Abstract
Missense mutations in TP53 comprise >75% of all p53 alterations in cancer, resulting in highly stabilized mutant p53 proteins that not only lose their tumor-suppressor activity, but often acquire oncogenic gain-of-functions (GOFs). GOF manifests itself in accelerated tumor onset, increased metastasis, increased drug resistance and shortened survival in patients and mice. A known prerequisite for GOF is mutant p53 protein stabilization, which itself is linked to aberrant protein conformation. However, additional determinants for mutant p53 stabilization likely exist. Here we show that in initially heterozygous mouse tumors carrying the hotspot GOF allele R248Q (p53Q/+), another necessary prerequisite for mutant p53 stabilization and GOF in vivo is loss of the remaining wild-type p53 allele, termed loss-of-heterozygosity (LOH). Thus, in mouse tumors with high frequency of p53 LOH (osteosarcomas and fibrosarcomas), we find that mutant p53 protein is stabilized (16/17 cases, 94%) and tumor onset is significantly accelerated compared with p53+/− tumors (GOF). In contrast, in mouse tumors with low frequency of p53 LOH (MMTV-Neu breast carcinomas), mutant p53 protein is not stabilized (16/20 cases, 80%) and GOF is not observed. Of note, human genomic databases (TCGA, METABRIC etc.) show a high degree of p53 LOH in all examined tumor types that carry missense p53 mutations, including sarcomas and breast carcinomas (with and without HER2 amplification). These data – while cautioning that not all genetic mouse models faithfully represent the human situation – demonstrate for the first time that p53 LOH is a critical prerequisite for missense mutant p53 stabilization and GOF in vivo.
Similar content being viewed by others
Main
Missense mutations in TP53 (mutp53) comprise >75% of all p53 alterations in cancer, resulting in highly stabilized mutant p53 proteins that not only lose their tumor-suppressor activity, but often acquire oncogenic gain-of-functions (GOFs).1, 2, 3, 4, 5 GOF activities promote cancer metabolism, stemness, and malignant progression and invasion. This results in accelerated tumor onset, increased metastasis, increased drug resistance and shortened survival in patients and mice.5, 6, 7 Accordingly, mutp53 knockin mice carrying the human hotspot missense R248Q mutation have significantly earlier tumor onset and shorter survival than p53-null mice.5 In agreement, in human patients with sporadic cancers across six major tumor entities, cancers with GOF mutp53 R282 and R248 alleles show a twofold higher hazard ratio (i.e., increased mortality) compared with cancers with loss-of-function (LOF) mutp53 alleles (nonsense, frameshift and deletion mutations).8 Similarly, germline Li–Fraumeni syndrome (LFS) patients carrying R248Q mutp53 exhibit markedly faster tumor onset by 10.5 years and higher tumor numbers per person than LFS patients carrying LOF mutp53.5
Conversely, mutp53 elimination significantly suppresses tumor growth and metastasis and markedly extends survival in various mouse models.7, 9, 10 For example, mutp53 depletion by RNAi has strong cytotoxic effects in human cancer cell lines in vitro and in xenografts.7 In allografts, knockdown of mutp53 in KrasG12D pancreatic cancer cells strongly reduces their metastatic ability.9 Finally, in a conditional inactivatable (‘floxable’) autochthonous mouse model, ablation of the R248Q knockin allele extends survival by 37%, induces regression or stagnation of advanced tumors and strongly suppresses metastasis.10
A known prerequisite for mutp53 GOF is its massive constitutive protein stabilization specifically in tumors – but not in normal cells – of knockin mice.6, 11, 12 Currently about 11 million patients worldwide live with cancers expressing highly stabilized mutp53, raising the question: what factors determine mutp53 stabilization leading to oncogenic GOF? One established determinant are the aberrant protein conformations of both the structural and DNA-contact classes of missense mutant p53 proteins, requiring constitutive chaperone complexing (with, e.g., Hsp90 and Hsp40) to protect them from their E3 ubiquitin ligases Mdm2 and CHIP and proteasomal degradation.10, 13, 14, 15, 16, 17, 18 Indeed, pharmacological inhibition of the HSP90 chaperone machinery destabilizes mutp53, leading to 48% and 59% prolonged survival in R175H and R248Q knockin mice, respectively.10 We hypothesized that besides aberrant conformation additional determinants of mutp53 stabilization likely exist. Here we show that loss of the remaining wild-type p53 (wtp53) allele, termed loss-of-heterozygosity (LOH), is also critical for missense mutp53 stabilization and GOF in vivo.
