Abstract
Transforming growth factor-β (TGF-β) induces apoptosis in many types of cancer cells and acts as a tumor suppressor. We performed a functional analysis of TGF-β signaling to identify a molecular mechanism that regulated survival in small cell lung cancer cells. Here, we found low expression of TGF-β type II receptor (TβRII) in most small cell lung cancer cells and tissues compared to normal lung epithelial cells and normal lung tissues, respectively. When wild-type TβRII was overexpressed in small cell lung cancer cells, TGF-β suppressed cell growth in vitro and tumor formation in vivo through induction of apoptosis. Components of polycomb repressive complex 2, including enhancer of zeste 2 (EZH2), were highly expressed in small cell lung cancer cells; this led to epigenetic silencing of TβRII expression and suppression of TGF-β-mediated apoptosis. Achaete-scute family bHLH transcription factor 1 (ASCL1; also known as ASH1), a Smad-dependent target of TGF-β, was found to induce survival in small cell lung cancer cells. Thus, EZH2 promoted small cell lung cancer progression by suppressing the TGF-β-Smad-ASCL1 pathway.
Similar content being viewed by others
Introduction
Lung cancer causes mortality more than any other type of cancer [1]. Lung cancer is mainly classified as either small cell lung cancer (SCLC) or non-small cell lung cancer (NSCLC), with incidences of ~15 and 84%, respectively [2]. SCLC, high-grade neuroendocrine tumors, has been reported to have the worst prognosis, with a 5-year survival rate of ~5% [3]. Those patients are mostly treated with anti-cancer drugs and/or radiation. However, a primary clinical issue is the acquisition of chemoresistance in SCLC cells [4]. Thus, it is essential to develop novel strategies for SCLC therapy. For successful drug discovery, it is important to find molecular mechanism(s) that maintain survival in SCLC cells.
Transforming growth factor (TGF)-β is a cytokine that exerts many biological functions. TGF-β binds to two different types of serine-threonine kinase receptors, termed type II (TβRII) and type I receptors (TβRI is also known as activin receptor-like kinase 5, ALK-5) expressed on the cell surface. Upon ligand binding, two TβRIIs and two TβRIs form a heterotetrameric complex, and this activated complex phosphorylates the receptor-regulated Smads (R-Smads), Smad2 and Smad3. The phosphorylated R-Smads form complexes with their common-partner Smad (Co-Smad), Smad4, and the R-Smad/Co-Smad complex translocates to the nucleus. Then, the complexes associate with various transcription factors and transcriptional co-activators or co-repressors, which in turn, regulate transcription of a wide spectrum of target genes [5–7].
TGF-β has been reported to have bi-directional roles in cancer progression [8]. TGF-β induces cell cycle arrest at G1 by regulating expression of cyclin-dependent kinase inhibitor 1 A (CDKN1A, also known as p21), CDKN2B (also known as p15), the v-myc avian myelocytomatosis viral oncogene homolog (MYC), and cell division cycle protein 25A (CDC25A) [9]. TGF-β also induces apoptosis in several types of cancer cells through multiple mechanisms [9, 10]. In ~80% of NSCLC tissues, expression of TβRII was lower than that of normal lung tissues [11]. It was also shown that restoration of TβRII into NSCLC cells inhibited their growth in vitro and in vivo. Conversely, TGF-β also plays critical roles in cancer metastasis via the epithelial-mesenchymal transition (EMT) [12]. Once EMT occurs in NSCLC cells, they acquire mesenchymal characteristics, which results in invasion and metastasis [13]. In NSCLC cells, thyroid transcription factor-1 (TTF-1) suppresses TGF-β-mediated EMT and inhibits cell migration and invasion [14]. Thus, the roles of TGF-β in the progression of NSCLC has been intensively studied. In contrast to NSCLC, the roles of TGF-β in the progression of SCLC have not been fully investigated. A few studies have reported that expression of TGFBR2 (the gene that encodes TβRII) was decreased in some SCLC cells, but the mechanisms were not detailed [15, 16]. Therefore, the present study aimed to clarify the roles of TGF-β in SCLC cells, to identify the mechanisms involved in the downregulation of TβRII, and to identify novel TGF-β target genes in this type of cancer.
Results
Downregulation of TβRII expression in SCLC cells
First, we investigated whether TGF-β signals were transduced in SCLC cells. Phosphorylation of Smad2 and induction of SMAD7, one of the direct targets of TGF-β, were examined in human SCLC cells (H146, H82, H209 and H345) and in NSCLC cells (A549 and H441) with immunoblotting and quantitative real-time reverse transcription-PCR (qRT-PCR) analyses. TGF-β-mediated phosphorylation of Smad2 was observed in H146 cells as well as in A549 and H441 cells (Figure 1a and data not shown, see Isogaya et al [17]). Induction of SMAD7 by TGF-β was also observed in H146, A549 and H441 cells (Figure 1b). However, in the other SCLC cells, these responses were not induced by TGF-β. A qRT-PCR analysis also showed that expression of TGFBR2 and SMAD3 was decreased in SCLC cells, but other TGF-β signaling components, including SMAD2, SMAD4 and TGFBR1 (the gene that encodes TβRI), were expressed at normal levels in these cells (Figure 1c). These expression profiles were confirmed with comprehensive gene expression analysis data from the gene expression omnibus (GEO) of the National Center for Biotechnology Information (NCBI) with statistically significant differences (Figure 1d, and Supplementary Figure S1). Since TGF-β signal is transduced even in the low expression levels of Smad3 if Smad2 is expressed in H146 cells (Figure 1b), we assumed that TGF-β signal transduction was attenuated in SCLC cells through the decreased expression of TβRII, and therefore, we decided to focus on the roles of TβRII in SCLC in the present study.
TGF-β signal transduction is attenuated in several SCLC cells due to decreased expression of TβRII. (a and b) SCLC and NSCLC cells were stimulated with TGF-β for 2 h. (a) Immunoblot of cell lysates probed with the indicated antibodies; (b) qRT-PCR analysis of SMAD7 expression. Data represent mean±s.d. **P<0.01; ***P<0.001. (c) qRT-PCR analysis shows expression of TGF-β signaling components in SCLC and NSCLC cells. Data represent mean±s.d. (d) Comprehensive gene expression analysis from the NCBI GEO database (GSE32036) shows expression profiles of TGF-β signaling components in normal lung epithelial cells (n=59), SCLC cells (n=29) and NSCLC cells (n=119). Raw data were normalized by quantile algorithm. The color indicates the distance from the median of each row. GEO, gene expression omnibus; NCBI, National Center for Biotechnology Information; NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer; TGF-β, transforming growth factor-β; qRT-PCR, quantitative real-time reverse transcription-PCR.
