Abstract
STAT1, which carries tumor suppressor functions in several models, consists of two isoforms, namely STAT1α and STAT1β. The biological function and significance of STAT1β has never been examined in human cancer. We examined STAT1β function in esophageal squamous cell carcinoma (ESCC) by transfecting a STAT1β gene into various ESCC cell lines. The interaction between STAT1α and STAT1β was examined by using co-immunoprecipitation and confocal microscopy. The prognostic significance of STAT1β expression, detectable by immunohistochemistry and western blot, was evaluated in a large cohort of ESCC patients. Enforced expression of STAT1β induced and prolonged the expression and phosphorylation of STAT1α in ESCC cells, and these effects were amplified by gamma-interferon (IFN-γ). We also found that STAT1β interacts with STAT1α and decreases STAT1α degradation by the proteasome. Moreover, STAT1β substantially increased the DNA binding and transcription activity of STAT1. STAT1β also sensitized ESCC cells to chemotherapeutic agents, including cisplatin and 5-flurouracil. Using western blot and immunohistochemistry, we found that STAT1β was frequently decreased in esophageal cancer, as compared to their adjacent benign esophageal epithelial tissue. Loss of STAT1β significantly correlated with lymph node metastasis, invasion and shorter overall survival in ESCC patients. Therefore, STAT1β plays a key role in enhancing the tumor suppressor function of STAT1α, in ESCC, in a manner that can be amplified by IFN-γ.
Similar content being viewed by others
Main
Signal transducer and activator of transcription 1 (STAT1), a key mediator of interferon (IFN) signaling, regulates a variety of cellular activities, such as apoptosis, proliferation and differentiation.1 In response to extracellular stimuli, such as IFN-γ, activation of STAT1 is achieved by Janus kinase-mediated phosphorylation of its conserved tyrosine and serine residues, present in the C-terminal transactivation domain, to result in STAT1 dimerization, nuclear translocation, DNA binding and eventually modulation of expression of its target genes.2 In a number of models, STAT1 has been shown to possess tumor suppressor functions, and the evidence can be summarized as follows: (1) pro-apoptotic effects are largely mediated through STAT1 signaling,2 (2) constitutively active STAT1 can effectively induce apoptosis and inhibit cell growth,3 (3) STAT1 is frequently downregulated in various human cancers, including breast cancer, head and neck cancer, multiple myeloma and leukemia.4, 5 In our previous studies, we have reported that STAT1 is an important tumor suppressor in esophageal squamous cell carcinoma (ESCC), where loss of STAT1 contributes to the pathogenesis of these tumors and correlates with a worse clinical outcome.6, 7
STAT1β, a naturally spliced isoform of STAT1, lacks a 38-amino acid segment that includes the conserved STAT1S727 phosphorylation site and most of the C-terminal transactivation domain. STAT1β has not been extensively studied, although one report has described that STAT1β in human B-cells is transcriptionally inactive and exerts a dominant-negative effect on STAT1α, the full-length STAT1 isoform.8 Specifically, overexpression of STAT1β was found to inhibit the phosphorylation, DNA binding and transcriptional activity of STAT1 in human B-cells.8 However, in another study using B-cells, STAT1β was reported to induce cell death via a mechanism that is independent of p53 and STAT1α.9 In a more recently published paper, STAT1β was found to be transcriptionally active and capable of eliciting IFN-γ-dependent immunity against infection in vivo.10 Nonetheless, the biological function and clinical significance of STAT1β in human cancers has never been examined, and whether STAT1β possesses tumor suppressor activity is unknown.
Using ESCC cell lines as a model, we examined the biological and clinical significance of STAT1β. Our results support the concept that STAT1β enhances the expression and tumor suppressor function of STAT1α, and this effect can be amplified by IFN-γ stimulation. within support of this concept, loss of STAT1β in ESCC tumors correlates with a significantly worse clinical outcome.
Results
STAT1β increases expression and tyrosine phosphorylation of STAT1α
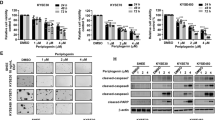
Both ESCC cell lines (EC1 and KYSE150) had no detectable expression of STAT1 or p-STAT1Y701 after the transfection of an empty vector (Figure 1a, left). At 12 h after the addition of varying doses of IFN-γ, there was also no appreciable change in the expression of STAT1 and p-STAT1Y701. In contrast, cells transfected with STAT1β showed detectable STAT1α expression (Figure 1a, right). At 12 h after the addition of various amounts of IFN-γ, the total STAT1α as well as p-STAT1Y701 (containing both STAT1α and STAT1β) levels increased in a dose-dependent manner. Time course experiments revealed similar results. As shown in the middle panel of Figure 1b, following STAT1β transfection, IFN-γ addition resulted in a rapid and dramatic increase in STAT1α as well as p-STAT1Y701. Importantly, the enhancement of p-STAT1α by STAT1β was almost as potent and sustained as by STAT1α transfection.
