. The relatively delayed S6K1 activation could merely be due to its different subcellular localization from that of Akt. The specific details notwithstanding, mTORC1 and mTORC2 elicits negative and positive Akt regulation, respectively, within each cell. Elevated Akt activity is often found in malignant tumor cells 52, 54, 118. Clarifying the regulations between Akt and mTOR complexes will be greatly appreciated for developing future therapeutics targeting mTOR pathway components.
mTORC2 and AGC kinase regulations
Akt belongs to the AGC kinase family, which also contains the p70 ribosomal S6 kinase (S6K), p90 ribosomal protein S6 kinase, serum- and glucocorticoid-induced protein kinase, and PKC. These kinases play distinctive roles in various vital physiological processes, but share great structural resemblances around the kinase domain. The activation of these kinases is often achieved by multiple phosphorylation events, normally including phosphorylation on an HM site and an activation loop site near or within the kinase domain 114, 123. Interestingly, despite the structural similarities, the HM sites of different AGC kinases are recognized and regulated by distinct upstream regulators. For example, S6K1 and Akt are exclusively phosphorylated by mTORC1 and mTORC2, respectively. Compared to Akt, S6K1 possesses an additional C-terminal domain, believed to cover the HM motif when S6K1 is in the inactive state 124. Because of this structure, only mTORC1 can recognize S6K1 and phosphorylate it on the HM site. One might postulate that mTORC2 would be able to phosphorylate S6K1 if the C-terminal domain was deleted. Indeed, S6K1δC has been reported as an mTORC2 substrate and can be phosphorylated by mTORC2 on the HM site both in vivo and in vitro 50, 125, 126. By deleting the C-terminal domain, S6K1δC becomes structurally similar to Akt. Curiously, most of the AGC kinases are similar to Akt, lacking the C-terminal domain. This possibly indicates a common regulatory mechanism involving mTORC2 for the majority of AGC kinases. In support of this notion, PKCα has been identified as an mTORC2 target 21, 104. The HM phosphorylation and the protein levels of PKCα are dramatically decreased in Rictor−/− MEFs (49 and unpublished observations). There are 10 known PKC family members, all of which share a conserved activation loop site, an HM site and a turn motif site. It would be interesting to learn whether mTORC2 regulates all the PKC members. It would be even more exciting to determine whether mTORC2 contributes to the regulation of AGC kinases in general.
mTORC2 upstream regulations
It has been established that Rheb positively and directly regulates mTORC1. This immediately caused us to examine whether Rheb also regulates mTORC2. We recently reported that Rheb has a negative effect on mTORC2, and the regulation is likely to be indirect 50. Knockdown of dRheb in cultured Drosophila S2 cells decreased dS6K phosphorylation but increased dAkt phosphorylation. This suggests that dRheb has positive and negative effects on dTORC1 and dTORC2 activity, respectively. In mammalian cells, over-expression of Rheb dramatically stimulated mTORC1 kinase activity both in vivo and in vitro, but had no positive effect on mTORC2. Moreover, we failed to detect any direct binding between mTORC2 and Rheb. These results suggest that Rheb may affect mTORC2 indirectly. It is known that the S6K1-dependent negative feedback attenuates insulin signaling. This prompted us to knock down Chico, the IRS homolog in Drosophila and examine dRheb RNAi effects. As expected, knockdown of Chico largely blocked the activation of dAkt caused by dRheb knockdown. This suggests that Rheb exerts a negative effect on mTORC2 probably through the S6K1-dependent negative feedback loop.
mTORC2 has been placed downstream of PI3K signaling because Akt S473 phosphorylation is stimulated by growth factors and inhibited by low concentrations of wortmannin, a specific PI3K inhibitor (108 and unpublished observations). Other than this, not much is known about the upstream regulation of mTORC2. The mechanism by which mTORC2 is activated remains an important question to be answered. Akt is likely to be phosphorylated and activated at the cell membrane due to the binding to PIP3 through its PH domain 114. Upon stimulation, PDK1, which phosphorylates Akt on T308, is also recruited to the cell membrane via its PH domain 127. Among all the known mTORC2 subunits, only Sin1 possesses a PH domain at its C-terminus 112. Can Sin1 bring the whole large complex to the cell membrane upon stimulation? Where and how mTORC2 phosphorylates Akt on S473? Through the identification of new mTOR-binding proteins, we may have more clues with which to answer these important questions.