TβRII suppresses SCLC tumor growth through TGF-β-induced apoptosis
To examine the roles of TGF-β in SCLC progression, wild-type TβRII was introduced into H82 cells (H82-TβRII cells) or H345 cells (H345-TβRII cells) with lentiviral vectors. Both phosphorylation of Smad2 and induction of SMAD7 by TGF-β were observed in TβRII-expressing cancer cells, but not in control SCLC cells that expressed green fluorescent protein (GFP) alone (H82-GFP cells and H345-GFP cells; Figure 2a and b). Thus, TGF-β signal transduction was successfully recovered by expressing TβRII. These cells were subcutaneously xenografted into nude mice to examine tumor growth in vivo. Tumor formation was decreased in mice injected with H82-TβRII cells and H345-TβRII cells, compared with mice xenografted with the control cells (Figure 2c). Although the expression of TGFBR2 mRNA was low (Figure 1c) and the TβRII protein was not detected by immunoblot analysis (data not shown), Smad-dependent TGF-β signal was transduced in H146 cells (Figures 1a and b), suggesting that a low level of TβRII protein may be functioning in these cells. Thus, a GFP-tagged dominant-negative form of TβRII (dnTβRII) was overexpressed in H146 cells (H146-dnTβRII cells; Supplementary Figure S2a). Both phosphorylation of Smad2 and induction of SMAD7 were inhibited by the introduction of dnTβRII (Supplementary Figures S2b and S2c). When these cells were subcutaneously xenografted into mice, tumor formation was accelerated in mice injected with H146-dnTβRII cells compared with those injected with H146-GFP cells (Supplementary Figure S2d). These results suggested that TGF-β may act as a tumor suppressor in vivo. We assessed angiogenesis in these tumor tissues by staining for CD31 expression (also known as platelet/endothelial cell adhesion molecule 1, PECAM1). However, there was no remarkable difference in CD31 expression between H146-GFP and H146-dnTβRII cells xenografted tissues (Supplementary Figure S2e). This finding suggested that tumor suppression was mediated by the effect of TGF-β on SCLC cells, not by its effect on angiogenesis in the tumor microenvironment.
TβRII suppresses SCLC tumor growth through TGF-β-induced apoptosis. (a and b) SCLC cells were infected with lentivirus vectors encoding GFP (H82-GFP and H345-GFP) or TβRII (H82-TβRII and H345-TβRII). Cells were stimulated with TGF-β for 2 h. (a) Immunoblots of cell lysates probed with the indicated antibodies. (b) qRT-PCR analysis of SMAD7 expression. Data represent mean±s.d. ***P<0.001. (c) Mice received subcutaneous transplantations of H82-GFP (n=15) and H82-TβRII cells (n=12) or H345-GFP (n=7) and H345-TβRII cells (n=9), and tumor volumes were measured 16 days (H82) or 17 days (H345) after transplantation. Data represent mean±s.e.m. *P<0.05; **P<0.01. (d) Cell proliferation assay. SCLC cells were stimulated with TGF-β for 6 days (H82) or 12 days (H345). Data represent mean±s.d. ***P<0.001. (e) Cell cycle analysis. (left panels) H345-GFP and H345-TβRII cells were unstimulated (top panels) or stimulated (bottom panels) with TGF-β for 12 days; the number of cells in each cell cycle stage is shown (color coding shown in right panel). (right panel) Percentage of cells in each cell cycle stage. (f) Immunoblots of cell lysates probed with the indicated antibodies. SCLC and control HaCaT cells were stimulated with TGF-β for 48 h (HaCaT) or 12 days (H345). (g) qRT-PCR analysis shows cell cycle-related gene expression. H345-GFP, H345-TβRII and HaCaT cells were stimulated with TGF-β for 2 h (MYC) or 24 h (CDKN1A, CDKN2B and CDC25A). Data represent mean±s.d. *P<0.05; **P<0.01; ***P<0.001. GFP, green fluorescent protein; SCLC, small cell lung cancer; TGF-β, transforming growth factor-β; qRT-PCR, quantitative real-time reverse transcription-PCR.
We postulated that TGF-β might suppress the proliferative activity of SCLC cells. When we restored TβRII expression in these cells, TGF-β significantly suppressed the in vitro proliferation of H82 cells and H345 cells (Figure 2d). Moreover, dnTβRII expression canceled TGF-β-mediated growth inhibition in H146 cells (Supplementary Figure S2f). Cell cycle analysis revealed that TGF-β increased the sub-G0/G1 population in H345-TβRII cells compared with H345-GFP cells (Figure 2e). TGF-β also induced the cleavage of poly (ADP-ribose) polymerase (PARP) in H345-TβRII cells (Figure 2f), which suggested that TGF-β decreased the number of SCLC cells by inducing apoptosis. TGF-β is also known to suppress proliferation of many types of cells by regulating CDK activators or inhibitors. We found that expression levels of CDKN1A, CDKN2B, MYC or CDC25A in H345-TβRII cells were not markedly altered by TGF-β (Figure 2g). However, in human keratinocyte HaCaT cells, TGF-β upregulated the expression of CDKN1A and CDKN2B and downregulated the expression of MYC and CDC25A. Moreover, expression of retinoblastoma protein (pRB) was not detected in H345 cells (Figure 2f). These results suggested that TGF-β suppressed proliferation of SCLC cells by inducing apoptosis, but not by regulating the cell cycle.
Importance of EZH2-mediated silencing of TβRII for SCLC tumor formation
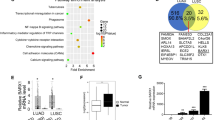
Histone methyltransferase mediates methylation on lysine or arginine residues of histones of the H3 and H4 families to regulate transcription of various genes [18]. We postulated that, in SCLCs, the expression of TβRII might be epigenetically silenced through histone modification by histone methyltransferases. Therefore, we investigated histone methyltransferase expression in SCLC cells. Comprehensive gene expression analyses from the NCBI GEO data set revealed that SCLC cells and SCLC tissues displayed higher expression of the enhancer of zeste 2 (EZH2), SUZ12 polycomb repressive complex 2 subunit (SUZ12), and embryonic ectoderm development (EED) than normal lung epithelial cells and normal lung tissues, respectively (Figure 3a, and Supplementary Figure S3). We also found that their expressions were increased in SCLC cells but not in NSCLC cells (Figure 3b and c). These molecules associate to form the polycomb repressive complex 2 (PRC2), which inhibits gene transcription through methylation of lysine 27 in histone H3 (H3K27me3). High expression of H3K27me3 was observed in SCLC cells, which was attenuated by the treatment with an EZH2 inhibitor, GSK343 (Figure 3c), suggesting that PRC2 is implicated in transcriptional regulation of several genes in SCLC cells. H146 cells showed different expression profiles of the PRC2 complex from those in the other SCLC cells. The expression level of EZH2 protein was similar to those in the other SCLC cells, while the expression levels of EZH2, EED and SUZ12 mRNAs were lower than those in the other SCLC cells (Figures 3b and c).