Results
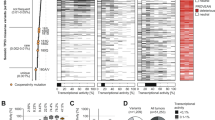
TCGA, METABRIC and other databases of sporadic human cancer show wtp53 allele loss (LOH) in the majority of missense mutp53 tumors, including ovarian cancer, breast cancer and sarcomas (Figure 1, Tables 1 and 2). Specifically, in human HER2 breast cancer with concomitant missense mutp53, wtp53 LOH occurs in 82.3% of patients (Table 1). Thus, we hypothesized that LOH is a second determinant of mutp53 stabilization and GOF in vivo.
To test this, we combined the heterozygous hotspot GOF allele R248Q (‘p53Q/+’)5, 10 with the MMTV-Neu (‘Neu’) oncogene19 expressing additional wild-type HER2 copies selectively in the mammary gland, as mutp53 has a strong prognostic value in HER2-positive breast cancer, that is, significantly increased mortality.20 Although p53Q/+;Neu mice developed breast cancer faster than p53+/+;Neu mice, surprisingly breast cancer latency between p53Q/+;Neu and p53−/+;Neu siblings was similar (Figure 2b), suggesting that mutp53 R248Q did not exert a dominant-negative (DN) effect over wtp53 but simply behaved as a LOF allele in Neu-driven breast tumorigenesis in vivo, hence the curves overlap.
Survival curves analyzing tumor onset of sarcomas and breast carcinomas in p53Q/+;Neu, p53−/+;Neu and p53+/+;Neu mouse cohorts. (a) Sarcoma onset is faster in p53Q/+;Neu compared with p53−/+;Neu mice. This indicates either a DN effect over wtp53 or, alternatively, p53 LOH resulting in mutp53 GOF specifically in sarcoma. (b) Breast cancer latency in p53Q/+;Neu and p53−/+;Neu siblings is similar, reflecting that the majority of p53Q/+ breast tumors did not undergo LOH (see Figure 3b) in contrast to human breast cancer, and also did not exert a DN effect over wtp53 but simply behaved as a LOF allele. Kaplan–Meier analysis;n, number of mice; P, log rank statistics
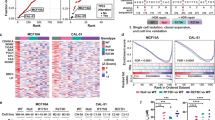
However, about half of p53Q/+;Neu and p53−/+;Neu mice did not develop breast cancer but instead developed osteosarcomas and fibrosarcomas, which originate from mesenchymal tissues where MMTV-Neu is not expressed (Figure 2a). Notably, sarcoma onset was faster in p53Q/+;Neu compared with p53−/+;Neu mice, indicating either a DN effect of mutp53 over wtp53 or, alternatively, wtp53 LOH resulting in mutp53 GOF specifically in sarcoma. Importantly, this survival difference between sarcoma and breast cancer correlated with mutp53 stabilization in nearly all examined sarcomas (94%, 16/17), but only in rare breast carcinomas (20%, 4/20), even within the same animal (Figure 3a, e.g., animal #1248).
