Abstract
Long noncoding RNAs (lncRNAs) have important roles in various biological processes. Our previous work has revealed that dedifferentiation of retinal pigment epithelium (RPE) cells contributes to the pathology of age-related macular degeneration (AMD). Herein, we show roles of lncRNAs in RPE differentiation. We used microarray to identify lncRNA expression profiles in human induced pluripotent stem cells (hiPSCs) and hiPSC-derived RPE cells. A total of 217 differentially expressed lncRNAs along with the differentiation were initially identified, among which 13 lncRNAs showed a consistent fold change of over 2. LncRNA ZNF503-AS1, located in the cytoplasm of RPE cells, was found consistently upregulated along with RPE differentiation, and downregulated in the RPE-choroid of AMD patients. In vitro study further suggested that ZNF503-AS1 insufficiency could inhibit RPE differentiation, and promote its proliferation and migration. As ZNF503-AS1 is transcribed from the antisense strand of the ZNF503 gene locus, we further revealed its regulatory role in ZNF503 expression. ZNF503-AS1 was reversely correlated with ZNF503 expression. Our results also suggested that ZNF503 could inhibit RPE differentiation, and promote its proliferation and migration. Thus, ZNF503-AS1 potentially promotes RPE differentiation through downregulation of ZNF503 expression. In addition, nuclear factor-κB was recognized as a potential upstream transcript factor for ZNF503-AS1, which might participate in promoting RPE differentiation by regulating the expression of ZNF503-AS1. Taken together, our study identifies a group of RPE differentiation relevant lncRNAs, and the potential role of ZNF503-AS1 in the pathology of atrophic AMD, which might help with the intervention of AMD patients.
Similar content being viewed by others
Main
Retinal pigment epithelium (RPE) is a monolayer of cuboidal, polarized, and pigmented epithelial cells located in the outer retina between photoreceptors and choroidal vessels.1, 2, 3 RPE forms a part of the blood/retina barrier, secrets multiple growth factors, and is crucial in maintaining regular retinal functions.2, 3, 4 Dysfunction and depletion of RPE cells are involved in multiple retinal degenerations, including age-related macular degeneration (AMD).2, 5, 6, 7 AMD is a universal leading cause for irreversible vision loss in people aged over 55.8, 9, 10 Clinically, AMD can be classified into two major categories, namely atrophic and exudative AMD.10 Our group has previously recognized RPE dedifferentiation as a crucial contribution factor to the pathogenesis of atrophic AMD.1, 3 Atrophic AMD is typified by subepithelial deposits and degeneration of RPE cells involving but not limited to the macular region.10 No efficient therapies have been developed for atrophic AMD. Therefore, better insights into its pathology and seeking for a putative therapeutic target have become the focus of current researches. On the basis of our previous findings, inhibiting RPE dedifferentiation may retard or block AMD disease course, thus helping with better control of AMD patients.
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides structurally homologous to protein-coding mRNAs, but has little or no protein-coding potential.11 LncRNAs can modulate gene expressions as competing endogenous RNAs (ceRNAs).11, 12, 13, 14 The ceRNA hypothesis defines a microRNA (miRNA)-mediated post-transcriptional regulatory network. In this network, protein-coding and non-protein-coding RNAs share one or more miRNA response elements and compete for miRNA binding, further modulating each other’s expressions.12, 15 LncRNAs have been reported to have regulatory roles in diverse biological processes, including cell differentiation, stem cell maintenance, and epigenetic regulation.16, 17, 18 Dysregulation of lncRNAs is found involved in multiple human diseases such as cancer, neurological problems, and cardiovascular diseases.19 Roles of lncRNAs in ocular diseases, such as diabetic retinopathy, proliferative vitreoretinopathy, glaucoma, ocular tumors, and ocular neovascularization, have also been identified.11, 14, 20, 21, 22, 23, 24, 25, 26 However, the association between lncRNAs and RPE differentiation is poorly understood. In this study, we aim to reveal the roles of a lncRNA, ZNF503-AS1, in RPE differentiation, and to seek for a lncRNA-based potential therapeutic target for dry AMD.
Results
Differently expressed lncRNAs during RPE differentiation
To reveal the roles of lncRNAs on RPE differentiation, we used microarrays to modulate the expression profiles of lncRNAs and mRNAs in human induced pluripotent stem cells (hiPSCs) and in hiPSC-derived RPE cells (hiPSC-RPE) at 30, 60, and 90 days post differentiation (d.p.d.), respectively. Pluripotency of the hiPSC colonies and competence of differentiation have been previously determined.3 Among all identified lncRNAs, only those recorded in Encyclopedia of DNA Elements (ENCODE) showed a consistent expressional change, and presented a fold change of over 2 in the 30 d.p.d. hiPSC-RPE compared to undifferentiated hiPSC were included. A total of 217 differentially expressed lncRNAs, including 116 upregulated and 101 downregulated lncRNAs, were sorted out with their chromosomal locations annotated (Figures 1a and b; Supplementary Table S1). Of all 217 identified lncRNAs, 13 showed a consistent fold change of over 2 among all time points along with the differentiation, indicating their crucial roles in RPE differentiation, and were selected for further analysis (Table 1; Figure 1a).