EZH2 is highly expressed in SCLC cells. (a) Expression profiles of PRC2 components in SCLC cells, based on the data in Figure 1d. The color indicates the distance from the median of each row. (b) qRT-PCR analysis shows expression of PRC2 components in SCLC and NSCLC cells. Data represent mean±s.d. (c) Immunoblot of cell lysates probed with the indicated antibodies. (left panels) SCLC and NSCLC cells were treated with GSK343 in a wide concentration range (0, 0.4, 2 and 10 μm) for 3 days. (right panel) Relative expression of EZH2 protein in each cell without GSK343 was quantified. NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer; qRT-PCR, quantitative real-time reverse transcription-PCR.
In order to directly examine whether EZH2 is involved in downregulation of TGFBR2, chromatin immunoprecipitation (ChIP)-qRT-PCR analysis using anti-EZH2 antibody was performed (Figure 4a). EZH2 bound to the several loci in TGFBR2 in H345 cells. Moreover, transcription of TGFBR2 mRNA was increased in GSK343-treated SCLC cells (Figure 4b). When EZH2 expression was silenced in H345 cells with a short hairpin RNA (shRNA) (H345-shEZH2), the knockdown of EZH2 led to an increase in TGFBR2 expression (Figure 4c); in turn, TGF-β induced Smad2 phosphorylation and SMAD7 expression (Figures 4d and e). These results suggested that EZH2 played a critical role in downregulating TβRII in SCLC cells.
EZH2-mediated silencing of TβRII is required for SCLC tumor formation. (a) qRT-PCR analysis post-immunoprecipitation with anti-EZH2 antibody shows EZH2 enrichment in the TGFBR2 locus of H345 cells. Hypoxanthine guanine phosphoribosyl transferase1 (HPRT1) was used as the negative control. Data represent mean±s.d. TGFBR2 locus 1, chromosome 3: 30606416–30606492 bp; TGFBR2 locus 2, chromosome 3: 30605676–30605771 bp; TGFBR2 locus 3, chromosome 3: 30604616–30604714 bp; TGFBR2 locus 4, chromosome 3: 30603967–30604061 bp. *P<0.05; **P<0.01. (b) qRT-PCR analysis shows TGFBR2 expression. SCLC cells were treated with GSK343 (10 μM) for 7 days. Data represent mean±s.d. (c) qRT-PCR analysis shows EZH2 and TGFBR2 expression. H345 cells were infected with lentivirus vectors with control shRNA (H345-shNTC) or shRNA that targeted EZH2 (H345-shEZH2). Data represent mean±s.d. ***P<0.001. (d) Immunoblot of cell lysates from cells in (c) stimulated with TGF-β for 2 h and probed with the indicated antibodies. H345-GFP cells and H345-TβRII cells were negative and positive controls, respectively. (e) qRT-PCR analysis shows SMAD7 expression. Cells in (c) were stimulated with TGF-β for 4 h. Data represent mean±s.d. H345-GFP cells and H345-TβRII cells were negative and positive controls, respectively. **P<0.01; ***P<0.001. (f) Cell cycle analysis of cells in (c) stimulated with TGF-β for 12 days. (left panels) The number of cells in each cell cycle stage is shown (color coding shown in right panel). (right panel) Percentage of cells in each cell cycle stage. (g) Mice received subcutaneous transplants of H345-shNTC cells (n=7) or H345-shEZH2 #1 cells (n=7). (left panels) Representative photographs 4 weeks after injection. Arrow heads indicate tumors. (right panel) Tumor volumes at the indicated time points. Data represent mean±s.e.m. **P<0.01. SCLC, small cell lung cancer; qRT-PCR, quantitative real-time reverse transcription-PCR.
We also investigated whether EZH2 was important for TGF-β-mediated apoptosis and tumor formation in H345 cells. Cell cycle analysis showed that TGF-β increased the sub-G0/G1 population in H345-shEZH2 cells, but not in H345-shNTC cells (Figure 4f). Moreover, the ability of H345-shEZH2 cells to form tumors was attenuated, compared to that of H345-shNTC cells (Figure 4g).
ASCL1 is negatively regulated by TGF-β in a Smad-dependent manner
We next attempted to identify TGF-β target genes that were involved in SCLC cell apoptosis. ChIP-sequencing (ChIP-seq) analysis was performed with an anti-Smad2/3 antibody in H345-TβRII cells to identify comprehensively Smad2/3-regulated genes. The characteristics of SCLC cells are thought to depend on the expression of several neuroendocrine-related genes, including the achaete-scute family basic helix-loop-helix transcription factor 1 (ASCL1, also known as ASH1), synaptophysin (SYP), neural cell adhesion molecule (NCAM) and v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog (MYCL) [19]. Therefore, we focused on Smad2/3 binding to these gene loci. ChIP-seq analysis showed that, in the presence of TGF-β, Smad2/3 significantly bound to two loci of the ASCL1 gene and several loci of the NCAM1 gene, but not to the SYP locus or MYCL locus, in H345-TβRII cells (Figure 5a). Among the binding regions in the NCAM1 locus, Smad2/3 strongly bound to the first intron. Comprehensive gene expression analysis from the NCBI GEO data sets showed that the mRNA levels of each of these neuroendocrine-related genes were elevated in SCLC cells and SCLC tissues, except for SYP in the SCLC tissue (Supplementary Figures S4a and S4b). Next, TGF-β regulation of these genes was assessed by qRT-PCR analysis (Figure 5b and Supplementary Figures S5a and S5b). In H345-TβRII cells, treatment with TGF-β caused ASCL1 expression to decrease within 1 h, and it reached a minimum at 4 h. TGF-β also decreased ASCL1 expression within 2 h in H146 cells. However, MYCL, NCAM1 and SYP were not regulated by TGF-β in these cells. Moreover, TGF-β suppressed ASCL1 protein expression in SCLC cells (Figure 5c). In accordance with a previous analysis (Supplementary Figure S4a), in NSCLC cells, ASCL1 was only weakly expressed, and it was not regulated by TGF-β (A549 and H441) (Supplementary Figure S5c). These results suggested that ASCL1 was a TGF-β target, and regulation of ASCL1 by TGF-β was specific for SCLC cells.