Loss of wtp53 allele is required for missense mutant p53 stabilization and GOF. (a) The vast majority of sarcomas (16/17 cases, 94%) have stabilized mutp53. In contrast, the majority of breast carcinomas (16/20 cases, 80%) do not. Immunohistochemistry for mutp53. Mouse identity in parentheses. Arrows indicate the osteoid in osteosarcoma. (b) Analysis of wtp53 copy number in sarcomas and breast carcinomas of p53Q/+;Neu mice by quantitative genotyping. Tumors with mutp53 stabilization (all sarcomas and three breast cancers tested) have significantly higher LOH than tumors without mutp53 stabilization (majority of breast cancers). Note, as sarcomas have high normal stroma contamination (top, blue mutp53-negative stromal cells, which do not have LOH), the actual LOH in sarcomas is most likely even higher because of dilution of the tumor genotype, causing LOH underestimation. For the same reason, copy numbers of the two highest sarcoma cases (two left red bars) are likely inflated. The wtp53 signal was normalized to the Rosa26 signal. Tail biopsies from p53+/+ (two wt alleles), p53Q/+, p53−/+ (one wt allele) and p53−/− mice (no wt alleles) were used as normal control tissues without LOH. Bars represent mean±S.D. of two technical replicas of individual cases. ***P<0.001. (c) Schematic diagram of the proposed mechanism for mutp53 stabilization and GOF in heterozygous tumors. Loss of the wtp53 allele (LOH) causes accumulation of highly stabilized mutp53 protein, which triggers tumor development and is the principle mechanism and prerequisite of GOF
Thus, we asked whether sarcomas are more prone to p53 LOH than breast tumors. Indeed, qPCR of genomic DNA showed that p53 LOH occurs in all sarcomas, but rarely in breast cancer (Figure 3b). Moreover, the few breast tumors that did stabilize mutp53 also underwent p53 LOH. Together, this strongly suggests that LOH is a critical prerequisite for mutp53 stabilization and GOF (Figure 3c). To corroborate our LOH data, we analyzed p53 target genes as another readout for the remaining wtp53 allele activity (Figure 4). Indeed, all tumors with stabilized mutp53, including the single ‘outlier’ breast cancer tested, had reduced or undetectable Mdm2 and p21 levels, respectively, and sarcomas also had reduced Bax and Puma expression correlating with their LOH.
Discussion
In sum, we propose that p53 LOH is a necessary prerequisite for mutp53 stabilization and GOF activity in vivo (Figure 3c). Indeed, we find that TP53 LOH is a frequent event in human cancers with missense mutp53, including sarcomas (61%), breast cancer with or without HER2 amplification (up to 82%) and ovarian cancer (75%) (Figure 1, Tables 1 and 2). This high LOH frequency coincides with mutp53 protein stabilization21, 22 and GOF in human cancers.5, 8 Our TP53 LOH data are in agreement with earlier reports finding 60% TP53 LOH in LFS patients,23 81% in sporadic breast cancer patients (all molecular subtypes pooled)20 and 93% across 10 sporadic human cancer types,24 all expressing missense mutp53. Note that the latter study with the highest frequency includes ‘copy neutral’ TP53 LOH (defined as reduced wtp53 mRNA expression but genomic copy present) and also corrects for dilutional effects from stromal contamination.24 This suggests that conventional and even quantitative real-time PCR – which we used in our analysis – likely underestimate true functional p53 LOH.
In full agreement with the human data, sarcomas in our mouse model also exhibit GOF because they undergo LOH, which enables mutp53 stabilization. Similarly, Shetzer et al.25 found that isolated mesenchymal stem cells from heterozygous R175H/+ mice form subcutaneous tumors only after they undergo wtp53 LOH. How mechanistically p53 LOH induces mutp53 stabilization awaits further investigation. A possible contributor could be reduced expression of the wtp53 target gene Mdm2 (Figure 4), the main ubiquitin ligase for both wtp53 and mutp53.11, 16
Although a few murine breast cancer cases in our MMTV-Neu model (4/20) did undergo LOH and exhibited mutp53 stabilization, for unknown reasons the majority (16/20) lacked LOH and therefore lacked mutp53 stabilization. We speculate that the pressure for p53 LOH is eliminated because of the Neu oncogene. This gives us pause that not all mouse models faithfully mimic the human genetic constellation for every tissue type, as the MMTV-Neu model contrasts with human breast cancer, which exhibit prominent LOH despite the presence of other oncogenic drivers (Figure 1a, Table 1).20
Materials and methods
TP53 LOH analysis in sporadic human cancers
TP53 LOH in sporadic human cancers was analyzed using the cBioPortal tool (www.cbioportal.org). The breast cancer data set included METABRIC and provisional TCGA databases, with 3014 samples with known mutant p53 status in total. The sarcoma data set included provisional TCGA, MSKCC/Broad Institute, Institut Curie and other databases, with 710 samples with known mutp53 status in total.