STAT1β increases and prolongs tyrosine 701 phosphorylation of STAT1α. (a) ESCC cell lines EC1 and KYSE150 were stimulated with IFN-γ, at the doses indicated, or left untreated (w/o) after empty vector or Flag-tagged STAT1β transfection. Total-protein extracts were used for detection of Tyr701-phosphorylated and total STAT1, flag and STAT1α by western blotting. (b) Both cell lines were stimulated with IFN-γ (10 ng/ml) for the time indicated, or left untreated (w/o) after empty vector or Flag-tagged STAT1β and STAT1α transfection. (c) EC1 and KYSE150 cells were stimulated with IFN-γ (10 ng/ml) for the indicated times after empty vector or Flag-tagged STAT1β and STAT1α transfection. Phospho-STAT1 was detected with an Alexa Fluor 568-conjugated secondary antibody (red). DAPI (1 μg/ml) was used for nuclear staining (blue). Fluorescence signals were analyzed with a Zeiss LSM 710 confocal microscope (scale bar 5 μm). (d) Tyrosine phosphorylation of STAT1α in EC1 cells was detected by immunoprecipitation and western blotting, after empty vector or Flag-tagged STAT1β or STAT1βY701F plasmid transfection, upon IFN-γ stimulation. Data are representative of three independent experiments
To further substantiate our finding that STAT1β increases the expression and phosphorylation of STAT1α, we performed confocal microscopy using EC1 and KYSE150. No nuclear p-STAT1Y701 was detectable at 1 h or 24 h after IFN-γ stimulation (Figure 1c). In contrast, in STAT1β-transfected cells, a strong nuclear p-STAT1Y701 signal was detectable at 1 h, after IFN-γ addition, then declined by 24 h. Similar findings were found following STAT1α transfection, although the p-STAT1Y701 signal was slightly more intense than that resulting from STAT1β transfection. These results correlated well with the western blot results illustrated in Figure 1b.
We then performed immunoprecipitation. Tyrosine phosphorylation of STAT1α in EC1 cells was increased in the presence of STAT1β, but not the empty vector (Figure 1d). Importantly, tyrosine phosphorylation of STAT1α was largely abrogated when the STAT1βY701F mutant, instead of wild-type STAT1β, was used for transfection. This finding highlights the importance of the activation/phosphorylation of STAT1β in potentiating the expression and phosphorylation of STAT1α.
STAT1β interacts with STAT1α and protects STAT1α from proteasome degradation
To investigate the mechanisms by which STAT1β enhances STAT1α expression and phosphorylation, we asked if STAT1β increased the expression of STAT1α mRNA. By quantitative RT-PCR, we found a significant decrease in STAT1α mRNA after STAT1β transfection in both ESCC cell lines, whereas transfection of the STAT1βY701F mutant did not have any appreciable effect (Figure 2a). Since the relatively low STAT1 expression in ESCC can be attributed to its degradation via the ubiquitin-proteasome pathway (manuscript submitted), we tested if this pathway is also involved in the upregulation of STAT1α mediated by STAT1β. In keeping this hypothesis, we performed immunoprecipitation using an antibody against STAT1α. As shown in Figure 2b, we found that transfection of STAT1β almost completely abrogated STAT1α ubiquitination. Consistent with our previous data, the Y701F mutation of STAT1β lacked this effect.
STAT1β interacts with STAT1α to protect STAT1α from proteasome degradation. (a) STAT1α mRNA expression was detected by real-time PCR after transfection of empty vector or Flag-tagged STAT1β or STAT1βY701F plasmids. Values were normalized to GAPDH and calculated relative to empty vector-transfected cells. Mean values and standard errors (SE) from at least three independent experiments are shown. (b) Immunoprecipitation–immunoblotting analysis was performed for STAT1α and ubiquitination in EC1 cells transfected with empty vector or Flag-tagged STAT1β or STAT1βY701F plasmids. (c) The interaction of STAT1α and STAT1β was investigated by immunoprecipitation and western blot analysis in EC1 cells with or without IFN-γ stimulation. Co-immunoprecipitaion was carried out with control IgG and anti-Flag or anti-STAT1α antibodies as indicated. Immunoprecipitated proteins were analyzed by western blot with anti-STAT1α and anti-Flag, respectively. (d) Co-localization of STAT1α and STAT1β in vivo. EC1 and KYSE150 cells were placed on coverslips and stained with the indicated antibodies (scale bar 5 μm)
To further investigate the relationship between STAT1α and STAT1β, we performed co-immunoprecipitation and western blot experiments. By co-immunoprecipitation experiments (Figure 2c), in the absence of IFN-γ (lanes 1 and 2), we found evidence of physical binding between Flag-tagged STAT1β and STAT1α in KYSE150 cells. This effect was amplified when IFN-γ was added (Figure 2c, lanes 3 and 4). The physical interaction between these two STAT1 isoforms was further supported by our confocal microscopy results (Figure 2d). In contrast, there was no detectable physical binding between Flag-tagged STAT1βY701F. Taken together, our results suggest a model in which STAT1β protects STAT1α from proteosomal degradation, thus increasing the total protein level of STAT1α.
STAT1β enhances DNA binding and transcription activity of STAT1α
Using a STAT1 luciferase reporter, we assessed the effect of STAT1β on the transcriptional activity of STAT1 in ESCC cell lines. As shown in Figure 3a, STAT1β transfection significantly increased the transcriptional activity of STAT1, as compared to that of empty vector (P<0.05). Again, transfection of the STAT1βY701F mutant yielded only a minimal increase, as compared to empty vector (P=0.71). Similar observations were observed when transfected cells were stimulated with IFN-γ, although the effects were more profound.