ASCL1 is negatively regulated by TGF-β in a Smad-dependent manner. (a) ChIP-seq analysis using anti-Smad2/3 antibody. H345-TβRII cells were stimulated with TGF-β for 1.5 h. Arrows indicate transcription start sites and direction. (b) qRT-PCR analysis shows SMAD7 and ASCL1 expression in H345-GFP and H345-TβRII cells after TGF-β stimulation for the indicated times. Data represent mean±s.d. (c) Immunoblot of cell lysates probed with the indicated antibodies. H345-GFP and H345-TβRII cells were stimulated with TGF-β for 12 days. (d) qRT-PCR analysis post-immunoprecipitation with anti-Smad2/3 antibody shows Smad2/3 enrichment. H345-GFP and H345-TβRII cells were stimulated with TGF-β for 1.5 h. Hemoglobin beta (HBB) was used as a negative control. Data represent mean±s.d. ASCL1 locus 1, chromosome 12: 103351326–103352062 bp; ASCL1 locus 2, chromosome 12: 103351326–103352062 bp. *P<0.05; **P<0.01. (e) qRT-PCR analysis shows ASCL1 expression in indicated cells. Data represent mean±s.d. ***P<0.001. (f) qRT-PCR analysis shows SMAD7 and ASCL1 expression. H345-GFP and H345-TβRII cells were stimulated with TGF-β for 4 h after pre-treatment with CHX (3 μM) for 24 h. Data represent mean±s.d. ***P<0.001. (g) qRT-PCR analysis shows SMAD4 expression. H345-GFP and H345-TβRII cells were infected with lentivirus vector with control shRNA (shNTC) or shRNA that targeted Smad4 (shSmad4). Data represent mean±s.d. (h) qRT-PCR analysis shows SMAD7 and ASCL1 expression. Cells in (g) were stimulated with TGF-β for 4 h. Data represent mean±s.d. *P<0.05; ***P<0.001. ChIP, chromatin immunoprecipitation; TGF-β, transforming growth factor-β; qRT-PCR, quantitative real-time reverse transcription-PCR.
We then focused on the molecular mechanism underlying TGF-β-mediated transcriptional regulation of ASCL1 in SCLC cells. ChIP-qRT-PCR analysis was performed to confirm whether Smad2/3 bound in the ASCL1 locus. We found that TGF-β-stimulated H345-TβRII cells showed a sixfold-enrichment of Smad2/3 binding to these loci, compared with those to hemoglobin beta (HBB) locus (Figure 5d). In addition to the recovery of TβRII expression and TGF-β signal transduction in H345-shEZH2 cells (see Figures 4d and e), ASCL1 expression was decreased in these cells (Figure 5e). Next, cycloheximide (CHX), a de novo protein synthesis inhibitor, was used to investigate whether ASCL1 expression was directly or indirectly regulated by TGF-β. Even in the presence of CHX, TGF-β could upregulate SMAD7 and downregulate ASCL1 expression (Figure 5f). This result suggested that, similar to SMAD7, the expression of ASCL1 was directly regulated by TGF-β.
TGF-β can activate both Smad and non-Smad pathways. To determine which pathway played a predominant role in regulating ASCL1 expression, we used a shRNA (shSmad4) to silence the expression of Smad4 in H345-TβRII cells. The knockdown of Smad4 attenuated the induction of SMAD7 by TGF-β (Figures 5g and h). Moreover, TGF-β-mediated downregulation of ASCL1 expression was canceled in H345-TβRII cells by shSmad4, but not in cells infected with negative control shRNA (shNTC). These results suggested that TGF-β directly suppressed ASCL1 expression in a Smad-dependent manner.
ASCL1 promotes survival of SCLC cells
Next, we determined whether negative regulation of ASCL1 transcription was important for TGF-β-mediated apoptosis of SCLC cells. When expression of ASCL1 was silenced in H345 cells with shRNA (shASCL1) (Figure 6a), cell growth was inhibited by the induction of apoptosis (Figures 6b and c). Conversely, when ASCL1 was exogenously introduced by transferring lentiviral vectors into H345-TβRII cells, cell cycle analysis revealed that the TGF-β-mediated increase in the sub-G0/G1 population was attenuated (Figures 6d and e). These results suggested that ASCL1 had an important role in enabling SCLC cells to escape from TGF-β-induced apoptosis.
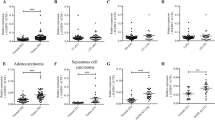
Downregulation of ASCL1 is important for TGF-β-mediated apoptosis in SCLC cells. (a) qRT-PCR analysis shows ASCL1 expression. H345 cells were infected with lentivirus vectors encoding control shRNA (shNTC) or shRNA that targeted ASCL1 (shASCL1). H345-TβRII cells stimulated with TGF-β for 48 h served as control. Data represent mean±s.d. *P<0.05; ***P<0.001. (b) Cell proliferation assay. Cells in (a) were incubated for 12 days. Data represent mean±s.d. ***P<0.001. (c) Cell cycle analysis of cells in (b). (left panels) The number of cells in each cell cycle stage is shown (color coding shown in right panel). (right panel) Percentage of cells in each cell cycle stage. (d) Immunoblot of cell lysates probed with the indicated antibodies. H345-GFP and H345-TβRII cells were infected with lentivirus vectors encoding GFP alone or ASCL1, and then stimulated with TGF-β for 12 days. (e) Cell cycle analysis of cells in (d). (left panels) The number of cells in each cell cycle stage is shown (color coding shown in right panel). (right panel) Percentage of each cell cycle stage in indicated cells is shown. (f) qRT-PCR analysis shows ASCL1 expression in H345 cells transfected with control siRNA (siNTC) or siRNA that targeted ASCL1 (siASCL1). ASCL1 expression was determined 72 h post-transfection. Data represent mean±s.d. ***P<0.001. (g) Mice received subcutaneous transplants of cells in (f) (siNTC, n=7, siASCL1, n=7). (Left panels) Representative photographs; (right panel) incidence of tumor formation 2 weeks after injection. Arrow heads indicate tumors. GFP, green fluorescent protein; SCLC, small cell lung cancer; TGF-β, transforming growth factor-β; qRT-PCR, quantitative real-time reverse transcription-PCR.
Next, we performed subcutaneous transplantations in mice to determine whether knocking down ASCL1 expression would suppress tumor growth or formation. We knocked down ASCL1 expression by treating cells with small interfering RNA (siRNA) that targeted ASCL1 (siASCL1). Control cells were treated with negative control siRNA (siNTC) (Figure 6f). The siNTC treated-H345 cells formed tumors in six out of seven mice, but siASCL1-treated H345 cells formed tumors in only one out of seven mice (Figure 6g). These results suggested that ASCL1 was involved in SCLC tumor formation.