Rictor/Sin1 complexes
As an essential cell growth controller, mTOR knockout studies are extremely challenging 15, 16, 17. Most of the in vivo data regarding mTORC2 function are obtained from Rictor−/−, Sin1−/−, and mLST8−/− MEFs 49, 105, 106. To some extent, these results only indirectly reflect mTORC2 functions. Curiously, transient knockdown of mTOR by RNAi in HeLa cells did not eliminate Akt phosphorylation on S473, although S6K1 phosphorylation on T389 was blocked 106. In addition, Rictor and Sin1 form a very stable complex in vivo. The association between these two proteins seems much more stable than their interactions with mTOR (106 and unpublished observations). The fact that knockdown of Rictor or Sin1 decreases mTORC2 kinase activity towards Akt in vitro cannot exclude the possibility that a Rictor or Sin1 associated kinase, other than mTOR, phosphorylates Akt in vivo. In addition, although purified mTORC2 can phosphorylate Akt in vitro, conducting the kinase assay appears to be difficult. Curiously, Protor-1 (protein observed with Rictor-1) and Protor-2 have been identified as Rictor-binding proteins, but are not essential for mTORC2 assembly 128. Their functions remain unknown. These facts raise the suspicion that Rictor and Sin1 may have much broader physiological functions independent of mTORC2. It is possible that Rictor and Sin1 form a scaffold for different complexes involving different kinases.
Dysregulation of mTOR in human diseases
Elevated mTORC1 activity may be a common cause underlying many hamartoma syndromes 87. For example, in TSC patients, mutation of either the TSC1 or TSC2 gene leads to hyperactive mTORC1. In PJS patients, mutation of LKB1 ultimately stimulates mTORC1 activity via the AMPK-TSC pathway. Upregulation of mTORC1 activity is pro-growth and provides the cells with growth advantages in multiple biological processes, including mass accumulation, proliferation, and survival 129. These cells, often with a larger cell size and a faster growth rate, are structurally disorganized from the tissue and lead to the formation of benign tumors. Therefore, inhibiting mTORC1 activity has been considered a promising option for hamartoma syndrome treatment. The most well-studied mTORC1 inhibitor is rapamycin. By binding to mTOR, rapamycin potently and specifically inhibits mTORC1 function. As mentioned at the beginning of this article, three rapamycin analogs, CCI-779 (Wyeth), RAD001 (Novartis), and AP23573 (Ariad Pharmaceuticals Inc.) are currently in clinical trials for treating cancers.
Hamartoma syndromes are characterized by benign tumors. Normally the benign nature of these tumors is partially explained by the S6K1-dependent negative feedback inhibition. Genetic mutations cause hyperactive mTORC1 that subsequently activates S6K1. Active S6K1 restrains Akt signaling by phosphorylating and inhibiting IRS1. Decreased Akt signaling not only in turn inhibits mTORC1 activity to limit tumor growth but also suppresses Akt downstream effectors, many of which are important for tumor malignancy 84, 130. Because of this S6K1-mediated feedback inhibition, it casts doubt on the feasibility of long-term rapamycin treatment in patients. Although prolonged rapamycin treatment blocks S6K1 activity and decreases tumor size, it also sensitizes the PI3K-Akt signaling via the feedback loop. It has been observed in TSC−/− cells that long-term rapamycin treatment inhibited S6K1 phosphorylation and restored cellular insulin response 50. Therefore, for hamartoma patients, especially those who have elevated PI3K-Akt signaling (i.e. with PTEN mutation 131), long-term administration of rapamycin may put them under higher risk of malignancy progression 54. For these patients, other drugs, such as PI3K inhibitors, have to be used in combination with rapamycin. Curiously, the identification of mTORC2 may provide an alternative to deal with such a dilemma. mTORC2 presents positive regulation of Akt. Drugs that inhibit mTORC2 may effectively suppress Akt signaling and thereby prevent tumor malignancy 132. It has also been reported that prolonged rapamycin treatment sequesters mTORC2 in some cell types 103. In these cells, decreased mTORC2 led to strong inhibition of Akt. Under this scenario, rapamycin-derived drugs would be able to inhibit tumor growth and prevent malignancy at the same time. It is not clear why mTORC2 responds to prolonged rapamycin treatment in a cell-type-dependent manner. Understanding the molecular mechanism could be the key to determine whether and when to use the rapamycin-derived drugs. Current advances in human genetics and micro-array technology make it possible to quickly collect the genetic information of individual patients. Such information will be valuable to develop personalized medical treatment plans.