STAT1β enhances transcription activity and DNA binding of STAT1. (a) EC1 and KYSE150 cell lines were transfected with either empty vector, or Flag-tagged STAT1β or STAT1βY701F plasmids, then stimulated with IFN-γ. After 48 h, the transcription activity of STAT1 was detected by a dual-luciferase reporter assay. (b) After transfection with STAT1β or STAT1βY701F plasmids, the DNA binding ability of STAT1 was detected, in both cell lines, with a STAT1 biotin-probe followed by western blotting. (c) Total RNA was extracted and used for RT-qPCR analysis for the genes indicated. GAPDH was used for normalization, and expression levels were calculated relative to empty vector-transfected cells. Triplicate experiments were performed and results from a representative experiment are shown. *P< 0.05; **P<0.01
To test whether the increased transcriptional activity of STAT1 mediated by STAT1β transfection was caused by an increase in STAT1-DNA binding, we performed pull-down experiments using a biotinylated probe containing the STAT1 DNA-binding consensus sequence. As shown in Figure 3b, upon IFN-γ stimulation, STAT1-DNA was markedly enhanced by STAT1β transfection, as compared to that of empty vector or STAT1βY701F.
To confirm the effect of STAT1β on gene transcription, we performed quantitative RT-PCR. ESCC cells were transiently transfected with either empty vector, wild-type STAT1β or STAT1βY701F, then the mRNA expression levels of several known STAT1 downstream targets (including IRF1, TAP1, CXCL10, GBP2 and ICAM10) were analyzed.11 Compared to transfection with empty vector, STAT1β transfection at 24 h significantly increased the mRNA expression of all five target genes examined (Figure 3c). As above, enforced expression of the STAT1Y701F mutant did not appreciably increase the expression of these five genes.
STAT1β enhances the tumor suppressor function of STAT1α
To evaluate the biological effect of STAT1β in ESCC cells, we performed colony formation assays using EC1 and KYSE150 cells. Transfection of STAT1β significantly decreased soft-agar colony formation of both cell lines (P<0.05), whereas STAT1βY701F did not show similar tumor suppressor effects (Figure 4a). To support the functional role of STAT1β, we knocked down STAT1 and then measured soft agar colony formation in the presence of IFN-γ (1 ng/ml) (Figure 4b). Upon STAT1 siRNA knockdown, both EC1 and KYSE150 cells showed a significant increase in the number of colonies. However, enforced STAT1β expression significantly attenuated the biological effect of STAT1 siRNA knockdown (lane 3). Furthermore, enforced expression of STAT1β in the absence of STAT1 siRNA knockdown (lane 4) brought the number of colonies to the lowest level in both cell lines.
Biological functions of STAT1β in ESCC. (a) Tumorigenicity, assessed by colony formation assay, was detected in EC1 and KYSE150 cells, transfected with STAT1β, STAT1βY701F or empty vector, at different doses of IFN-γ stimulation after 10 days. (b) In both cell lines, colony formation was performed in the cells with siRNA knockdown of STAT1 and transfection of STAT1β. (c) Using western blot analysis, cleaved and total PARP expression was detected in EC1 and KYSE150 cells transfected with STAT1β or empty vector. (d) The chemosensitivity to 5-FU and cisplatin of ESCC cells was assessed by colony formation after transfection of STAT1β for 10 days. (e) Cell growth, as assessed by MTS assay, was detected after STAT1β transfection in EC1 and KYSE150 cells after a 4-day exposure to IFN-γ. Triplicate experiments were performed and results from a representative experiment are shown (*P<0.05)
Western blotting revealed that transfection of STAT1β led to the expression of cleaved poly-ADP ribose polymerase (PARP) in ESCC cells stimulated with IFN-γ (Figure 4c). Also, STAT1β transfection synergized with 5-fluorouracil (5-FU) and cisplatin in decreasing colony formation (Figure 4d), as well as the number of viable cells in an MTS assay (Figure 4e).
Expression of STAT1β in ESCC tumors and its prognostic significance
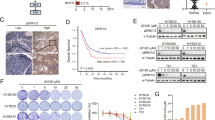
By western blot, we detected the expression of p-STAT1Y701 and total STAT1 in 12 ESCC tumors and their case-matched, adjacent normal tissues. As illustrated in Figure 5a, all 12 cases showed low levels of expression of the STAT1α isoform and p-STAT1αY701 (the upper bands). Most (8/12, 66.6%) tumors showed lower expression of p-STAT1βY701 and STAT1β (the lower bands); only (16.7%) cases showed a slight increase and another 2 (16.7%) cases showed no appreciable difference.