EZH2-mediated silencing of TβRII in SCLC tissues
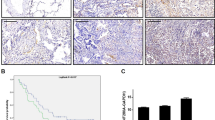
Finally, we compared the expression of EZH2, TβRII and ASCL1 in SCLC tissues and NSCLC tissues to that in normal lung tissues (Supplementary Table S1). Immunohistochemical analysis showed that EZH2 was strongly expressed in the nuclei of all SCLC cells (Figure 7a). However, its expression was not observed in those of normal lung epithelial cells. In contrast, TβRII was weakly expressed in SCLC cells, while it was expressed on the surface of normal lung epithelial cells (Figure 7a). Although ASCL1 was also detected in nuclei of some SCLC cells, it was not observed in most of normal lung epithelial cells and other lung cancer cells (Figure 7a and Supplementary Figure S6). The expression profiles of EZH2, TβRII and ASCL1 are shown in Figure 7b. The number of positive samples, grouped according to expression frequency, is shown in Supplementary Table S2. When all lung tissues in Figure 7 were considered, there was a negative correlation between EZH2 and TβRII and a positive correlation between EZH2 and ASCL1 (Supplementary Table S3). These results supported the notion that EZH2 had a role in epigenetic silencing of TβRII in human SCLC tissues, and that the loss of TβRII upregulated the expression of ASCL1.
Expression profiles of EZH2, TβRII and ASCL1 in human normal lung tissues and human lung cancer tissues. (a) Lung tissues were stained with HE, anti-EZH2 antibody, anti-TβRII antibody and anti-ASCL1 antibody. Representative images show normal tissues (top and third rows) and SCLC tissues (second and bottom rows) from two patients, as indicated. (Insets) ASCL1 staining in boxed region is shown at high magnification. Scale bars are 30 μm. (b) Expression profiles of samples in (a) and Supplementary Figure S6 were analyzed by defining scores (s) that corresponded to the frequency of positive cells (f) in each sample, as follows: s=4 for 80⩽f⩽100; s=3 for 50⩽f<80; s=2 for 20⩽f<50; s=1 for 0<f<20; s=0 for f=0. Data represent means. *P<0.05; ***P<0.001. HE, hematoxylin-eosin; LCNEC, large cell neuroendocrine carcinoma; normal, normal lung; SCLC, small cell carcinoma; Ad, adenocarcinoma; Sq, squamous cell carcinoma.
Discussion
The present study clarified the tumor suppressive role of TGF-β in SCLCs. We showed that TβRII was expressed in normal lung epithelial cells, and that it inhibited abnormal cell growth by downregulating ASCL1 in a Smad-dependent manner (Figures 8a and b). However, EZH2 was highly expressed in SCLC cells, which epigenetically attenuated the expression of TβRII; thus, TGF-β signaling was suppressed, which resulted in high ASCL1 levels and progression of SCLC. These results suggested new therapeutic strategies for targeting EZH2 and ASCL1 in SCLC therapy.
Disruption of TGF-β-mediated tumor suppression in SCLC cells. (a) In normal lung epithelial cells, TGF-β induces apoptosis through the suppression of ASCL1 expression in a Smad-dependent manner. (b) In SCLC cells, high EZH2 expression attenuates TβRII expression through histone H3K27 tri-methylation. Disruption of TGF-β signaling elevates ASCL1 expression, which in turn protects SCLC cells from apoptosis. SCLC, small cell lung cancer; TGF-β, transforming growth factor-β.
We demonstrated that TGF-β inhibited proliferation of SCLC cells in vivo and in vitro. The SCLC cells used in this study carried mutations in the DNA binding region of TP53, and they did not express pro-apoptotic pRB, except for the H209 cells (Figure 2f, data not shown) [20]. Thus, TGF-β-mediated apoptosis in SCLC cells may occur independently of p53 and pRB. TGF-β causes cell cycle arrest at the G1 phase or apoptosis in many types of cancer cells [9]. For cell cycle arrest at the G1 phase, TGF-β regulates p21, p15, c-Myc and CDC25A in various types of cells, but it did not regulate these genes in SCLC cells. TGF-β was also reported to induce apoptosis by inducing expression of BCL2-like 11 (BCL2L11, also known as Bim), growth arrest and DNA-damage-inducible beta (GADD45B) and inositol polyphosphate-5-phosphatase 145 kDa (INPP5D, also known as SHIP) [9]. In contrast, our comprehensive gene expression analysis and ChIP-seq analysis showed that the TGF-β-mediated apoptosis in SCLC cells appeared to be independent of those genes (data not shown). Based on these observations, we attempted to identify novel target(s) for TGF-β, which could regulate survival in SCLC cells.
In many types of cancers, TβRII is dysfunctional through either genetic mutation or transcriptional repression [21–23]. The TGFBR2 locus was shown to be mutated in the 10-adenine (A10) tract of exon 3 and serine-threonine kinase domain in some cancers [21–24]. Previous studies demonstrated that genetic mutations in the TGFBR2 A10 tract or expression of a truncated TβRII were not common in SCLC [15, 25]. Moreover, loss of heterozygosity in chromosome 3, including in the TGFBR2 locus, was not observed in SCLC [25]. Those studies indicated that genetic mutation was not a common mechanism for the dysfunction of TβRII in SCLC. In contrast, transcriptional repression of TGFBR2 was reported in retinoblastoma and hematopoietic malignancies [23, 26].
In this study, we showed that PRC2 components were highly expressed in most of the SCLC cells, and that increased EZH2 expression caused silencing of TβRII expression in SCLC cells. Sato et al [27]. reported that many kinds of genes, such as JUB, PTRF, DMKN, AXL and EPHB4, were identified as targets for EZH2 in SCLC cells by ChIP-seq analysis. They also found that introduction of JUB inhibited cellular growth, suggesting that suppression of these genes by EZH2 other than TGFBR2 might be involved in the growth of SCLC. Genetic alternations in TP53 and RB1 are commonly observed in patients with SCLC; consequently, these were considered as early events that triggered SCLC development [28–30]. EZH2 expression was upregulated in Rb1 knockout MEF cells [31]. Based on those studies, our finding that EZH2 was highly expressed in SCLC suggests that EZH2 is an oncogenic factor. EZH2 was also reported to be highly expressed in breast cancer and prostate cancer [32, 33]; moreover, EZH2 inhibitors have been considered a promising therapy for certain types of tumors [34, 35]. A specific EZH2 inhibitor induced TGFBR2 expression (Figure 4b); therefore, EZH2 inhibitors may also effectively eradicate SCLC cells by restoring TβRII expression, and thus, enabling TGF-β-mediated apoptosis.