Besides the involvement in tumorigenesis, dysregulation of mTORC1 has been detected in many human metabolic disorders, including obesity and diabetes. In S6K1 knockout mice, fat accumulation and adipose tissues were decreased 83. Correspondingly, defective dTOR signaling led to loss of fat in Drosophila 133. These observations imply that mTORC1 promotes adipogenesis, and elevated mTORC1 activity may contribute to obesity. In addition, adipose tissues secrete many hormones that regulate a variety of physiological processes. For example, leptin produced by adipose tissues directly regulates appetite through the central nervous system 134, 135. Chronically high levels of leptin in blood may induce leptin resistance that increases food intake and contributes to obesity. Meanwhile, hyperactive mTORC1 downregulates IRS1 through the S6K1-dependent negative feedback inhibition. Loss of IRS1 desensitizes the cells to insulin, causing insulin resistance. Long-term insulin resistance often causes type II diabetes 79. Thus, dysregulation of mTORC1 may provide one explanation for why type-II diabetes is always associated with obesity, although these two disorders are multifactorial.
In addition to the physiological importance of mTOR in peripheral tissues, emerging evidence suggests that hypothalamic mTOR in the central nervous system is a key factor in regulating food intake 136. Cota et al. recently discovered that in rat, activated mTOR and its downstream effector S6K preferably co-localize at the arcuate nucleus (ARC) of the hypothalamus, a region important for the brain to receive leptin signal and maintain energy homeostasis for the entire animal. Interestingly, when directly injected to the ARC, leucine, the amino acid known to activate peripheral mTOR signaling, increases hypothalamic mTOR activity, based on the upregulation of S6K1 and S6 phosphorylation, and consequently decreases food intake and body weight. Complementarily, prior to leucine stimulation, administration of rapamycin that specifically inhibits mTOR activity prevents the amino-acid-induced anorexia. These results suggest that hypothalamic mTOR senses nutrient availability and regulates food intake. This implies a mechanism by which the brain monitors the body energy status and exerts proper physiological actions. In addition, inhibition of the mTOR activity attenuates leptin's ability to reduce appetite. Leptin is a hormone secreted by the adipose tissues and, along with insulin, relays the information of adipogenesis to the central nervous system 134, 135. Therefore, this finding provides evidence for a direct connection between fat accumulation and the hypothalamic mTOR-dependent energy sensing. In summary, these findings suggest that hypothalamic mTOR is a sensor for both nutrient and fat signals in the central nervous system. This function of hypothalamic mTOR provides the organism with protective regulation. Overabundance of nutrients in peripheral tissues activates mTOR and promotes adipogenesis. At the same time, the energy overflow and the leptin from the fat cells activate hypothalamic mTOR in the central nervous system to suppress food intake and prevent metabolic imbalance. It is conceivable that dysregulation of mTOR may disrupt the balance and cause metabolic disorders. Therefore, unraveling the regulation mechanisms of the peripheral and hypothalamic mTOR is vital for developing future therapeutic interventions.