Expression of STAT1β in ESCC patient samples. (a). STAT1β expression in ESCC tumors was examined by western blot. Compared to benign esophageal tissue harvested at the surgical margins in the same specimens (labeled N), cancerous tissue (labeled Ca) often expressed a lower level of STAT1β (e.g. cases 1–3). A small subset of tumors (e.g. case 4) had high levels of STAT1β. (b) Immunohistochemistry of formalin-fixed, paraffin-embedded tissues showed variable levels of predominantly cytoplasmic STAT1β were detectable in esophageal epithelial and ESCC tissues. Based on the staining intensity, normal epithelia and tumors in our cohort were categorized into STAT1-negative, STAT1-weak or STAT1-strong (IHC stain, scale bar, 20 μm). (c) The immunohistochemistry scores of STAT1β showed that expression of STAT1β is higher in normal tissues compared to cancer tissues. (d) By Kaplan–Meier analysis, a significant correlation between overall survival and the expression level of STAT1β was found between patients with STAT1β-strong and STAT1β-weak/negative staining (*P<0.05)
We then performed immunohistochemistry (IHC) using 33 ESCC tumors and the case-matched, adjacent normal tissues. As illustrated in Figures 5b and c, STAT1β immunoreactivity was found in 29 (87.9%) cases of benign esophageal epithelial tissues, but only 19 (57.6%) of ESCC tumors. Using our IHC scoring method, we found that ESCC tumors had lower STAT1β expression than the benign epithelia, in keeping with the concept that STAT1β is a tumor suppressor and its expression is frequently lost during the carcinogenesis of ESCC.
In view of its tumor suppressor function, the prognostic significance of STAT1β was evaluated in a cohort of 201 ESCC tumors. We found that STAT1β was undetectable in 94 (46.8%) cases, weakly expressed in 47 (23.3%), and strongly expressed in 60 (29.9%) cases. By comparing the STAT1β staining intensity (negative/weak or strong) and total STAT1 staining obtained from one of our previous studies,6 we found a positive correlation (R=0.765, P<0.0001, illustrated in Table 1). The correlation of STAT1β and various clinical parameters was also assessed (Table 2), Negative/weak STAT1β staining significantly correlated with depth of invasion (P<0.001), lymph node metastasis (P=0.045) and a high clinical stage (P=0.026). Clinical follow-up data were available in 130 of 201 cases included in this study (median follow-up, 21.5 months, and range 5–92 months). The prognostic value of STAT1β was analyzed by Kaplan–Meier survival analysis and the cut-point was determined by X-tile software. As shown in Figure 5d, patients with expression of total STAT1βstrong (n=43) had a significantly better clinical outcome compared to the STAT1βnegative/weak (n=87, P=0.025). STAT1strong/STAT1βstrong patients (n=27) had longer survival than both the STAT1strong/STAT1βnegative/weak patients (n=12) and STAT1negative/weak/STAT1βnegative/weak patients (n=35) (Supplementary Figure 1).
Discussion
We previously demonstrated that STAT1 is a tumor suppressor in ESCC.6, 7 Similar to STAT3 and STAT4, STAT1 has two isoforms, namely STAT1α and STAT1β. STAT1α is the full-length isoform and is considered to be the transcriptionally active form of STAT1, which is also known to form complexes with other transcription factors to modulate gene expression in normal cells.10 STAT1α has been reported to mediate various cellular activities, including inhibition of cell growth, cell cycle arrest and apoptosis.2 In comparison, the function of STAT1β has not been extensively studied. STAT1β is the truncated form of STAT1 and lacks a 38-amino acid segment that includes most of the transactivation domain and the functionally important serine 727.12 Corresponding to its truncated structure, it has been shown that STAT1β can bind to the promoter of genes such as IRF1, LMP2 and TAP1, but is transcriptionally inactive.13, 14
As mentioned previously, STAT1β is believed to exert dominant negative effect on STAT1α, due to its lack of the transactivation domain and regulatory serine 727 phosphorylation site. This concept came from a study of human B cells, in which enforced expression of STAT1β was found to inhibit STAT1α activation by decreasing the tyrosine 701 phosphorylation, DNA binding and transcriptional activity of STAT1α, as well as protecting the cells from fludarabine-induced apoptosis.8 In keeping with this concept, infection with Mycobacterium tuberculosis and Leishmania Mexicana has been found to increase the expression and phosphorylation of STAT1β, leading to an inhibition of STAT1 signaling.15, 16 In another study, there is evidence that the ratio of STAT1α/STAT1β may affect the sensitivity of cells to viral infection.17 The mechanism of how STAT1β inhibits STAT1α is incompletely understood. One possible explanation is that STAT1β competes with STAT1α for the same DNA binding sites, but is incapable of activating the gene expression.8, 15, 16 Another possible explanation is that STAT1β may compete with STAT1α for the same receptor sites, thus interfering with STAT1α activation.
Results from a more recent study challenge the view that STAT1β is simply an inhibitor of STAT1α. Specifically, STAT1β has been found to induce death in human B cells, independently of p53 or STAT1α.9 In a recently published paper, it was reported that STAT1β is transcriptionally activated in response to IFN-γ, and IFN-γ-induced tyrosine phosphorylation and promoter binding of STAT1 is prolonged in the absence of STAT1α.10 In the same study, it also was reported that STAT1β can induce the expression of many gene targets of STAT1 upon IFN-γ stimulation.10
Our findings from this current study also challenge the view that STAT1β is an inhibitor of STAT1α. We find that STAT1β can increase the expression of STAT1α and potentiate its activation/phosphorylation mediated by IFN-γ. Indeed, we found that STAT1β substantially enhances the DNA binding and transcription activity of STAT1α in ESCC cells and STAT1β can modulate the expression of known STAT1 gene targets, including IRF1, TAP1 and GPB2. Probably through effects on STAT1α, STAT1β exerts tumor suppressor effects in ESCC. It is clear that our findings, and others, contradict previous studies regarding STAT1β, as discussed above. While the explanations for this discrepancy are not clear, we consider that cell-type specificity is likely a contributing factor. Researchers also demonstrated prolonged phosphorylation of STAT1 after STAT1β transfection, and that this effect is related to a reduction of SOCS1,10 which is a negative regulator of the JAK-STAT1 pathway.18 However, in our study, we did not find a change in SOCS1 expression by ESCC cells after STAT1β transfection.
