Abstract
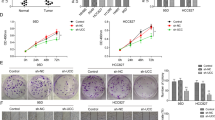
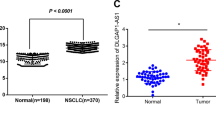
Lung cancer is the leading cause of cancer-related death around the world. Long noncoding RNA (lncRNA) has pivotal roles in cancer occurrence and development. However, only a few lncRNAs have been functionally characterized. In the present study, we investigated the effects of lncRNA-HIT (HOXA transcript induced by TGFβ) expression on non-small cell lung cancer (NSCLC) cell phenotype with the gain-of-function and loss-of-function assays. We found that ectopic expression or knockdown of lncRNA-HIT markedly increased or decreased NSCLC cell proliferation, respectively. Moreover, we also showed that lncRNA-HIT interacted with E2F1 to regulate its target genes, such as Survivin, FOXM1, SKP2, NELL2 and DOK1. Collectively, our findings indicated that lncRNA-HIT affected the proliferation of NSCLC cells at least in part via regulating the occupancy of E2F1 in the promoter regions of its target genes. The lncRNA-HIT–E2F1 complex may be a potential target for NSCLC treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A . Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108.
McErlean A, Ginsberg MS . Epidemiology of lung cancer. Semin Roentgenol 2011; 46: 173–177.
Schmitz SU, Grote P, Herrmann BG . Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 2016; 73: 2491–2509.
Goff LA, Rinn JL . Linking RNA biology to lncRNAs. Genome Res 2015; 25: 1456–1465.
Huang JL, Zheng L, Hu YW, Wang Q . Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis 2014; 35: 507–514.
Fatica A, Bozzoni I . Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014; 15: 7–21.
Geisler S, Coller J . RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013; 14: 699–712.
Mercer TR, Dinger ME, Mattick JS . Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10: 155–159.
El Azzouzi H, Doevendans PA, Sluijter JP . Long non-coding RNAs in heart failure: an obvious lnc. Ann Transl Med 2016; 4: 182.
Wang K, Liu F, Liu CY, An T, Zhang J, Zhou LY et al. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ 2016; 23: 1394–1405.
Yang J, Lin J, Liu T, Chen T, Pan S, Huang W et al. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer 2014; 85: 110–115.
Richards EJ, Zhang G, Li ZP, Permuth-Wey J, Challa S, Li Y et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) beta: LncRNA-hit-mediated TGFbeta-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem 2015; 290: 6857–6867.
Jia X, Wang Z, Qiu L, Yang Y, Wang Y, Chen Z et al. Upregulation of LncRNA-HIT promotes migration and invasion of non-small cell lung cancer cells by association with ZEB1. Cancer Med 2016; 5: 3555–3563.
Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011; 54: 1679–1689.
Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest 2012; 122: 1469–1486.
Chen HZ, Tsai SY, Leone G . Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer 2009; 9: 785–797.
Laine A, Westermarck J . Molecular pathways: harnessing E2F1 regulation for prosenescence therapy in p53-defective cancer cells. Clin Cancer Res 2014; 20: 3644–3650.
Siouda M, Yue J, Shukla R, Guillermier S, Herceg Z, Creveaux M et al. Transcriptional regulation of the human tumor suppressor DOK1 by E2F1. Mol Cell Biol 2012; 32: 4877–4890.
Xu F, You X, Liu F, Shen X, Yao Y, Ye L et al. The oncoprotein HBXIP up-regulates Skp2 via activating transcription factor E2F1 to promote proliferation of breast cancer cells. Cancer Lett 2013; 333: 124–132.
Kim DH, Roh YG, Lee HH, Lee SY, Kim SI, Lee BJ et al. The E2F1 oncogene transcriptionally regulates NELL2 in cancer cells. DNA Cell Biol 2013; 32: 517–523.
Johnson JL, Pillai S, Chellappan SP . Genetic and biochemical alterations in non-small cell lung cancer. Biochem Res Int 2012; 2012: 940405.
Huang CL, Liu D, Nakano J, Yokomise H, Ueno M, Kadota K et al. E2F1 overexpression correlates with thymidylate synthase and survivin gene expressions and tumor proliferation in non small-cell lung cancer. Clin Cancer Res 2007; 13: 6938–6946.
Gorgoulis VG, Zacharatos P, Mariatos G, Kotsinas A, Bouda M, Kletsas D et al. Transcription factor E2F-1 acts as a growth-promoting factor and is associated with adverse prognosis in non-small cell lung carcinomas. J Pathol 2002; 198: 142–156.
Escote X, Fajas L . Metabolic adaptation to cancer growth: from the cell to the organism. Cancer Lett 2015; 356(2 Pt A): 171–175.
Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreaticcancer. Oncogene 2013; 32: 1616–1625.
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 2014; 26: 344–357.
Acknowledgements
This research is supported by a grant from the projects of science and technology development plan of Jilin province (No. 20140311050YY) and the projects of health and family planning commission of Jilin province (No. 20152009).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Yu, L., Fang, F., Lu, S. et al. lncRNA-HIT promotes cell proliferation of non-small cell lung cancer by association with E2F1. Cancer Gene Ther 24, 221–226 (2017). https://doi.org/10.1038/cgt.2017.10
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cgt.2017.10
This article is cited by
-
Biological roles of SLC16A1-AS1 lncRNA and its clinical impacts in tumors
Cancer Cell International (2024)
-
LncRNA MEG8 promotes NSCLC progression by modulating the miR-15a-5p-miR-15b-5p/PSAT1 axis
Cancer Cell International (2021)
-
LINC00319 promotes cancer stem cell-like properties in laryngeal squamous cell carcinoma via E2F1-mediated upregulation of HMGB3
Experimental & Molecular Medicine (2021)
-
The multifaceted roles of FOXM1 in pulmonary disease
Cell Communication and Signaling (2019)
-
Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells
Molecular Cancer (2017)