TGF-β-target genes have been comprehensively identified in many kinds of tumors, including liver cancer, pancreatic cancer, NSCLC and breast cancer [17, 36–38], but rarely in neuroendocrine tumors. Identification of novel target(s) may improve our understanding of SCLC cell characteristics. In the present study, our comprehensive gene expression analysis and ChIP-seq analysis demonstrated that the TGF-β-induced apoptosis in SCLC cells could be attributed to negative regulation of ASCL1 by TGF-β. It was previously shown that ASCL1 expression was inhibited by Notch signaling in SCLC cells [39]. This report was the first to reveal the mechanism underlying TGF-β regulation of ASCL1 expression. ASCL1, a member of the basic helix-loop-helix family transcription factors, has a crucial role in the differentiation of neural stem cells into neuronal lineages [40]. ASCL1 is also expressed in neuroendocrine tumors, especially in cases with poor prognoses [41–48]. In addition, several genes were identified as targets for ASCL1 [49]. Among them, miRNA-375 (miR-375) was supposed to inactivate Yes-associated protein (YAP)1 in SCLC [50]. In H345-TβRII cells, TGF-β exhibited a downgulation of ASCL1, followed by upregulation of primary-miR 375 (pri-miR-375) (data not shown). Thus, our present findings suggested that the TGF-β-Smad-ASCL1 pathway is important in SCLC progression, and that it may also have an important role in other neuroendocrine tumors.
Materials and Methods
Cell culture and reagents
Human SCLC H82, H146, H209, H345 cells and human NSCLC A549 and H441 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured as recommended. Human skin keratinocyte HaCaT cells were previously described [51]. TGF-β3 (R & D Systems, Minneapolis, MN, USA) was reconstituted in 4 mm HCl and 0.1% bovine serum albumin (BSA, Sigma-Aldrich, St Louis, MO, USA) and used at a concentration of 1 ng ml−1. GSK343 (Sigma-Aldrich) and CHX (Sigma-Aldrich) were reconstituted in dimethyl sulfoxide. See also Supplementary Information.
Cell proliferation assay
H82 cells (1×104 cells), H146 cells (3×104 cells) and H345 cells (3×104 cells) were seeded on 12-well plates, and then stimulated with TGF-β. Cell proliferation was evaluated with Cell Count Reagent SF (Nacalai Tesque, Kyoto, Japan). Absorbance at 450 nm was measured with a Model 680 Microplate Reader (Bio-Rad, Melville, NY, USA), followed by subtraction of absorbance at 595 nm.
In vivo tumor growth assay
In vivo experiments were performed as previously described [52]. The protocols were approved by the Animal Ethics Committee of The University of Tokyo (approval number: 2186). BALB/c nu/nu mice (4 weeks, male) were purchased from Charles River Laboratories (Yokohama, Japan). Cells were resuspended in culture media supplemented with 50% BD Matrigel (BD Bioscience, San Jose, CA, USA), and then subcutaneously injected into mice (3×106 cells in 100 μl per mouse). For xenograft transplantations, H345 cells were treated with siRNA against ASCL1 for 72 h in vitro, followed by subcutaneous transplantation of ASCL1-silenced cells into mice.
Gene expression analysis
Total RNA was extracted with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Complementary DNA was synthesized with the random hexamer protocol described in the PrimeScript II 1st strand complementary DNA Synthesis Kit (Takara, Otsu, Japan). For qRT-PCR analysis, gene expression was quantified with the StepOne Plus Real time-PCR System (Life Technologies, Tokyo, Japan) and the Fast SYBR Green Master Mix (Life Technologies). The expression level of each gene was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primer sequences are shown in Supplementary Table S4.
Immunoblotting
Immunoblotting was previously described [53, 54]. Cells were lysed in radio-immunoprecipitation assay buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, and 0.5% sodium deoxycholate) with the Complete Protease Inhibitor Cocktail (Roche Diagnostics, Tokyo, Japan) and an EDTA-free phosphatase inhibitor cocktail (Nacalai Tesque). Protein concentrations were quantified with the BCA Protein Assay (ThermoFisher Scientific, Yokohama, Japan). Equal amounts of total protein were applied to SDS-polyaclylamide gel electrophoresis, and transferred to Fluoro Trans W membrane (Pall, East Hills, NY, USA). Chemiluminescence images were captured on ImageQuant LAS4000 (Fujifilm, Tokyo, Japan). Image J software (NIH) was used to quantify blot band intensities in Figure 3c. See also Supplementary Information.
Lentiviral vector construction and lentivirus production
A lentiviral vector system (provided by Dr Hiroyuki Miyoshi, RIKEN) was used to induce specific gene introduction and knockdown. For gene introduction, we inserted complementary DNAs encoding the human wild-type TGFBR2, TGFBR2 with a truncated intracellular domain and a carboxy-terminal GFP tag (dnTβRII), and human wild-type ASCL1, into the entry vector, pENTR201 [55]. Then, pENTR201 vectors were inserted into the lentiviral destination vector, pCSII-EF-RfA or pCSII-CMV-RfA, as previously described [56]. Vectors encoding GFP were also generated as controls.
Similarly, shRNAs designed to knockdown a specified gene were inserted into the entry vector pENTR4-H1. Then, pENTR4-H1 vectors that carried shRNAs specific for human ASCL1 or EZH2 were inserted into the lentiviral destination vector, pCS-RfA-EG. The shRNA target sequences for gene knockdowns were obtained from Dharmacon siDESIGN Center (GE Healthcare, Piscataway, NJ, USA; Supplementary Table S5). Lentiviral vectors were produced as described previously [53]. Culture supernatant was concentrated with Lenti-X Concentrator (Clontech, Palo Alto, CA, USA), then used for lentiviral vector infections.
siRNA
An Accell-siRNA SMARTpool specific for human ASCL1 was purchased from Dharmacon (GE Healthcare), and reconstituted in 1×siRNA buffer (100 μm, GE Healthcare). The siRNA target sequences are shown in Supplementary Table S6. Cells were treated with siRNA at a final concentration of 1 μm.
Cell cycle analysis
After washing with phosphate-buffered saline, cells were fixed with ice-cold 70% EtOH in phosphate-buffered saline and stored for more than 16 h at −20 °C. The fixed cells were resuspended in phosphate-buffered saline containing 0.25 mg ml−1 RNase A, and then incubated at 37 °C for 1 h. Cells were labeled with 50 μg ml−1 propidium iodide (PI, Life Technologies) for 30 min at 4 °C; then, cell cycle analysis was performed with a Gallios Flow Cytometer (Beckman Coulter, Miami, FL, USA). The distribution of each cell cycle stage was analyzed with the FlowJo (Tomy Digital Biology, Tokyo, Japan) Watson Pragmatic cell cycle analysis program.
ChIP-qRT-PCR analysis and ChIP-seq analysis
ChIP was previously described [51, 57]. See also Supplementary Information. ChIP-qRT-PCR analyses were performed with the StepOne Plus Real time-PCR System and FastStart Universal SYBR Green Master (Rox) (Roche). Primer sequences for each gene locus are shown in Supplementary Table S4.