Conclusion
Despite its humble beginning two decades ago, mTOR is now recognized as a central regulator in a diverse array of vital cellular processes, including proliferation, growth, differentiation, and survival. The physiological functions of mTOR continue to expand. The regulation of mTOR has progressed from a signaling pathway to a network (Figure 2). Due to space limitations, we could not cover every aspect of mTOR upstream regulation, including the input from cellular stressors, the contribution of mitogen-activated protein kinases, p53-related apoptotic signals, and so on. We also left out quite a bit of mTOR downstream regulations, such as transcription and autophagy. We only briefly touched upon the involvement of mTOR in tumorigenesis and metabolic disorders. In fact, mTOR has much broader implications related to human pathophysiology, including cardiovascular diseases, autoimmune disorders, aging, and neuronal-related diseases. In recent years, many outstanding research groups have dedicated their efforts to elucidating mTOR functions and regulations. Exciting progress has been achieved, and the discoveries have begun to show promising clinical implications. Nevertheless, our understanding of mTOR, especially regarding mTORC2, is far from complete. How is mTORC2 activated and regulated? What are the undiscovered functions of mTORC2? How do mTORC1 and mTORC2 coordinate with each other to regulate distinct cellular processes? What are the relationships between the signaling cascades upstream or downstream of mTOR complexes? The answers to these questions will not only advance our understanding of many vital physiological processes but also help us develop new strategies for treating cancer and improving the quality of human life.
mTOR signaling network in mammalian cells. mTORC1, the rapamycin-sensitive complex, consists of mTOR, Raptor, mLST8, and PRAS40. TSC1/2-Rheb is the major upstream regulator of mTORC1. Through the TSC1/2-Rheb axis, mTORC1 integrates cellular energy levels, growth factors, and Wnt signals to regulate protein translation by phosphorylating S6K1 and 4EBP1. Phosphorylated S6K1 (active) inhibits IRS1 function and thus attenuates insulin/PI3K signaling. hVPS34 has been reported to sense nutrient availabilities for mTORC1. The mTORC2 subunits include mTOR, Rictor, Sin1, and mLST8. mTORC2 controls cell structure and survival by regulating PKCα and Akt. The upstream regulation of mTORC2 remains unknown. Arrows represent activation, bars represent inhibition, and dots represent binding.
References
Morice MC, Serruys PW, Sousa JE, et al. A randomized comparison of a sirolimus-eluting stent with a standard stent for coronary revascularization. N Engl J Med 2002; 346:1773–1780.
Moses JW, Leon MB, Popma JJ, et al. Sirolimus-eluting stents versus standard stents in patients with stenosis in a native coronary artery. N Engl J Med 2003; 349:1315–1323.
Marks AR . Sirolimus for the prevention of in-stent restenosis in a coronary artery. N Engl J Med 2003; 349:1307–1309.
Heitman J, Movva NR, Hall MN . Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991; 253:905–909.
Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN . Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 1993; 73:585–596.
Harding MW, Galat A, Uehling DE, Schreiber SL . A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 1989; 341:758–760.
Siekierka JJ, Hung SH, Poe M, Lin CS, Sigal NH . A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature 1989; 341:755–757.
Siekierka JJ, Wiederrecht G, Greulich H, et al. The cytosolic-binding protein for the immunosuppressant FK-506 is both a ubiquitous and highly conserved peptidyl-prolyl cis-trans isomerase. J Biol Chem 1990; 265:21011–21015.
Brown EJ, Albers MW, Shin TB, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994; 369:756–758.
Chiu MI, Katz H, Berlin V . RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci USA 1994; 91:12574–12578.
Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH . RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994; 78:35–43.
Sabers CJ, Martin MM, Brunn GJ, et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem 1995; 270:815–822.
Fingar DC, Blenis J . Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 2004; 23:3151–3171.
Wullschleger S, Loewith R, Hall MN . TOR signaling in growth and metabolism. Cell 2006; 124:471–484.