Animals
Hotspot knockin mice harboring human exons 4–9 and the p53 R248Q missense mutation (‘Q’ allele) and p53−/− mice, both on pure C57Bl6 background, were previously described.5, 10 MMTV-Neu (‘Neu’) transgenic mice on pure FVBN background were from Jackson Laboratories (Bar Harbor, ME, USA) (FVB/N-Tg(MMTVneu)202Mul/J).19 To obtain p53Q/+;Neu/+ and control p53+/−;Neu/+ mice, parental p53 R248Q/+ and p53−/+ strains were first crossed to obtain p53 R248Q/- mice, followed by cross with Neu/Neu mice. Only female mice were used for all experiments. Animals were monitored weekly to determine their breast cancer and sarcoma onset and were promptly killed when their tumors reached 2 cm3 in volume or when animals appeared moribund. All animals were treated humanely and according to the guidelines issued by the Institutional Animal Care and Use Committee (IACUC) at Stony Brook University.
Immunohistochemistry and histology
For immunohistochemical analysis, freshly dissected tissues were formalin fixed, paraffin embedded and sectioned (5 μm). Slides were deparaffinized and boiled in citrate buffer (10 mM, pH 6.0, 35 min) for antigen retrieval, blocked in 10% goat serum and incubated with the primary antibody (mutp53, Santa Cruz, Dallas, TX, USA, FL393, sc-6243, dilution 1:500) for 2 h at room temperature. After PBS washing, slides were incubated with biotinylated secondary antibody and HRP-streptavidin using the Histostain SP Broad Spectrum kit (Invitrogen, Carlsbad, CA, USA, 959943B), stained with DAB substrate with hematoxylin counterstain and coverslipped. In addition, cancer type (breast cancer versus osteosarcoma or fibrosarcoma) was determined by H&E staining (data not shown).
Quantitative LOH analysis
Genomic DNA was isolated from sarcomas, breast carcinomas and control tails using DNeasy Blood and Tissue kit (Qiagen, 69506) and quantified by spectrophotometer. Quantitative real-time PCR was performed in duplicates with QuantiTect SYBR Green Mix (Qiagen, Germantown, MD, USA, 204143) on the MJ Research DNA Engine Opticon 2 machine, using 8 ng genomic DNA and the following mouse wtp53 allele-specific primer pairs: 5′-ACAGCGTGGTGGTACCTTAT-3′ (forward) and 5′-TATACTCAGAGCCGGCCT-3′ (reverse). These wtp53 primers anneal to mouse exons 5 and 6 and do not recognize the humanized mutp53 allele. For all samples, the wtp53 signal was normalized to the Rosa26 signal measured by the following primers: 5′-AAAGTCGCTCTGAGTTGTTAT-3′ (forward) and 5′-GGAGCGGGAGAAATGGATATG-3′ (reverse).
Statistical analysis
Kaplan–Meier analysis and log rank statistics was used to analyze tumor onset. Unpaired two-tailed Student’s t-test was used to analyze p53 LOH and expression of p53 target genes. *P<0.05, ***P<0.001.
References
Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004; 119: 861–872.
Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004; 119: 847–860.
Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA 2010; 107: 246–251.
Doyle B, Morton JP, Delaney DW, Ridgway RA, Wilkins JA, Sansom OJ . p53 mutation and loss have different effects on tumourigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma. J Pathol 2010; 222: 129–137.
Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U . Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ 2013; 20: 898–909.
Brosh R, Rotter V . When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713.
Muller PA, Vousden KH . Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014; 25: 304–317.
Xu J, Wang J, Hu Y, Qian J, Xu B, Chen H et al. Unequal prognostic potentials of p53 gain-of-function mutations in human cancers associate with drug-metabolizing activity. Cell Death Dis 2014; 5: e1108.