Expression profiles of lncRNAs in RPE differentiation. (a) Hierarchical clustering of all 217 lncRNAs with a consistent expressional change along with the differentiation. Thirteen lncRNAs with consistent fold change of over 2 are highlighted. Blue: downregulated lncRNAs; yellow: upregulated lncRNAs. (b) Chromosomal locations of the 217 differentially expressed lncRNAs are shown as a Circos plot. (c–o) The 13 lncRNAs with consistent fold change of over 2 are selected for validation of the microarray results in hiPSC-RPE at 0, 30, 60, and 90 d.p.d., including upregulated RP11-367G18.1 (c), CTD-2319I12.1 (d), RP3-395M20.8 (e), RP11-1020M18.10 (f), H19 (g), RP11-195B3.1 (h), RP11-554I8.2 (i), and ZNF503-AS1 (j), and downregulated ESRG (k), LINC00617 (l), RP11-469A15.2 (m), LINC01162 (n), and LINC00173 (o)
To validate the microarray data, we next confirmed expressions of the 13 lncRNAs using real-time PCR. Agreed with the microarray data, lncRNAs RP11-367G18.1 (ENST00000452675), CTD-2319I12.1 (ENST00000590346), RP3-395M20.8 (ENST00000416860), RP11-1020M18.10 (ENST00000548135), H19 (ENST00000414790), RP11-195B3.1 (ENST00000436340), RP11-554I8.2 (ENST00000417112), and ZNF503-AS1 were found consistently upregulated (Figures 1c–j), while ESRG (ENST00000583516), LINC00617 (ENST00000503525), RP11-469A15.2 (ENST00000427825), LINC01162 (ENST00000447262), and LINC00173 (ENST00000480237) were downregulated (Figures 1k–o).
ZNF503-AS1 is mainly localized in cytoplasm
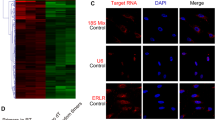
ZNF503-AS1 is an intergenic lncRNA located on chromosome 10q22.2, and is partly conserved among multiple species (Supplementary Figure S1). However, the biological function of ZNF503-AS1 is largely unknown. Our above results confirmed that ZNF503-AS1 was consistently upregulated along with the differentiation from hiPSC to RPE (Figure 1j). RNA-fluorescent in situ hybridization (FISH) suggested that ZNF503-AS1 was expressed in human RPE (ARPE19) cells, and was mainly localized in cytoplasm (Figure 2a).
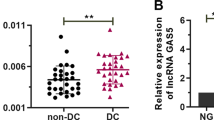
Intracellular localization and clinical relevance of ZNF503-AS1. (a) RNA-FISH suggested that ZNF503-AS1 was mainly localized in the cytoplasm of ARPE19 cells. U6 was taken as a representative for nuclear localization, and 18S for cytoplasmic localization. (b) Relative expression of ZNF503-AS1 in the macular RPE of an aged donor compared to a young donor. (c and d) ZNF503-AS1 expressions compared between the extramacular RPE-choroid tissue of the control group and AMD patients (c) and dry AMD patients (d), respectively
Clinical relevance of ZNF503-AS1
Taken that RPE dedifferentiation was involved in the pathogenesis of retinal degenerative diseases,1, 3 we assumed that ZNF503-AS1 expression would be reduced in dysfunctional RPE. To test our hypothesis, we compared ZNF503-AS1 expression between macular RPE-choroid of a 70-year-old male and a 30-year-old female donor. Our previous study suggested that the male donor had RPE dysfunction.3 As expected, ZNF503-AS1 expression was significantly reduced in the aged donor with RPE dysfunction when compared with the young female donor (Figure 2b). To further tell the role of ZNF503-AS1 in AMD pathogenesis, we compared its expression among RPE-choroid samples of 38 AMD patients and 46 healthy controls. Personal particulars and clinical diagnosis for each participant were detailed before.27 As indicated in Figure 2c, ZNF503-AS1 was downregulated in AMD patients when compared with controls. Subgroup analysis was then performed to see with which AMD subtype ZNF503-AS1 was mostly correlated. AMD diagnostic criterion was described previously.27 Our findings suggested that ZNF503-AS1 expression was only found reduced in RPE-choroid of dry AMD patients (n=16), a branch of atrophic AMD (Figure 2d). Taken together, our findings suggested that dysregulation of ZNF503-AS1 is involved in RPE dysfunction, especially in atrophic AMD.