Martin PM, Sutherland AE . Exogenous amino acids regulate trophectoderm differentiation in the mouse blastocyst through an mTOR-dependent pathway. Dev Biol 2001; 240:182–193.
Murakami M, Ichisaka T, Maeda M, et al. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol 2004; 24:6710–6718.
Gangloff YG, Mueller M, Dann SG, et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol 2004; 24:9508–9516.
Huang S, Houghton PJ . Targeting mTOR signaling for cancer therapy. Curr Opin Pharmacol 2003; 3:371–377.
Peterson RT, Beal PA, Comb MJ, Schreiber SL . FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J Biol Chem 2000; 275:7416–7423.
Edinger AL, Linardic CM, Chiang GG, Thompson CB, Abraham RT . Differential effects of rapamycin on mammalian target of rapamycin signaling functions in mammalian cells. Cancer Res 2003; 63:8451–8460.
Jacinto E, Loewith R, Schmidt A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 2004; 6:1122–1128.
Nave BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR . Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J 1999; 344 Part 2: 427–431.
Sekulic A, Hudson CC, Homme JL, et al. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res 2000; 60:3504–3513.
Reynolds THt, Bodine SC, Lawrence JC Jr . Control of Ser2448 phosphorylation in the mammalian target of rapamycin by insulin and skeletal muscle load. J Biol Chem 2002; 277:17657–17662.
Cheng SW, Fryer LG, Carling D, Shepherd PR . Thr2446 is a novel mammalian target of rapamycin (mTOR) phosphorylation site regulated by nutrient status. J Biol Chem 2004; 279:15719–15722.
Holz MK, Blenis J . Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem 2005; 280:26089–26093.
Holz MK, Ballif BA, Gygi SP, Blenis J . mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 2005; 123:569–580.
Jeno P, Ballou LM, Novak-Hofer I, Thomas G . Identification and characterization of a mitogen-activated S6 kinase. Proc Natl Acad Sci USA 1988; 85:406–410.
Thomas G . The S6 kinase signaling pathway in the control of development and growth. Biol Res 2002; 35:305–313.
Gingras AC, Raught B, Sonenberg N . Regulation of translation initiation by FRAP/mTOR. Genes Dev 2001; 15:807–826.
Fingar DC, Salama S, Tsou C, Harlow E, Blenis J . Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 2002; 16:1472–1487.
Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J . mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol 2004; 24:200–216.
Kwiatkowski DJ . Tuberous sclerosis: from tubers to mTOR. Ann Hum Genet 2003; 67:87–96.
Gao X, Zhang Y, Arrazola P, et al. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol 2002; 4:699–704.
Potter CJ, Pedraza LG, Xu T . Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol 2002; 4:658–665.
Goncharova EA, Goncharov DA, Eszterhas A, et al. Tuberin regulates p70 S6 kinase activation and ribosomal protein S6 phosphorylation. A role for the TSC2 tumor suppressor gene in pulmonary lymphangioleiomyomatosis (LAM). J Biol Chem 2002; 277:30958–30967.
Inoki K, Li Y, Zhu T, Wu J, Guan KL . TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 2002; 4:648–657.
Potter CJ, Huang H, Xu T . Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 2001; 105:357–368.
Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK . The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 2001; 105:345–355.
Gao X, Pan D . TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev 2001; 15:1383–1392.
Radimerski T, Montagne J, Hemmings-Mieszczak M, Thomas G . Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev 2002; 16:2627–2632.
Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J . Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci USA 2002; 99:13571–13576.
Manning BD, Cantley LC . United at last: the tuberous sclerosis complex gene products connect the phosphoinositide 3-kinase/Akt pathway to mammalian target of rapamycin (mTOR) signalling. Biochem Soc Trans 2003; 31:573–578.
Loewith R, Jacinto E, Wullschleger S, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 2002; 10:457–468.
Hara K, Maruki Y, Long X, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002; 110:177–189.
Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002; 110:163–175.
Kim DH, Sarbassov dos D, Ali SM, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell 2003; 11:895–904.
Oshiro N, Yoshino K, Hidayat S, et al. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells 2004; 9:359–366.