Weissmueller S, Manchado E, Saborowski M, Morris JP 4th, Wagenblast E, Davis CA et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell 2014; 157: 382–394.
Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 2015; 523: 352–356.
Terzian T, Suh Y-A, Iwakuma T, Post SM, Neumann M, Lang GA et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev 2008; 22: 1337–1344.
Suh YA, Post SM, Elizondo-Fraire AC, Maccio DR, Jackson JG, El-Naggar AK et al. Multiple stress signals activate mutant p53 in vivo. Cancer Res 2011; 71: 7168–7175.
Blagosklonny MV, Toretsky J, Bohen S, Neckers L . Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc Natl Acad Sci USA 1996; 93: 8379–8383.
Whitesell L, Sutphin PD, Pulcini EJ, Martinez JD, Cook PH . The physical association of multiple molecular chaperone proteins with mutant p53 is altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol 1998; 18: 1517–1524.
Li D, Marchenko ND, Moll UM . SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ 2011; 18: 1904–1913.
Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res 2011; 9: 577–588.
Parrales A, Ranjan A, Iyer SV, Padhye S, Weir SJ, Roy A, Iwakuma T . DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat Cell Biol 2016; 18: 1233–1243.
Alexandrova EM, Moll UM . Depleting stabilized GOF mutant p53 proteins by inhibiting molecular folding chaperones: a new promise in cancer therapy. Cell Death Differ 2017; 24: 3–5.
Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ . Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA 1992; 89: 10578–10582.
Silwal-Pandit L, Vollan HK, Chin SF, Rueda OM, McKinney S, Osako T et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res 2014; 20: 3569–3580.
Yemelyanova A, Vang R, Kshirsagar M, Lu D, Marks MA, Shih IM et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol 2011; 24: 1248–1253.
Köbel M, Piskorz AM, Lee S, Lui S, LePage C, Marass F et al. Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J Pathol Clin Res 2016; 2: 247–258.
Varley JM, Evans DG, Birch JM . Li-Fraumeni syndrome—a molecular and clinical review. Br J Cancer 1997; 76: 1–14.
Parikh N, Hilsenbeck S, Creighton CJ, Dayaram T, Shuck R, Shinbrot E et al. Effects of TP53 mutational status on gene expression patterns across 10 human cancer types. J Pathol 2014; 232: 522–533.
Shetzer Y, Kagan S, Koifman G, Sarig R, Kogan-Sakin I, Charni M et al. The onset of p53 loss of heterozygosity is differentially induced in various stem cell types and may involve the loss of either allele. Cell Death Differ 2014; 21: 1419–1431.
Acknowledgements
This work was supported by NCI grant R01CA176647, TRO grant Walk for Beauty (Stony Brook Foundation) and Wilhelm Sander Stiftung grant 2011.029.1 to UMM, NCI grant K22CA190653-01A1 to EMA, and DOD grant W81XWH-16-1-0448 (BC151569) to NDM.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Alexandrova, E., Mirza, S., Xu, S. et al. p53 loss-of-heterozygosity is a necessary prerequisite for mutant p53 stabilization and gain-of-function in vivo. Cell Death Dis 8, e2661 (2017). https://doi.org/10.1038/cddis.2017.80
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2017.80
This article is cited by
-
P53 loss of heterozygosity (LOH) in formalin-fixed paraffin-embedded leiomyosarcoma (LMS): a novel report
Irish Journal of Medical Science (1971 -) (2024)
-
Multidimensional quantitative phenotypic and molecular analysis reveals neomorphic behaviors of p53 missense mutants
npj Breast Cancer (2023)
-
Modelling aggressive prostate cancers of young men in immune-competent mice, driven by isogenic Trp53 alterations and Pten loss
Cell Death & Disease (2022)
-
Is loss of p53 a driver of ductal carcinoma in situ progression?
British Journal of Cancer (2022)
-
Differences in somatic TP53 mutation type in breast tumors by race and receptor status
Breast Cancer Research and Treatment (2022)