ZNF503-AS1 promotes RPE differentiation
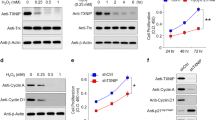
We next used transfection assays to modulate expression levels of ZNF503-AS1 in cells to evaluate its role in RPE differentiation. Exogenous ZNF503-AS1 was significantly upregulated in cells transfected with Ac-ZNF503-AS1 plasmid (Figure 3a), and endogenous ZNF503-AS1 was downregulated in cells transfected with ZNF503-AS1 scramble small interfering RNA (siRNA; Figure 3b). Noteworthy, three pairs of siRNAs oligos targeting different regions of ZNF503-AS1 were initially designed and tested, and the siRNA showing best efficiency and stability was chosen for further studies (Supplementary Figure S2A). Increased expressions of RPE dedifferentiation-related markers, including microphthalmia-associated transcription factor (MITF; NM_198159), SOX2 (NM_003106), POU domain class 5 transcription factor 1 (POU5F1; NM_002701), and homeobox protein NANOG (NANOG; NM_024865), were detected in 30 days hiPSC-RPE cells transfected with ZNF503-AS1 siRNA (Figure 3c). In addition, mRNA expression levels of RPE markers, including retinoid isomerohydrolase (RPE65; NM_000329), retinaldehyde-binding protein 1 (RLBP1; NM_000326), lecithin retinol acyltransferase (LRAT; NM_004744), tyrosine-protein kinase Mer (MERTK; NM_006343), bestrophin-1 (BEST1; NM_001139443), cytokeratin-18 (KRT18; NM_000224), tight junction protein ZO-1 (TJP1; NM_003257), and catenin beta-1 (CTNNB1; NM_001904), were tested by real-time PCR. In both 30 d.p.d. hiPSC-RPE and ARPE19 cells, ectopic overexpression of ZNF503-AS1 elevated mRNA expressions of those RPE markers (Figures 3d and f), while its insufficiency inhibited their expressions (Figures 3e and g).
ZNF503-AS1 promotes RPE differentiation. (a and b) Relative miRNA expression of ZNF503-AS1 in hiPSC-RPE at 30 d.p.d. transfected with Ac-ZNF503-AS1 plasmid compared to empty vector (a), and in cells transfected with ZNF503-AS1 siRNA compared to scramble siRNA (b). (c) Relative mRNA expressions of RPE dedifferentiation-related markers, including MITF, SOX2, POU5F1, and NANOG, in hiPSC-RPE at 30 d.p.d. transfected with ZNF503-AS1 siRNA compared to scramble siRNA. (d–g) Relative mRNA expressions of RPE65, RLBP1, LRAT, MERTK, BEST1, KRT18, TJP1, and CTNNB1 in hiPSC-RPE at 30 d.p.d. (d) and ARPE19 cells (e) transfected with Ac-ZNF503-AS1 compared with empty vector, and in hiPSC-RPE at 30 d.p.d. (f) and ARPE19 cells (g) transfected with ZNF503-AS1 siRNA compared to scramble siRNA. (h–j) Relative protein expressions of Mertk, cytokeratin-18, ZO-1, and β-catenin in ARPE19 cells transfected with Ac-ZNF503-AS1 compared with empty vector (h and i), and in cells transfected with ZNF503-AS1 siRNA compared to scramble siRNA (h and j)
Immunoblotting further revealed that protein expression levels of Mertk (NP_006334), cytokeratin-18 (NP_000215), ZO-1 (NP_003248), and β-catenin (NP_001895) were increased in ZNF503-AS1-overexpressed group (Figures 3h and i) while decreased in ZNF503-AS1-interfered group (Figures 3h and j). Meanwhile, immunofluorescent staining was used to monitor the intracellular expression and localization of bestrophin-1 (NP_004174), ZO-1, and β-catenin. Enhanced expressions of all three proteins were observed in ARPE19 cells overexpressing ZNF503-AS1 (Figures 4a–c), while decreased protein expressions were detected in cells with ZNF503-AS1 downregulated. No obvious mislocalization of proteins was identified in all transfected groups. Taken together, our results indicated that ZNF503-AS1 might contribute to the differentiation of RPE cells.
Immunofluorescence of RPE relevant proteins, including bestrophin-1 (a), ZO-1 (b), and β-catenin (c). Increased expressions of all three proteins were observed in ARPE19 cells transfected with Ac-ZNF503-AS1 when compared with cells transfected with empty vector, while decreased protein expressions were detected in cells transfected with ZNF503-AS1 siRNA compared to cells transfected with scramble siRNA. No obvious mislocalization of proteins was identified in all transfected groups
ZNF503-AS1 suppresses RPE proliferation and migration
Reportedly, cell proliferation and migration can follow the dedifferentiation of postmitotic tissues, including RPE.3, 28, 29 To better reveal the role of ZNF503-AS1 in RPE function, we tried to determine its effects on RPE proliferation and migration in ARPE19 cells. Rates of cell proliferation and migration were automatically monitored till 72 h post transfection. On the basis of our findings, both proliferation and migration were consistently inhibited in cells transfected with Ac-ZNF503-AS1 plasmid at all time points post transfection (Figures 5a and b). However, rates of both proliferation and migration elevated in ZNF503-AS1 siRNA-transfected group (Figures 5c and d). Thus, our data indicated an inhibitory role of ZNF503-AS1 in RPE proliferation and migration.