Guertin DA, Stevens DM, Thoreen CC, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 2006; 11:859–871.
Yang Q, Inoki K, Kim E, Guan KL . TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc Natl Acad Sci USA 2006; 103:6811–6816.
Cantley LC . The phosphoinositide 3-kinase pathway. Science 2002; 296:1655–1657.
Luo J, Manning BD, Cantley LC . Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell 2003; 4:257–262.
Brazil DP, Yang ZZ, Hemmings BA . Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem Sci 2004; 29:233–242.
Hay N . The Akt-mTOR tango and its relevance to cancer. Cancer Cell 2005; 8:179–183.
Vanhaesebroeck B, Alessi DR . The PI3K-PDK1 connection: more than just a road to PKB. Biochem J 2000; 346Part 3: 561–576.
Cantrell DA . Phosphoinositide 3-kinase signalling pathways. J Cell Sci 2001; 114:1439–1445.
Toker A, Newton AC . Cellular signaling: pivoting around PDK-1. Cell 2000; 103:185–18.
Belham C, Wu S, Avruch J . Intracellular signalling: PDK1 – a kinase at the hub of things. Curr Biol 1999; 9:R93–R96.
McManus EJ, Collins BJ, Ashby PR, et al. The in vivo role of PtdIns(3,4,5)P3 binding to PDK1 PH domain defined by knockin mutation. EMBO J 2004; 23:2071–2082.
Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC . Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 2002; 10:151–162.
Dan HC, Sun M, Yang L, et al. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J Biol Chem 2002; 277:35364–35370.
Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA . Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 2003; 5:566–571.
Stocker H, Radimerski T, Schindelholz B, et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol 2003; 5:559–565.
Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J . Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 2003; 13:1259–1268.
Garami A, Zwartkruis FJ, Nobukuni T, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 2003; 11:1457–1466.
Inoki K, Li Y, Xu T, Guan KL . Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003; 17:1829–1834.
Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J . Rheb binds and regulates the mTOR kinase. Curr Biol 2005; 15:702–713.
Manning BD, Cantley LC . Rheb fills a GAP between TSC and TOR. Trends Biochem Sci 2003; 28:573–576.
Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW . Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 2007; 445:785–788.
Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH . Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol 2007; 9:316–323.
Sancak Y, Thoreen CC, Peterson TR, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 2007; 25:903–915.
Kovacina KS, Park GY, Bae SS, et al. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem 2003; 278:10189–10194.
Oshiro N, Takahashi R, Yoshino K, et al. The proline-Rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mTOR complex 1. J Biol Chem 2007; 282:20329–20339.
Wang L, Harris TE, Roth RA, Lawrence JC . PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem 2007; 282:20036–20044.
Jaeschke A, Hartkamp J, Saitoh M, et al. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J Cell Biol 2002; 159:217–224.
Kwiatkowski DJ, Zhang H, Bandura JL, et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet 2002; 11:525–534.
Haruta T, Uno T, Kawahara J, et al. A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol Endocrinol 2000; 14:783–794.
Saltiel AR, Kahn CR . Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001; 414:799–806.
White MF . IRS proteins and the common path to diabetes. Am J Physiol Endocrinol Metab 2002; 283:E413–E422.
Greene MW, Sakaue H, Wang L, Alessi DR, Roth RA . Modulation of insulin-stimulated degradation of human insulin receptor substrate-1 by Serine 312 phosphorylation. J Biol Chem 2003; 278:8199–8211.
Harrington LS, Findlay GM, Gray A, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol 2004; 166:213–223.
Shah OJ, Wang Z, Hunter T . Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol 2004; 14:1650–1656.
Um SH, Frigerio F, Watanabe M, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 2004; 431:200–205.
Manning BD . Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J Cell Biol 2004; 167:399–403.
Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G . Mammalian TOR: a homeostatic ATP sensor. Science 2001; 294:1102–1105.
Inoki K, Zhu T, Guan KL . TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003; 115:577–590.