For ChIP-seq analysis, total amounts of double stranded DNA were quantified with Qubit dsDNA HS Assay Kits (Life Technologies). Libraries were prepared with IonXpress Plus Fragment Library Kit (Life Technologies). Libraries were quantified with Ion Library Quantification Kit (Life Technologies). Emulsion PCR and product purification were performed with Ion PGM Template OT2 400 Kit (Life Technologies). The amplified samples were sequenced with Ion PGM Sequencer (Life Technologies) with Ion PGM Hi-Q Sequencing Kit. The acquired read tags were mapped onto the NCBI hg19 human genome assembly. Analyses of ChIP-seq data were previously described [58, 59]. The significant Smad2/3 binding region was calculated using CisGenome version 2 using default parameters except for window size (400 bp), cut-off counts (⩾10 reads (P=0.023516)), and step size (25 bp). Raw ChIP-seq and peak call data are available at GEO (GSE63871).
Immunohistochemistry
Immunohistochemistry was previously described [52]. Formalin-fixed, paraffin-embedded human clinical tissues and a tissue microarray were obtained from patients at the University of Tokyo Hospital with informed consent. The protocol was approved by the Research Ethics Committee at the University of Tokyo, Graduate School of Medicine (approval number: G2211-(8) and 2381-(4)). Immunohistochemistry was performed with the Ventana (Roche) or the VECTASTAIN Elite ABC Kit (Vector Laboratories, Burlingame, CA, USA) for hand-staining. In hand-staining, sections were deparaffinized in xylene, and autoclaved in 10 mm citrate buffer (pH 6.0) for 10 min at 121 °C for antigen retrieval. Endogenous peroxidase was inactivated in 3% H2O2 diluted with methanol for 20 min. The sections were immunostained with primary antibodies, then with secondary antibody, and subjected to the avidin/biotinylated peroxidase complex reaction. The immunodetection substrate was 3,3′-Diaminobenzidine (DAB, Vector Laboratories). Quantification was performed by counting the number of positive cells in each sample.
Statistical analysis
Comparisons between samples were performed with the Student’s t test after the F-test. Comparisons between groups were performed with the analysis of variance; one-way analysis of variance (Tukey’s method) was applied to comprehensive gene expression analyses and immunohistochemical analyses. The repeated measure analysis of variance was applied to in vivo experiments. Significant differences were defined as P<0.05.
References
Sun S, Schiller JH, Gazdar AF . Lung cancer in never smokers--a different disease. Nat Rev Cancer 2007; 7: 778–790.
Herbst RS, Heymach J V, Lippman SM . Lung cancer. N Engl J Med 2008; 359: 1367–1380.
Harris K, Khachaturova I, Azab B et al. Small cell lung cancer doubling time and its effect on clinical presentation: a concise review. Clin Med Insights Oncol 2012; 6: 199–203.
Jackman DM, Johnson BE . Small-cell lung cancer. Lancet 2005; 366: 1385–1396.
Heldin C-H, Miyazono K, ten Dijke P . TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 1997; 390: 465–471.
Massagué J . TGFβ signaling: receptors, transducers, and mad proteins. Cell 1996; 85: 947–950.
Feng X-H, Derynck R . Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol 2005; 21: 659–693.
Bierie B, Moses HL . TGF-β and cancer. Cytokine Growth Factor Rev 2006; 17: 29–40.
Pardali K, Moustakas A . Actions of TGF-β as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta 2007; 1775: 21–62.
Ikushima H, Miyazono K . TGFβ signalling: a complex web in cancer progression. Nat Rev Cancer 2010; 10: 415–424.
Anumanthan G, Halder SK, Osada H et al. Restoration of TGF-β signalling reduces tumorigenicity in human lung cancer cells. Br J Cancer 2005; 93: 1157–1167.
Xu J, Lamouille S, Derynck R . TGF-β-induced epithelial to mesenchymal transition. Cell Res 2009; 19: 156–172.
De Craene B, Berx G . Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13: 97–110.
Saito RA, Watabe T, Horiguchi K et al. Thyroid transcription factor-1 inhibits transforming growth factor-β-mediated epithelial-to-mesenchymal transition in lung adenocarcinoma cells. Cancer Res 2009; 69: 2783–2791.
Hougaard S, Nørgaard P, Abrahamsen N, Moses HL, Spang-Thomsen M, Skovgaard Poulsen H . Inactivation of the transforming growth factor β type II receptor in human small cell lung cancer cell lines. Br J Cancer 1999; 79: 1005–1011.
De Jonge RR, Garrigue-Antar L, Vellucci VF, Reiss M . Frequent inactivation of the transforming growth factor beta type II receptor in small-cell lung carcinoma cells. Oncol Res 1997; 9: 89–98.
Isogaya K, Koinuma D, Tsutsumi S et al. A Smad3 and TTF-1/NKX2-1 complex regulates Smad4-independent gene expression. Cell Res 2014; 24: 994–1008.
Greer EL, Shi Y . Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13: 343–357.
Calbo J, van Montfort E, Proost N et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 2011; 19: 244–256.
Ianari A, Natale T, Calo E et al. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell 2009; 15: 184–194.
Markowitz S, Wang J, Myeroff L et al. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science 1995; 268: 1336–1338.
Chowdhury S, Ammanamanchi S, Howell GM . Epigenetic targeting of transforming growth factor β receptor II and implications for cancer therapy. Mol Cell Pharmacol 2009; 1: 57–70.
Kim S-J, Im Y-H, Markowitz SD, Bang Y-J . Molecular mechanisms of inactivation of TGF-β receptors during carcinogenesis. Cytokine Growth Factor Rev 2000; 11: 159–168.
Lynch MA, Nakashima R, Song H et al. Mutational analysis of the transforming growth factor β receptor type II gene in human ovarian carcinoma. Cancer Res 1998; 58: 4227–4232.
Tani M, Takenoshita S, Kohno T, Hagiwara K, Nagamachi Y . Infrequent mutations of the transforming growth factor beta-type II receptor gene at chromosome 3p22 in human lung cancers with chromosome 3p deletions. Carcinogenesis 1997; 18: 1119–1121.
Malkoski SP, Haeger SM, Cleaver TG et al. Loss of transforming growth factor beta type II receptor increases aggressive tumor behavior and reduces survival in lung adenocarcinoma and squamous cell carcinoma. Clin Cancer Res 2012; 18: 2173–2183.
Sato T, Kaneda A, Tsuji S et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep 2013; 3: 1911.
Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A . Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003; 4: 181–189.
Sutherland KD, Proost N, Brouns I, Adriaensen D, Song J-Y, Berns A . Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 2011; 19: 754–764.
Peifer M, Fernández-Cuesta L, Sos ML et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012; 44: 1104–1110.
Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K . EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003; 22: 5323–5335.