Rates of cell proliferation and migration in different transfected groups. (a and b) Both proliferation (a) and migration (b) were inhibited in ARPE19 cells transfected with Ac-ZNF503-AS1 when compared with cells transfected with empty vector. (c and d) Rates of proliferation (c) and migration (d) were increased in ARPE19 cells transfected with ZNF503-AS1 siRNA compared to cells transfected with scramble siRNA. (e and f) In cells transfected with ZNF503 siRNA, rates of both proliferation (e) and migration (f) were decreased when compared with cells transfected with scramble siRNA
ZNF503-AS1 is regulated by nuclear factor-κB
We applied PROMO online program to predict transcription factor binding sites (TFBS) in the promoter region of ZNF503-AS1. Nuclear factor-κB (NF-κB) was revealed as its putative transcript factor with three potential TFBS in its promoter region (Figure 6a). We then used ammonium pyrrolidinedithiocarbamate (PDTC) to inhibit the activation of NF-κB, and tumor necrosis factor-α (TNF-α) to stimulate its activation in ARPE19 cells. Expression of NF-κB p65 (activated form of NF-κB) was upregulated in cells treated with PDTC (Figures 6c and d), and was downregulated in cells treated with TNF-α (Figures 6b and d). To better understand the regulatory role of NF-κB, we next tested ZNF503-AS1 expression in different treated groups. Our data supported that expression of ZNF503-AS1 was reduced in the PDTC-treated group (Figure 6e) and was elevated in the TNF-α-treated group (Figure 6f).
ZNF503-AS1 is regulated by NF-κB. (a) PROMO program predicts NF-κB as a putative transcript factor for ZNF503-AS1 with three potential TFBS in its promoter region. (b–d) Protein expression of NF-κB p65, the activated form of NF-κB, was increased in ARPE19 cells treated with PDTC (b and c) and was decreased in cells treated with TNF-α (b and d) when compared to cells treated with DMSO. (e and f) mRNA expression of ZNF503-AS1 was upregulated in ARPE19 cells treated with PDTC (e) and was downregulated in cells treated TNF-α (f) when compared to cells treated with DMSO
ZNF503-AS1 regulates ZNF503 expression
ZNF503-AS1 is transcribed from the antisense strand of the ZNF503 gene locus. We then assessed whether ZNF503-AS1 could affect the expression of ZNF503. We observed that ZNF503 mRNA was decreased in cells overexpressing ZNF503-AS1 (Figure 7a), and was increased in cells with ZNF503-AS1 knocked down (Figure 7b). Our data suggested a potential regulatory role of ZNF503-AS1 in ZNF503 expression.
ZNF503 promotes RPE differentiation, and inhibits cell proliferation and migration. (a and b) ZNF503 expression was elevated in ARPE19 cells transfected with Ac-ZNF503-AS1 when compared to cells transfected with empty vector (a), and was reduced in cells transfected with ZNF503-AS1 siRNA when compared with cells transfected with scramble siRNA (b). (c) ZNF503 expression was remarkably decreased in ARPE19 cells transfected with ZNF503 siRNA. (d) In ARPE19 cells transfected with ZNF503 siRNA, mRNA expressions of RPE relevant markers, including RPE65, RLBP1, LRAT, KRT18, and CTNNB1, were upregulated. (e and f) Immunoblotting indicated that protein expressions of Mertk, cytokeratin-18, and β-catenin were increased in ARPE19 cells transfected with ZNF503 siRNA compared to cells transfected with scramble siRNA. (g and h) Immunofluorescent staining suggested that ZO-1 (g) and β-catenin (h) expressions were elevated in ARPE19 cells transfected with ZNF503 siRNA when compared to cells transfected with scramble siRNA. No obvious mislocalization of proteins was identified in all transfected groups
ZNF503 inhibits RPE differentiation, and promotes RPE proliferation and migration
As ZNF503-AS1 regulates ZNF503 expression, we next detected whether ZNF503 insufficiency would improve the regular function of RPE. Endogenous expression of ZNF503 was remarkably decreased in cells transfected with ZNF503 siRNA (Figure 7c). Three pairs of siRNA oligos targeting different regions of ZNF503 were initially designed and tested. We only selected the siRNA showing best efficiency and stability for further investigations (Supplementary Figure S2B). In ARPE19 cells transfected with ZNF503 siRNA, mRNA expressions of RPE differentiation relevant markers, including RPE65, RLBP1, LRAT, KRT18, and CTNNB1, were elevated (Figure 7d). Immunoblotting further indicated that ZNF503 insufficiency in ARPE19 cells would increase the protein expressions of Mertk, cytokeratin-18, and β-catenin (Figures 7e and f). Enhanced expressions of ZO-1 and β-catenin were also revealed in cells transfected with ZNF503 siRNA by immunofluorescence (Figures 7g and h). No obvious protein mislocalization was found. Thus, our data indicated an inhibitory role of ZNF503 in RPE differentiation.