Inoki K, Corradetti MN, Guan KL . Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet 2005; 37:19–24.
Shaw RJ, Bardeesy N, Manning BD, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 2004; 6:91–99.
Inoki K, Ouyang H, Zhu T, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 2006; 126:955–968.
Molenaar M, van de Wetering M, Oosterwegel M, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 1996; 86:391–399.
Brunner E, Peter O, Schweizer L, Basler K . pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature 1997; 385:829–833.
Tetsu O, McCormick F . Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999; 398:422–426.
Zhang X, Gaspard JP, Chung DC . Regulation of vascular endothelial growth factor by the Wnt and K-ras pathways in colonic neoplasia. Cancer Res 2001; 61:6050–6054.
Easwaran V, Lee SH, Inge L, et al. Beta-catenin regulates vascular endothelial growth factor expression in colon cancer. Cancer Res 2003; 63:3145–3153.
Hay N, Sonenberg N . Upstream and downstream of mTOR. Genes Dev 2004; 18:1926–1945.
Long X, Ortiz-Vega S, Lin Y, Avruch J . Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem 2005; 280:23433–23436.
Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG . The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem 2005; 280:18717–18727.
Nobukuni T, Joaquin M, Roccio M, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci USA 2005; 102:14238–14243.
Byfield MP, Murray JT, Backer JM . hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem 2005; 280:33076–33082.
Wullschleger S, Loewith R, Oppliger W, Hall MN . Molecular organization of target of rapamycin complex 2. J Biol Chem 2005; 280:30697–30704.
Reinke A, Anderson S, McCaffery JM, et al. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem 2004; 279:14752–14762.
Sarbassov DD, Ali SM, Sabatini DM . Growing roles for the mTOR pathway. Curr Opin Cell Biol 2005; 17:596–603.
Sarbassov DD, Ali SM, Sengupta S, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 2006; 22:159–168.
Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 2004; 14:1296–1302.
Jacinto E, Facchinetti V, Liu D, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 2006; 127:125–137.
Yang Q, Inoki K, Ikenoue T, Guan KL . Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev 2006; 20:2820–2832.
Shiota C, Woo JT, Lindner J, Shelton KD, Magnuson MA . Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev Cell 2006; 11:583–589.
Sarbassov DD, Guertin DA, Ali SM, Sabatini DM . Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005; 307:1098–1101.
Bayascas JR, Alessi DR . Regulation of Akt/PKB Ser473 phosphorylation. Mol Cell 2005; 18:143–145.
Wilkinson MG, Pino TS, Tournier S, et al. Sin1: an evolutionarily conserved component of the eukaryotic SAPK pathway. EMBO J 1999; 18:4210–4221.
Schroder W, Cloonan N, Bushell G, Sculley T . Alternative polyadenylation and splicing of mRNAs transcribed from the human Sin1 gene. Gene 2004; 339:17–23.
Schroder WA, Buck M, Cloonan N, et al. Human Sin1 contains Ras-binding and pleckstrin homology domains and suppresses Ras signalling. Cell Signal 2007; 19:1279–1289.
Scheid MP, Marignani PA, Woodgett JR . Multiple phosphoinositide 3-kinase-dependent steps in activation of protein kinase B. Mol Cell Biol 2002; 22:6247–6260.
Scheid MP, Woodgett JR . Unravelling the activation mechanisms of protein kinase B/Akt. FEBS Lett 2003; 546:108–112.
Biondi RM, Kieloch A, Currie RA, Deak M, Alessi DR . The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J 2001; 20:4380–4390.
Collins BJ, Deak M, Arthur JS, Armit LJ, Alessi DR . In vivo role of the PIF-binding docking site of PDK1 defined by knock-in mutation. EMBO J 2003; 22:4202–4211.
Alessi DR, Andjelkovic M, Caudwell B, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J 1996; 15:6541–6551.
Lawlor MA, Alessi DR . PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J Cell Sci 2001; 114:2903–2910.
Woodgett JR . Recent advances in the protein kinase B signaling pathway. Curr Opin Cell Biol 2005; 17:150–17.