Our findings also lead us to believe that the key mechanisms underlying the effects of STAT1β on STAT1α are related to its binding to STAT1α, thereby protecting it from being degraded via the proteasomal pathway. In support of the concept that STAT1β potentiates the tumor suppressor effect of STAT1α, we find that loss of STAT1β expression significantly correlates with a worse clinical outcome in a large cohort of ESCC patients. Our hypothetical model is summarized in Figure 6.
Schematic diagram showing STAT1β interacts with STAT1α. Upon IFN-γ stimulation, STAT1 can form three different dimers: α:α and β:β homodimers and α:β heterdimers. Dimers can be phosphorylated followed by translocation into the nucleus. Once STAT1 is released from the target gene promoter, it returns to the cytoplasm and is degraded by the proteasome. However, the STAT1β can bind to STAT1α to protect STAT1α from degradation, resulting in prolongation of the half-life of STAT1α and a concomitant increase in transcription activity
Similar to STAT1β, both STAT3β and STAT4β have been reported to be transcriptionally active and able to carry their unique functions.19 There are at least two mechanisms underlying the transcription activity and biological function of STAT1β. One mechanism is that the beta isoforms of STATs can interact with other transcription factors that provide a C-terminal transactivation domain. For example, STAT3β can cooperate with c-Jun to activate the α-macroglobulin promoter.20 Thus, gene transfection of STAT3β results in increasing STAT3α transcription activity.21, 22 Another mechanism is that STAT1β still retains the most important functional domain. For example, the abbreviated transactivation domain contains the tyrosine 701 phosphorylation site that is essential for various STAT1 functions. The N-terminal domain can interact with CBP, with low affinity, to exert low-level transcriptional activation,22, 23 and the SH2 domains can participate in dimerization of STAT1 to bind to receptors.24 Interaction of the coiled-coil domain and the DNA binding domain can form antiparallel STAT1 dimers.25 Although STAT1β lacks the serine 727 phosphorylation site, it is likely that STAT1β can mediate some degree of transcription activity of STAT1, since serine 727 has been reported to be necessary for the maximal activation of STAT1.22
The biological function and clinical significance of STAT1β has yet to be reported in human cancers. Moreover, no paper has described the expression of STAT1β in human normal or cancer tissues Therefore, in our paper, we firstly used ESCC cells as a model to determine the biological function of STAT1β. We show cells transfected with STAT1β are more sensitive to IFN-γ and anti-tumor drugs, such as 5-FU and cisplatin. These findings suggest that, in ESCC, STAT1β also exerts a tumor suppressor function. Analysis of ESCC patient samples also supports our hypothesis. In a previous paper, we found that expression of STAT1β is significantly higher in immortalized esophageal cell lines than in ESCC cell lines, indicating that STAT1β is downregulated in ESCC tumorigenesis.6 We further show that STAT1β is downregulated in ESCC tissues compared to case-matched normal esophageal epithelia, which is similar to our previous findings on STAT1 in ESCC. STAT1β expression is strongly related to the total expression of STAT1, which supports our hypothesis that STAT1β may stabilize STAT1α to modulate and increase total STAT1 expression. Another key finding of our study is that low STAT1β expression correlates with greater tumor invasion and lymph node metastasis, and worse clinical outcome.
In conclusion, we investigated the biological function and clinical significance of STAT1β in human cancer. In ESCC cells, STAT1β enhances the tumor suppressor function of STAT1 by increasing the expression and activation of STAT1α. The in vivo results also indicate that STAT1β is downregulated in the ESCC carcinogenesis, and correlates with worse clinical outcome. Our study challenges the concept that STAT1β is simply a dominant negative regulator of STAT1 and provides a new therapeutic target for treating ESCC patients.
Materials and methods
Cells and patient samples
Human ESCC cell lines, EC1 and KYSE 150, were used in this study. Cells were maintained in RPMI 1640 (KYSE150), or Dulbecco’s modified Eagle’s medium supplemented (EC1) with 10% fetal bovine serum, and 1 × antibiotic mixture (Invitrogen, Carlsbad, CA, USA). All cells were cultured at 37 °C in a humidified incubator containing 5% CO2.
We randomly collected 201 consecutive ESCC samples at the Shantou Tumor Hospital between 2005 and 2012. All patients underwent potentially curative surgery without preoperative chemotherapy or radiotherapy. In this cohort, 150 were men and 51 were women; the range of ages was 36–81 years, with a median of 57 years. Follow-up data were available for 130 patients; most (113, 86.9%) died during the follow-up period (median survival, 21.5 months). Of the 201 ESCC tumors, 33 case-matched normal esophageal tissues adjacent to the tumors were included in the study. Written informed consents were obtained from the patients, and the study was reviewed and approved by the institutional ethics committee of Shantou University Medical College.