Impacts of ZNF503 on cell proliferation and migration were also monitored. Our data demonstrated that rates of both proliferation and migration elevated in cells transfected with ZNF503 siRNA (Figures 5e and f), suggesting that ZNF503 could promote RPE proliferation and migration. Taken together, our findings indicated that ZNF503-AS1 promoted RPE differentiation and inhibited its proliferation and migration by interfering with ZNF503 expression.
Discussion
RPE dedifferentiation has been typified as a crucial contributing factor to the pathology of atrophic AMD.1, 3 Therefore, blocking RPE dedifferentiation is a promising strategy in the treatment of dry AMD. Our group has previously demonstrated that AKT2/mTOR pathway and a group of miRNAs are potentially involved in RPE differentiation.1, 3 However, the molecular mechanism underlying RPE differentiation is not fully understood. LncRNAs are important regulators in gene expression. Roles of lncRNA dysregulation in ocular diseases have been well established.11 However, its role in RPE differentiation or pathology of atrophic AMD has never been reported. In the present study, we used microarray to obtain lncRNA expression profiles in hiPSC and hiPSC-RPE at different stages. A total of 217 differentially expressed lncRNAs along with the differentiation were initially sorted out. Among all identified lncRNAs, 13 presented a consistent fold change of over 2, including 8 upregulated and 5 downregulated lncRNAs.
ZNF503-AS1 is an intergenic lncRNA with unrecognized biological interplay. Our data suggest that ZNF503-AS1 is expressed in the cytoplasm of RPE cells. We have demonstrated that ZNF503-AS1 is consistently upregulated along with RPE differentiation, and is downregulated in RPE-choroid of atrophic AMD patients and a senior donor with RPE dysfunction. In vitro data have also indicated the role of ZNF503-AS1 in promoting RPE differentiation, and inhibiting RPE proliferation and migration. ZNF503-AS1 is transcribed from the antisense strand of the ZNF503 gene locus. ZNF503 has been recognized as a transcriptional repressor that promotes mammary epithelial cell proliferation and migration, while its role in maintaining RPE function has not been fully elucidated.30, 31 RPE cell is a type of mammary epithelial cell. Similar to previous findings, our data indicated that ZNF503 could inhibit RPE differentiation, and promote its proliferation and migration. We have also revealed that ZNF503-AS1 expression is reversely correlated with the expression of ZNF503. Thus, our findings suggest that ZNF503-AS1 potentially promotes RPE differentiation by downregulation of ZNF503 expression. However, how ZNF503 dysregulation affect RPE function is still poorly understood. Therefore, more investigations are warranted. Thus, our results indicate that ZNF503-AS1 dysregulation is involved in RPE dedifferentiation and pathology of atrophic AMD, implying its application as a biomarker and therapeutic target for atrophic AMD.
The NF-κB pathway is involved in many cellular processes, including immunity and inflammation. Roles of NF-κB in inflammatory signaling and AMD pathogenesis have been well established.32, 33, 34 Activation of the NF-κB pathway has been previously suggested to stimulate pigmentation of cultured primary hRPE cells.35 In this study, NF-κB is recognized as a potential upstream transcript factor for ZNF503-AS1, which may participate in promoting RPE differentiation by regulating the expression of ZNF503-AS1.
Taken together, our study identifies a group of RPE differentiation relevant lncRNAs. We also reveal a potential role of ZNF503-AS1 in AMD pathogenesis. ZNF503-AS1 may become a biomarker and a potential therapeutic target for atrophic AMD. However, more investigations are still needed to better illustrate the roles of lncRNAs in RPE differentiation and AMD pathology.
Materials and methods
Samples
Postmortem specimens of a 30-year-old female donor and a 70-year-old male donor were provided by Lions New South Wales Eye Bank through Save Sight Institute, the University of Sydney, Australia.3 All procedures followed standard procedures of eye donation for research and were approved by the institutional ethical committees conformed to Declaration of Helsinki. Written informed consents were obtained from all donors before their donations.