Hietakangas V, Cohen SM . Re-evaluating AKT regulation: role of TOR complex 2 in tissue growth. Genes Dev 2007; 21:632–637.
Leevers SJ, Weinkove D, MacDougall LK, Hafen E, Waterfield MD . The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J 1996; 15:6584–6594.
Goberdhan DC, Paricio N, Goodman EC, Mlodzik M, Wilson C . Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev 1999; 13:3244–3258.
Newton AC . Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J 2003; 370:361–371.
Cheatham L, Monfar M, Chou MM, Blenis J . Structural and functional analysis of pp70S6k. Proc Natl Acad Sci USA 1995; 92:11696–11700.
Schalm SS, Tee AR, Blenis J . Characterization of a conserved C-terminal motif (RSPRR) in ribosomal protein S6 kinase 1 required for its mammalian target of rapamycin-dependent regulation. J Biol Chem 2005; 280:11101–11106.
Ali SM, Sabatini DM . Structure of S6 kinase 1 determines whether raptor-mTOR or rictor-mTOR phosphorylates its hydrophobic motif site. J Biol Chem 2005; 280:19445–19448.
Alessi DR, James SR, Downes CP, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol 1997; 7:261–269.
Pearce LR, Huang X, Boudeau J, et al. Identification of Protor as a novel Rictor-binding component of mTOR-complex-2. Biochem J 2007; 405:513–522.
Inoki K, Ouyang H, Li Y, Guan KL . Signaling by target of rapamycin proteins in cell growth control. Microbiol Mol Biol Rev 2005; 69:79–100.
Cheng JQ, Lindsley CW, Cheng GZ, Yang H, Nicosia SV . The Akt/PKB pathway: molecular target for cancer drug discovery. Oncogene 2005; 24:7482–7492.
Maehama T, Taylor GS, Dixon JE . PTEN and myotubularin: novel phosphoinositide phosphatases. Annu Rev Biochem 2001; 70:247–279.
Zeng Z, Sarbassov dos D, Samudio IJ, et al. Rapamycin derivatives reduce mTORC2 signaling and inhibit AKT activation in AML. Blood 2007; 109:3509–3512.
Teleman AA, Chen YW, Cohen SM . Drosophila melted modulates FOXO and TOR activity. Dev Cell 2005; 9:271–281.
Friedman JM, Halaas JL . Leptin and the regulation of body weight in mammals. Nature 1998; 395:763–770.
Schwartz MW, Woods SC, Porte D Jr, Seeley RJ, Baskin DG . Central nervous system control of food intake. Nature 2000; 404:661–671.
Cota D, Proulx K, Smith KA, et al. Hypothalamic mTOR signaling regulates food intake. Science 2006; 312:927–930.
Acknowledgements
We apologize to all the authors whose primary research we were unable to cite. We also thank Chung-Han Lee, Pamela Wong, June Pais, Beverly Piggot (University of Michigan, Ann Arbor, MI 48109, USA) and Jean Guan (Brown University, Providence, RI 02912, USA) for their critical reading and comments on the manuscript. We acknowledge the funding support from NIH (CA108941, GM62694) and Department of Defense (DOD, TS060032).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is cited by
-
The Dominant Mechanism of Cyclophosphamide-Induced Damage to Ovarian Reserve: Premature Activation or Apoptosis of Primordial Follicles?
Reproductive Sciences (2024)
-
Enhanced pro-apoptosis gene signature following the activation of TAp63α in oocytes upon γ irradiation
Cell Death & Disease (2022)
-
Signaling pathways and therapeutic interventions in gastric cancer
Signal Transduction and Targeted Therapy (2022)
-
Synergistic effects of Rapamycin and Fluorouracil to treat a gastric tumor in a PTEN conditional deletion mouse model
Gastric Cancer (2022)
-
Nutrient sensing, signaling transduction, and autophagy in podocyte injury: implications for kidney disease
Journal of Nephrology (2022)