Western blot analysis
A tissue lyser (Qiagen, Valencia, CA, USA) was used to prepare frozen tumor samples for western blot analysis. Cell lines and tumor samples were lysed in RIPA buffer containing protease inhibitor cocktail and Set II phosphatase inhibitor cocktail. Total tissue extracts were stored on ice for 20 min and then centrifuged at 13 000 r.p.m. at 4 °C for 15 min. Proteins from the supernatant were measured using a bicinchoninic acid assay (Thermo scientific, Rockford, IL, USA). Equal amounts of cell lysate were separated by 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis and evaluated by western blot analysis as described previously.26 Antibodies reactive with human β-actin (1:1000), STAT1 (1:1000), phospho-Tyr 701 STAT1 (or p-STAT1) (1:1000), Flag (1:1000), and PARP (1:1000) were purchased from Cell Signalling (Danvers, MA, USA). Anti-STAT1α (1:1000) was purchased from Santa Cruz (Dallas, TX, USA), and anti-STAT1β (1:200) was purchased from SignalChem (Richmond, BC, Canada).
Cell growth assay
The ESCC cell lines were transfected with STAT1β, mutant STAT1βY701A or empty vector. Then, 1 × 104 transfected cells were grown in each well of a 96-well microplate and treated with anti-tumor drug, either cisplatin or 5-flurouracil (5-FU), for 0–8 days. At different times, the cells were incubated with 10 μl MTS reagent (Invitrogen) for 1 h, and the increase in absorbance at 490 nm relative to the blank well control was measured using a microplate spectrophotometer.
Plasmid constructs and gene transfection
Plasmids including Flag-STAT1β and STAT1α were purchased from Addgene (Cambridge, MA, USA). Mutant STAT1βY701F plasmid was engineered in our laboratory. For each experiment, 1 × 106 ESCC cells were transiently transfected with 10 μg of STAT1-expressing plasmid or empty vector, in six-well plates, using the lipofectamine 2000 reagent (Invitrogen) as per the manufacturer’s suggested protocol.
Co-immunoprecipitation of STAT1α and STAT1β
To detect the interaction between STAT1α and STAT1β, whole-cell extracts were prepared by lysing the cells in an immunoprecipitation buffer. A total of 2 μg of anti-STAT1α monoclonal antibody (Santa Cruz Biotechnology, Dallas, TX, USA) was added to 500 μg of protein lysate isolated in cell lysis buffer (Sigma Aldrich, St Louis, MD, USA) and the samples were rotated overnight at 4 oC. Subsequently, 30 μl of protein G Plus/A beads (EMD Millipore, Billerica, MA, USA) was added to the samples and rocked overnight at 4 oC. The beads were then washed three times with cold phosphate-buffered saline (PBS), followed by a final wash using cold cell lysis buffer or RIPA buffer. Western blot analysis was then performed using standard techniques as previously described.26
Immunofluorescence and confocal microscopy
Cells were grown on cover slips coated with poly-L-lysine (Sigma Aldrich) in a six-well plate and fixed with 3% paraformaldehyde in PBS (pH 7.4). Cells were rinsed three times with PBS, permeabilized with Triton X-100, washed again with PBS, and incubated with 200 μl of anti-p-STAT1 antibody (1:50, Sigma Aldrich) overnight at room temperature in a humidified chamber. The cover slips were rinsed three times in PBS and incubated with secondary antibody conjugated with Alexa Fluor 488 or 568 (Invitrogen) at a 1:250 dilution for 1 h at room temperature. After three rinses in PBS, the coverslips were mounted on a slide using mounting media (Dako, Mississauga, ON, Canada). Cells were visualized with a Zeiss LSM 710 confocal microscope at the Core Cell Imaging Facility, Cross Cancer Institute (Edmonton, AB, Canada).
Colony formation assay
After STAT1β or empty vector transfection, cells were inoculated in six-well plates, at a density of 500 cells/well and incubated for 10 days at 37 °C. The cells were fixed with 4% buffered formalin for 15 min and then stained with 1% crystal violet (Sigma Aldrich) for 30 min. The plates were gently washed with PBS and dried before microscopic evaluation. Cell clusters with >30 cells were considered as a colony.
Luciferase activity assay
STAT1 transcription activity was measured with a STAT1 luciferase reporter. Luciferase activity was measured with the Dual-Luciferase reporter assay system (Thermo Scientific) according to the manufacturer’s instructions as described. Data were normalized for transfection efficiency by division of firefly luciferase activity with that of Renilla luciferase.
Subcellular fractionation and DNA binding assays
Nuclear and cytoplasmic protein of ESCC cells was extracted using an NE-PER protein extraction kit (Thermo Scientific) according to the manufacturer’s instructions. For western blot analysis, tubulin and histone deacetylase 1 (HDAC1) were used for the cytoplasmic and nuclear loading control, respectively.
Oligonucleotide pull-down assays were performed with an annealed nucleotide comprising the STAT1 consensus site (5′-CATGTTATGCATATTCCTGTAAGTG-3′) with a 5-biotin label. Nuclear extracts (50–100 μg) were incubated for 1 h at 4 °C with 1 μg oligonucleotide in binding buffer, then Sepharose–streptavidin (50 μl; Sigma) was added for 2 h at 4 °C. After three washes in PBS, the complexes were suspended in SDS sample buffer and processed for western blotting, as described above, and probed with anti-STAT1 and anti-pSTAT1 antibodies (Cell Signalling).