Culture and differentiation of hiPSC-RPE
HiPSC (IMR90-57) were cultured on mouse embryonic fibroblasts (SiDan-Sai Biotechnology Co., Ltd, Shanghai, China) in six-well tissue culture plates as detailed elsewhere.3 The hiPSCs were then differentiated into RPE cells according to the SFEB/CS method using low-molecular-weight compounds CKI-7 (5 μM) and SB-431542 (5 μM).36
Microarray profiling and computational analysis
Total RNA was isolated from hiPSC-RPE cells at different stages using TRIzol reagent (Invitrogen, Carlsbad, CA, USA).4, 37 Qualities and concentrations of RNA samples were determined using Nano-Drop ND-1000 spectrophotometer (Nano-Drop Technologies, Wilmington, DE, USA). Agilent Sureprint G3 Human GE 8 × 60 K Microarray (Agilent Co., Palo Alto, CA, USA) covering 27958 Entrez Gene RNAs and 7419 human lncRNAs was used to generate the lncRNA expression profiles from the following four groups in duet: hiPSC; and hiPSC-RPE at 30, 60, and 90 d.p.d. An Aglient G3 scanner was then used to determine the immunofluorescence intensities of the arrays. Data were further analyzed using feature extraction software. Multiples of differentially expressed genes were calculated. miRNA expression profiles of the four groups were generated as described before.3 In addition, microarray data (GSE29801) and sample information (GSM738433–GSM738607) of 175 independent RPE-choroid samples were downloaded from Gene Expression Omnibus data sets and analyzed as indicated previously.3, 27 Among the 175 samples, 88 were obtained from normal individuals, including 50 macular and 38 extramacular samples, and 87 were from AMD patients, including 41 macular and 46 extramacular samples.
Reverse transcription-PCR and real-time PCR
Reverse transcription-PCR (RT-PCR) and real-time PCR were performed using a previously defined protocol.4, 37 One microgram of the extracted total RNA was used for cDNA synthesis using PrimeScript RT Kit (Takara, Otsu, Japan). Real-time PCR was conducted using FastStart Universal SYBR Green Master (ROX; Roche, Basel, Switzerland) with StepOne Plus Real-time PCR System (Applied Biosystems, Darmstadt, Germany). Information of primers was provided in Supplementary Table S2.
Bioinformatics analysis
To enhance data credibility, only lncRNAs whose sequences had been recorded in ENCODE were included in the present study.38 Genomic location and conservation of lncRNA, ZNF503-AS1 (ENST00000416398), was obtained from UCSC genome browser (http://genome.ucsc.edu/index.html).39 We used PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) to identify putative TFBS in the promoter region of the lncRNA.40, 41 Results were demonstrated using WebLogo 3 (http://weblogo.threeplusone.com/).42, 43
Fluorescent in situ hybridization
ZNF503-AS1, U6, and 18 S FISH probes were synthesized by RiboBio (Guangzhou, China). FISH was conducted using the FISH kit per the manufacturer’s protocol (RiboBio). Briefly, ARPE19 cells were collected, fixed with 4% paraformaldehyde, permeabilized in 0.5% Triton X-100 on ice, and then treated with pre-hybridization buffer. We next hybridized cells with Cy3-labeled RNA of ZNF503-AS1 probe mix in a moist chamber. Cells were then stained with 4′,6-diamidino-2-phenylindole (DAPI). Images were collected using a confocal microscope (LSM 510; Carl Zeiss, Jena, Germany).
Reagents and cell transfection
ZNF503-AS1 and ZNF503 siRNAs were purchased from RiboBio Co. Ltd. Open reading frame sequence of ZNF503-AS1 was synthesized, amplified, and inserted into the expression vector pcDNA3.1 (Sigma, St. Louis, MO, USA) using XhoI and KpnI restriction sites to generate the Ac-ZNF503-AS1 plasmids. Sequences of the constructed plasmids were confirmed using Sanger sequencing.
For transfection assay, cells were seeded into six-well plates and transfected with 100 pmol siRNA or 4 μg expression vector using Lipofectamine 2000 transfection reagent (Invitrogen) per the manufacturer’s protocol. Cells were collected at 48 h post transfection for RNA isolation, and at 72 h post transfection for protein extraction and immunofluorescent staining. PDTC (Beyotime, Shanghai, China; concentration: 100 μM)44 and TNF-α (Sigma; concentration: 100 ng/μl) were used for cell stimulation. Cells were collected at 48 h after treatment.
Immunoblotting
Immunoblotting was performed per previously described protocols.3, 45 Briefly, cells were collected at 72 h post transfection in ice-cold RIPA buffer (Beyotime) containing protease inhibitors cocktail (Roche) for protein extraction. Extracted proteins were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). Membranes were then blocked, incubated with primary antibodies at 4 °C overnight (Supplementary Table S3), washed, and probed with corresponding horseradish peroxidase-conjugated secondary antibodies (1:10 000 diluted in 1 × PBS; ICL Inc., Newberg, OR, USA) for 1 h at room temperature. Autoradiography with the ECL-Western blotting system (BioRad, Hercules, CA, USA) was then applied to develop the blots per manufacturers’ protocols. ImageJ software (http://rsb.info.nih.gov/ij/index.html) was used to determine and quantify protein expressions.