Kleer CG, Cao Q, Varambally S et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 2003; 100: 11606–11611.
Varambally S, Dhanasekaran SM . The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–629.
McCabe MT, Ott HM, Ganji G et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012; 492: 108–112.
Helin K, Dhanak D . Chromatin proteins and modifications as drug targets. Nature 2013; 502: 480–488.
Hoshida Y, Nijman SM, Kobayashi M et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 2009; 69: 7385–7392.
Maupin KA, Sinha A, Eugster E et al. Glycogene expression alterations associated with pancreatic cancer epithelial-mesenchymal transition in complementary model systems. PLoS ONE 2010; 5: e13002.
Hiemer SE, Szymaniak AD, Varelas X . The transcriptional regulators TAZ and YAP direct transforming growth factor β-induced tumorigenic phenotypes in breast cancer cells. J Biol Chem 2014; 289: 13461–13474.
Sriuranpong V, Borges MW, Christopher L et al. Notch signaling induces rapid degradation of Achaete-Scute Homolog 1. Mol Cell Biol 2002; 22: 3129–3139.
Imayoshi I, Kageyama R . bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron 2014; 82: 9–23.
Borges M, Linnoila RI, van de Velde HJ et al. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature 1997; 386: 852–855.
Di Sant’Agnese PA . Neuroendocrine differentiation in prostatic carcinoma: An update on recent developments. Ann Oncol 2001; 12: S135–S140.
Shida T, Furuya M, Nikaido T et al. Aberrant expression of human achaete-scute homologue gene 1 in the gastrointestinal neuroendocrine carcinomas. Clin Cancer Res 2005; 11: 450–458.
Rapa I, Ceppi P, Bollito E et al. Human ASH1 expression in prostate cancer with neuroendocrine differentiation. Mod Pathol 2008; 21: 700–707.
Isogai E, Ohira M, Ozaki T, Oba S, Nakamura Y, Nakagawara A . Oncogenic LMO3 collaborates with HEN2 to enhance neuroblastoma cell growth through transactivation of Mash1. PLoS ONE 2011; 6: e19297.
Fujiwara T, Hiramatsu M, Isagawa T et al. ASCL1-coexpression profiling but not single gene expression profiling defines lung adenocarcinomas of neuroendocrine nature with poor prognosis. Lung Cancer 2012; 75: 119–125.
Kosari F, Ida CM, Aubry M-C et al. ASCL1 and RET expression defines a clinically relevant subgroup of lung adenocarcinoma characterized by neuroendocrine differentiation. Oncogene 2014; 33: 3776–3783.
Augustyn A, Borromeo M, Wang T et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci USA 2014; 111: 14788–14793.
Castro DS, Martynoga B, Parras C et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev 2011; 25: 930–945.
Nishikawa E, Osada H, Okazaki Y et al. miR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Res 2011; 71: 6165–6173.
Koinuma D, Tsutsumi S, Kamimura N et al. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor β signaling. Mol Cell Biol 2009; 29: 172–186.
Shirai YT, Ehata S, Yashiro M, Yanagihara K, Hirakawa K, Miyazono K . Bone morphogenetic protein-2 and -4 play tumor suppressive roles in human diffuse-type gastric carcinoma. Am J Pathol 2011; 179: 2920–2930.
Ehata S, Hanyu A, Hayashi M et al. Transforming growth factor-β promotes survival of mammary carcinoma cells through induction of antiapoptotic transcription factor DEC1. Cancer Res 2007; 67: 9694–9703.
Arase M, Horiguchi K, Ehata S et al. Transforming growth factor-β-induced lncRNA-Smad7 inhibits apoptosis of mouse breast cancer JygMC(A) cells. Cancer Sci 2014; 105: 974–982.
Wieser R, Attisano L, Wrana JL, Massague J . Signaling activity of transforming growth factor β type receptors lacking specific domains in the cytoplasmic region. Mol Cell Biol 1993; 13: 7239–7247.
Kawabata KC, Ehata S, Komuro A et al. TGF-β-induced apoptosis of B-cell lymphoma Ramos cells through reduction of MS4A1/CD20. Oncogene 2013; 32: 2096–2106.
Hoshino Y, Nishida J, Katsuno Y et al. Smad4 decreases the population of pancreatic cancer-initiating cells through transcriptional repression of ALDH1A1 . Am J Pathol 2015; 185: 1457–1470.
Mizutani A, Koinuma D, Tsutsumi S et al. Cell type-specific target selection by combinatorial binding of Smad2/3 proteins and hepatocyte nuclear factor 4α in HepG2 cells. J Biol Chem 2011; 286: 29848–29860.
Morikawa M, Koinuma D, Tsutsumi S et al. ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res 2011; 39: 8712–8727.
Acknowledgements
We thank Yasuyuki Morishita and Kei Sakuma (The University of Tokyo) for technical assistance, and Hiroyuki Miyoshi (RIKEN) for providing the lentiviral vectors. This work was supported by a KAKENHI Grant-in-Aid for scientific research on Innovative Area (Integrative Research on Cancer Microenvironment Network; grant number 22112002) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, a KAKENHI Grant-in-Aid for Young Scientists (B) (grant number 22700967) from the Japan Society for the Promotion of Science (JSPS) (SE), the Strategic Basic Research Program from Japan Science and Technology Agency (KM), and a Specific Research Grant from The Cell Science Research Foundation (SE). This study was performed in part as a research program for the Project for Development of Innovative Research on Cancer Therapeutics (P-Direct), MEXT.
Author information
Authors and Affiliations
Contributions
FM, SE and KM designed the study, analyzed the data and wrote the manuscript. FM and SE performed experiments. DK assisted with and performed the ChIP-seq analysis. ASU and MF assisted with the immunohistochemical analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Murai, F., Koinuma, D., Shinozaki-Ushiku, A. et al. EZH2 promotes progression of small cell lung cancer by suppressing the TGF-β-Smad-ASCL1 pathway. Cell Discov 1, 15026 (2015). https://doi.org/10.1038/celldisc.2015.26
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/celldisc.2015.26
This article is cited by
-
The emerging role of the gut microbiome in cancer cell plasticity and therapeutic resistance
Cancer and Metastasis Reviews (2023)
-
Classical epithelial-mesenchymal transition (EMT) and alternative cell death process-driven blebbishield metastatic-witch (BMW) pathways to cancer metastasis
Signal Transduction and Targeted Therapy (2022)
-
Heterogeneity of neuroendocrine transcriptional states in metastatic small cell lung cancers and patient-derived models
Nature Communications (2022)
-
Update 2021: Management of Small Cell Lung Cancer
Lung (2021)
-
The great escape: tumour cell plasticity in resistance to targeted therapy
Nature Reviews Drug Discovery (2020)