Quantitative RT-PCR
Using an RNeasy Mini Kit (Qiagen), total cellular RNA was extracted from cells following the manufacturer’s protocol. Primer sequences were: human interferon regulatory factor 1 (IRF1) forward: 5′-ATGCCCATCACTCGGATGC-3′, reverse: 5′-CCCTGCTTTGTATCGGCCTG-3′; human chemokine (C-X-C motif) ligand 10 (CXCL10) forward: 5′-GTGGCATTCAAGGAGTACCTC-3′, reverse: 5′-TGATGGCCTTCGATTCTGGATT-3′; STAT1α forward: 5′-CCAATGGAACTTGATGGCCC-3′, reverse: 5′-CAGAGCCCACTATCCGAGAC-3′; guanylate binding protein 2 (GBP2) forward: 5′-CTATCTGCAATTACGCAGCCT-3′, reverse: 5′-TGTTCTGGCTTCTTGGGATGA-3′; intercellular adhesion molecule 10 (ICAM10) forward: 5′-ATGCCCAGACATCTGTGTCC-3′, reverse: 5′-GGGGTCTCTATGCCCAACAA-3′; transporter associated with antigen processing 1 (TAP1) forward: 5′-GCAAGACGACTTACTCTGGGT-3′, reverse: 5′-GGATCTGACACCACTGGACC-3′.
Immunohistochemistry
Formalin-fixed, paraffin-embedded ESCC tumors were used for this study. All cases were retrieved from the file at the Department of Pathology, Shantou University Medical College. The diagnosis of these cases was based on criteria established by the World Health Organization classification scheme. Immunohistochemistry to detect STAT1 expression was performed using a method similar to that described previously.11 Briefly, formalin-fixed, paraffin-embedded tissue sections of 4 μm thickness were deparaffinized in xylene and hydrated in graded ethanol (100–50%). Antigen retrieval was performed by immersing tissue in citrate buffer (pH 6.0), microwaving in a pressure cooker for 20 min, and leaving to cool for 20 min at room temperature. After antigen retrieval, tissue sections were incubated with 10% hydrogen peroxide (H2O2) and methanol for 10 min to block endogenous peroxidase activity, followed by washing in running tap water for 5 min. Subsequently, the sections were incubated for 20 min in antibody diluent (Dako), followed by incubation overnight at 4 °C with a rabbit polyclonal antibody reactive with anti-STAT1β (1:200 dilution, Cellchemo, MA, USA). Immunostaining was visualized with a labeled streptavidin-biotin method (20 min in biotinylated link and 20 min in streptavidin-conjugated horse radish peroxidase, both from Dako) using (3, 3-diaminobenzidine/H2O2) DAB as a chromogen (Dako). Hematoxylin was used as a counter stain. Following staining, sections were dehydrated in graded ethanol (50–100%), followed by xylene incubation. Coverslips were applied using Permount solution (Fisher Scientific, Pittsburgh, PA, USA).
Staining was independently evaluated by two pathologists who were blinded to the clinical data. The percentages of positively stained cells were assigned the following scores: 0 (<5% positive), 1 (6–25% positive), 2 (26–50% positive), 3 (51–75% positive), or 4 (>75% positive). Staining intensity was scored on a scale of 0–3 as follows: 0, negative; 1, buff; 2, yellow; and 3, brown. The percentage of positive cells and the staining intensities were then multiplied to generate the immunoreactivity score for each case. Overall staining scores from 0 to 2, 3–6 and ⩾7 were considered negative, weak and strong expression, respectively. Both weak and strong expressions were considered positive.
Statistical analyses
Data were expressed as mean±standard deviation. The prognostic significance of the expression of various markers was analyzed using the Kaplan–Meier method. The correlation between STAT1β and other clinical parameters was evaluated using a chi-square or Student’s t-test. Differences among the treatment groups were assessed using ANOVA and the appropriate statistical software (SPSS, IBM, USA). A P-value of ⩽0.05 was considered as statistically significant.
Publisher’s Note:
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Adámková L, Soucková K, Kovarík J . Transcription protein STAT1: biology and relation to cancer. Folia Biol (Praha) 2007; 53: 1–6.
Kim HS, Lee MS . STAT1 as a key modulator of cell death. Cell signal 2007; 19: 454–465.
Sironi JJ, Ouchi T . STAT1-induced apoptosis is mediated by caspases 2, 3, and 7. J Biol Chem 2004; 279: 4066–4074.
Chan SR, Vermi W, Luo J, Lucini L, Rickert C, Fowler AM et al. STAT1 deficient mice spontaneously develop estrogen receptor α-positive luminal mammary carcinomas. Breast Cancer Res 2012; 14: 16.
Leibowitz MS, Andrade Filho PA, Ferrone S, Ferris RL . Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic Tlymphocytes. Cancer Immunol Immunother 2011; 60: 525–535.
Zhang Y, Molavi O, Su M, Lai R . The clinical and biological significance of STAT1 inesophageal squamous cell carcinoma. BMC Cancer 2014; 14: 791.
Zhang Y, Zhang Y, Yun H, Lai R, Su M . Correlation of STAT1 with apoptosis and cell-cycle markers in esophageal squamous cell carcinoma. PLoS One. 2014; 9: e113928.