Immunofluorescent staining
Immunofluorescence was performed per previously described protocols.3, 45 Cells were collected, fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, blocked in bovine serum albumin, and incubated with primary antibodies (Supplementary Table S3) at 4 °C overnight. Cells were then washed and incubated with corresponding fluorescence-conjugated secondary antibodies (1:1000 diluted in 1 × PBS; Invitrogen) for 1 h at room temperature. Cell nuclei were counterstained by DAPI (Sigma). Images were collected using a confocal microscope (LSM 510).
Monitoring cell proliferation and migration
Rates of cell proliferation and migration were detected in real-time using xCELLigence system E-Plate (Roche) according to the manufacturer’s protocol. To monitor cell proliferative rates, ~5000 cells were seeded in each well of the E-Plate. Transfection was performed at 24 h post plantation. For migration assay, 40 000 post-transfection cells were planted into each well. Cells were culture with fresh DME/F12 medium. Impedance value for each well was automatically determined by the xCELLigence system and expressed as a CI value. Rates of cell proliferation and migration were determined by calculating the slope of the line between two given time points.
Statistics
We used GraphPad Prism (version 4.0; GraphPad Software, San Diego, CA, USA) for statistical analysis. Student’s t-test was used for comparisons between groups. All presented data were based on biological triplicates. Data were shown as mean±S.D., and P-value <0.05 was considered as statistically significant.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Zhao C, Yasumura D, Li X, Matthes M, Lloyd M, Nielsen G et al. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest 2011; 121: 369–383.
Strauss O . The retinal pigment epithelium in visual function. Physiol Rev 2005; 85: 845–881.
Jiang C, Qin B, Liu G, Sun X, Shi H, Ding S et al. MicroRNA-184 promotes differentiation of the retinal pigment epithelium by targeting the AKT2/mTOR signaling pathway. Oncotarget 2016; 7: 52340–52353.
Zhao C, Bellur DL, Lu S, Zhao F, Grassi MA, Bowne SJ et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet 2009; 85: 617–627.
Ambati J, Fowler BJ . Mechanisms of age-related macular degeneration. Neuron 2012; 75: 26–39.
Langton KP, McKie N, Smith BM, Brown NJ, Barker MD . Sorsby's fundus dystrophy mutations impair turnover of TIMP-3 by retinal pigment epithelial cells. Hum Mol Genet 2005; 14: 3579–3586.
Gehrs KM, Anderson DH, Johnson LV, Hageman GS . Age-related macular degeneration—emerging pathogenetic and therapeutic concepts. Ann Med 2006; 38: 450–471.
Bressler NM . Age-related macular degeneration is the leading cause of blindness. JAMA 2004; 291: 1900–1901.
Jager RD, Mieler WF, Miller JW . Age-related macular degeneration. N Engl J Med 2008; 358: 2606–2617.
Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY . Age-related macular degeneration. Lancet 2012; 379: 1728–1738.
Li F, Wen X, Zhang H, Fan X . Novel insights into the role of long noncoding RNA in ocular diseases. Int J Mol Sci 2016; 17: 478.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP . A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011; 146: 353–358.
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010; 38: 5366–5383.
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 2015; 116: 1143–1156.
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep 2014; 4: 6088.
Rapicavoli NA, Poth EM, Blackshaw S . The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol 2010; 10: 49.
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477: 295–300.
Mercer TR, Dinger ME, Mattick JS . Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10: 155–159.
Wapinski O, Chang HY . Long noncoding RNAs and human disease. Trends Cell Biol 2011; 21: 354–361.
Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q . Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci 2014; 55: 941–951.
Huang J, Li YJ, Liu JY, Zhang YY, Li XM, Wang LN et al. Identification of corneal neovascularization-related long noncoding RNAs through microarray analysis. Cornea 2015; 34: 580–587.
Congrains A, Kamide K, Ohishi M, Rakugi H . ANRIL: molecular mechanisms and implications in human health. Int J Mol Sci 2013; 14: 1278–1292.
Zhou RM, Wang XQ, Yao J, Shen Y, Chen SN, Yang H et al. Identification and characterization of proliferative retinopathy-related long noncoding RNAs. Biochem Biophys Res Commun 2015; 465: 324–330.
Su S, Gao J, Wang T, Wang J, Li H, Wang Z . Long non-coding RNA BANCR regulates growth and metastasis and is associated with poor prognosis in retinoblastoma. Tumour Biol 2015; 36: 7205–7211.
Vu PY, Toutain J, Cappellen D, Delrue MA, Daoud H, El Moneim AA et al. A homozygous balanced reciprocal translocation suggests LINC00237 as a candidate gene for MOMO (macrosomia, obesity, macrocephaly, and ocular abnormalities) syndrome. Am J Med Genet A 2012; 158A: 2849–2856.
Fan J, Xing Y, Wen X, Jia R, Ni H, He J et al. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol 2015; 16: 139.
Newman AM, Gallo NB, Hancox LS, Miller NJ, Radeke CM, Maloney MA et al. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med 2012; 4: 16.
Adijanto J, Castorino JJ, Wang ZX, Maminishkis A, Grunwald GB, Philp NJ . Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J Biol Chem 2012; 287: 20491–20503.
Liu Y, Xin Y, Ye F, Wang W, Lu Q, Kaplan HJ et al. Taz-tead1 links cell-cell contact to zeb1 expression, proliferation, and dedifferentiation in retinal pigment epithelial cells. Invest Ophthalmol Vis Sci 2010; 51: 3372–3378.
Shahi P, Wang CY, Lawson DA, Slorach EM, Lu A, Yu Y et al. ZNF503/Zpo2 drives aggressive breast cancer progression by down-regulation of GATA3 expression. Proc Natl Acad Sci USA 2017; 114: 3169–3174.
Shahi P, Slorach EM, Wang CY, Chou J, Lu A, Ruderisch A et al. The transcriptional repressor ZNF503/Zeppo2 promotes mammary epithelial cell proliferation and enhances cell invasion. J Biol Chem 2015; 290: 3803–3813.
Cao S, Wang JC, Gao J, Wong M, To E, White VA et al. CFH Y402H polymorphism and the complement activation product C5a: effects on NF-kappaB activation and inflammasome gene regulation. Br J Ophthalmol 2016; 100: 713–718.
Kaarniranta K, Salminen A . NF-kappaB signaling as a putative target for omega-3 metabolites in the prevention of age-related macular degeneration (AMD). Exp Gerontol 2009; 44: 685–688.
Arjamaa O, Nikinmaa M, Salminen A, Kaarniranta K . Regulatory role of HIF-1alpha in the pathogenesis of age-related macular degeneration (AMD). Ageing Res Rev 2009; 8: 349–358.
Eidet JR, Reppe S, Pasovic L, Olstad OK, Lyberg T, Khan AZ et al. The silk-protein sericin induces rapid melanization of cultured primary human retinal pigment epithelial cells by activating the NF-kappaB pathway. Sci Rep 2016; 6: 22671.
Osakada F, Jin ZB, Hirami Y, Ikeda H, Danjyo T, Watanabe K et al. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci 2009; 122 (Pt 17): 3169–3179.
Chen X, Liu Y, Sheng X, Tam PO, Zhao K, Rong W et al. PRPF4 mutations cause autosomal dominant retinitis pigmentosa. Hum Mol Genet 2014; 23: 2926–2939.
Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012; 22: 1760–1774.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM et al. The human genome browser at UCSC. Genome Res 2002; 12: 996–1006.
Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM . PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002; 18: 333–334.
Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 2003; 31: 3651–3653.
Crooks GE, Hon G, Chandonia JM, Brenner SE . WebLogo: a sequence logo generator. Genome Res 2004; 14: 1188–1190.
Schneider TD, Stephens RM . Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 1990; 18: 6097–6100.
Mou L, Kang Y, Zhou Y, Zeng Q, Song H, Wang R . Neurokinin-1 receptor directly mediates glioma cell migration by up-regulation of matrix metalloproteinase-2 (MMP-2) and membrane type 1-matrix metalloproteinase (MT1-MMP). J Biol Chem 2013; 288: 306–318.
Liu Y, Chen X, Xu Q, Gao X, Tam PO, Zhao K et al. SPP2 mutations cause autosomal dominant retinitis pigmentosa. Sci Rep 2015; 5: 14867.
Acknowledgements
We thank all donors for their donations. We thank Prof Joshua L. Dunaief and Dr Delu Song from University of Pennsylvania for their technical support. This study was supported by National Natural Science Foundation of China (81525006, 81670864, 81730025, and 81700877); Jiangsu Province’s Innovation Team (to CZ); the Fundamental Research Funds of the State Key Laboratory of Ophthalmology (to CZ); Natural Science Foundation of Jiangsu Province (BK20171087 to XC); Open Foundation of State Key Laboratory of Reproductive Medicine (Nanjing Medical University, SKLRM-KA201607); and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, JX10231801). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by E Candi
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chen, X., Jiang, C., Qin, B. et al. LncRNA ZNF503-AS1 promotes RPE differentiation by downregulating ZNF503 expression. Cell Death Dis 8, e3046 (2017). https://doi.org/10.1038/cddis.2017.382
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2017.382
This article is cited by
-
The landscape of the long non-coding RNAs in developing mouse retinas
BMC Genomics (2023)
-
FGD5-AS1 facilitates the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells via targeting the miR-506-3p/BMP7 axis
Journal of Orthopaedic Surgery and Research (2021)
-
Epigenetic regulation of retinal development
Epigenetics & Chromatin (2021)
-
Research updates on the clinical implication of long noncoding RNA in digestive system cancers and chemoresistance
3 Biotech (2021)
-
lncRNA MEG3, Acting as a ceRNA, Modulates RPE Differentiation Through the miR-7-5p/Pax6 Axis
Biochemical Genetics (2021)