Baran-Marszak F, Feuillard J, Najjar I, Le Clorennec C, Béchet JM, Dusanter-Fourt I et al. Differential roles of STAT1alpha and STAT1beta in fludarabine-induced cell cycle arrest and apoptosis in human B cells. Blood. 2004; 104: 2475–2483.
Najjar I, Schischmanoff PO, Baran-Marszak F, Deglesne PA, Youlyouz-Marfak I, Pampin M et al. Novel function of STAT1beta in B cells: induction of cell death by a mechanism different from that of STAT1alpha. J Leukoc Biol 2008; 84: 1604–1612.
Semper C, Leitner NR, Lassnig C, Parrini M, Mahlakõiv T, Rammerstorfer M et al. STAT1β is not dominant negative and is capable of contributing to gamma interferon-dependent innate immunity. Mol Cell Biol 2014; 34: 2235–2248.
Satoh J, Tabunoki H . A comprehensive profile of ChIP-Seq based STAT1 target genes suggests the complexity of STAT1-mediated gene regulatory mechanisms. Gene Regul Syst Bio 2013; 7: 41–56.
Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE Jr . Proteins oftranscription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci USA 1992; 89: 7836–7839.
Zakharova N, Lymar ES, Yang E, Roeder RG, Malik S, Zhang JJ et al. Distinct transcriptional activation functions of STAT1a and STAT1b on DNA and chromatin templates. J Biol Chem 2003; 278: 43067–43073.
Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR . How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J 2000; 19: 4111–4122.
Alvarez GR, Zwilling BS, Lafuse WP . Mycobacterium avium inhibition of IFN-gamma signaling in mouse macrophages: Toll-like receptor 2 stimulation increases expression of dominant-negative STAT1 beta by mRNA stabilization. J Immunol 2003; 171: 6766–6773.
Bhardwaj N, Rosas LE, Lafuse WP, Satoskar AR . Leishmania inhibits STAT1-mediated IFN-gamma signaling in macrophages: increased tyrosine phosphorylation of dominant negative STAT1beta by Leishmania mexicana. Int J Parasitol 2005; 35: 75–82.
Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell JE Jr et al. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci USA 1996; 93: 15092–15096.
Dai X, Sayama K, Yamasaki K, Tohyama M, Shirakata Y, Hanakawa Y et al. SOCS1-negative feedback of STAT1 activation is a key pathway in the dsRNA-induced innate immune response of human keratinocytes. J Invest Dermatol 2006; 126: 1574–1581.
O'Malley JT, Eri RD, Stritesky GL, Mathur AN, Chang HC, Hogenesch H et al. STAT4 isoforms differentially regulate Th1 cytokine production and the severity of inflammatory bowel disease. J Immunol 2008; 181: 5062–5070.
Schaefer TS, Sanders LK, Nathans D . Cooperative transcriptional activity of Jun and Stat3 beta, a short form of Stat3. Proc Natl Acad Sci USA 1995; 92: 9097–9101.
Schaefer TS, Sanders LK, Park OK, Nathans D . Functional differences between Stat3alpha and Stat3beta. Mol Cell Biol 1997; 17: 5307–5316.
Pilz A, Ramsauer K, Heidari H, Leitges M, Kovarik P, Decker T et al. Phosphorylation of the Stat1 transactivating domain is required for the response to type I interferons. EMBO Rep 2003; 4: 368–373.
Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W et al. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell 2005; 17: 761–771.
Horvath CM, Stark GR, Kerr IM, Darnell JE Jr . Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcriptioncomplex. Mol Cell Biol 1996; 16: 6957–6964.
Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE Jr, Kuriyan J . Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 1998; 93: 827–839.
Lai R, Rassidakis GZ, Medeiros LJ, Leventaki V, Keating M, McDonnell TJ et al. Expression of the phosphorylated (active) forms of STAT3 in mantle cell lymphoma cell lines and tumors. J Pathol 2003; 199: 84–89.
Acknowledgements
We gratefully acknowledge Professor Liyan Xu at the Shantou University Medical College for sharing KYSE150 cells with us. This study was sponsored by Guangdong Innovation University Program (Certificates 2016KQNCX050) awarded to YZ, NSFC (31240077) awarded to MS and an operating research grant from the Canadian Institute of Health Research awarded to RL. The study was approved by the ethical review committees of the Medical College of Shantou University. The patients involved in our study provided written informed consents.
Author contributions
Concepts and experimental design: YZ, RL. Acquisition of data: YZ, HY,YC. Analysis and interpretation of data: YZ, MS, RL,YeC. Writing of the manuscript: YZ, MS, RL, ZL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Supplementary Information accompanies this paper on Cell Death and Disease website .
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, Y., Chen, Y., Yun, H. et al. STAT1β enhances STAT1 function by protecting STAT1α from degradation in esophageal squamous cell carcinoma. Cell Death Dis 8, e3077 (2017). https://doi.org/10.1038/cddis.2017.481
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2017.481
This article is cited by
-
PABPC1-induced stabilization of IFI27 mRNA promotes angiogenesis and malignant progression in esophageal squamous cell carcinoma through exosomal miRNA-21-5p
Journal of Experimental & Clinical Cancer Research (2022)