Abstract
Aberrant DNA hypermethylation in human cancer has been associated with Polycomb target genes in embryonic stem (ES) cells, but a functional link of the Polycomb-targeted differentiation program to tumorigenesis remains to be established. Here, through epigenome analysis correlating DNA hypermethylation in colon cancer with ES cell pluripotency and differentiation, we identified a set of DNA hypermethylated genes in cancer cells that are Polycomb targets strongly associated with ES cell differentiation, including HAND1, a developmental regulator. Intriguingly, HAND1 is silenced in over 90% of human primary colorectal tumors, and re-expression of HAND1 in colon cancer cells induces terminal differentiation, inhibits proliferation and prevents xenograft tumor formation. Moreover, hypermethylated HAND1 has a minimum enrichment of EZH2-H3K27me3 in cancer cells, but becomes EZH2 bound and bivalent upon the loss of DNA methylation, suggesting a sequential gene silencing event during oncogenesis. These findings established a functional role of Polycomb-targeted differentiation program as a tumor-suppressor event epigenetically inactivated in human cancer.
Similar content being viewed by others
Main
In addition to well-documented DNA hypermethylation, Polycomb-based epigenetic mechanisms are also emerging as new factors in tumorigenesis. The Polycomb group (PcG) protein EZH2 is a histone methyltransferase specific for histone H3 lysine trimethylation (H3K27me3)1 and its deregulation has been found frequently in aggressive human cancer.2, 3, 4 Although Polycomb-mediated gene repression is traditionally associated with early stage differentiation/development, evidence supporting EZH2 or H3K27me3-mediated gene silencing as a critical cancer-relevant epigenetic event is rapidly emerging. To date, several EZH2 target genes have been functionally linked to various cancer pathways,5, 6, 7, 8 further supporting its role in tumorigenesis. Currently, there is growing interest in defining the functional relationship between repressive histone modifications and DNA methylation in cancer development. Technological advances in high-resolution, genome-wide mapping of histone modifications, such as chromatin immunoprecipitation (ChIP)-PET or ChIP-seq, are now spurring systematic efforts to characterize epigenome modifications and their potential relationships.9, 10, 11, 12
Loss of differentiation is an important component in the pathogenesis of many cancers.13 It has been previously reported that many differentiation genes are transcriptionally repressed by Polycomb repressive complex 2 (PRC2) through bivalent chromatin modifications (carrying both H3K27me3 and H3K4me3 histone marks) to maintain the self-renewal features of embryonic stem (ES) cells.14, 15 Moreover, genes silenced by DNA hypermethylation in adult human tumors have been found to be preferentially targeted by PRC2 in human ES cells.16, 17, 18 Although these studies are provocative and suggest a possibility that the two gene silencing events may lead to inactivation of a differentiation/developmental program important for tumor surveillance, a functional link between Polycomb-targeted differentiation program with oncogenesis has not yet been established. Therefore, we aim to investigate the relationship between Polycomb-mediated histone modification and DNA methylation in cell differentiation and cancer development.
In this study, we performed integrative epigenome analysis in both human cancer cells and ES cells, and have identified a cancer gene silencing event associated with ES cell differentiation that is targeted by both DNA hypermethylation and Polycomb-mediated histone methylation in colon cancer cells. We further establish the critical role of the differentiation regulator HAND1, a key gene in this process, as a putative tumor suppressor epigenetically lost in colorectal cancer.
Results
Gene silencing associated with promoter DNA hypermethylation in colon cancer
As a first step to identify genes whose expression is affected by DNA hypermethylation, we compared the gene expression profiles of colorectal cancer line HCT116 with its counterpart HCT116-DKO in which both DNA methyltransfease 1 (DNMT1) and DNMT3B are genetically disrupted or HCT116 cells treated with DNA methylation inhibitor 5-Aza-2'-deoxycitidine (5-Aza-dC). Using the Illumina array, we have identified 1275 genes whose expression is upregulated (using threefold cutoff) in either condition (Figure 1a and Supplementary Table S1A). Further profiling of these genes across a variety of colon cancer cell lines and normal colon epithelium identified a subset of 753 genes consistently repressed in the cancer cell lines (Supplementary Table S1B). To determine whether the above gene set identified in established cell lines are of clinical relevance, we compared their expression profiles in 24 pairs of clinical colon tumor samples matched to normal colon tissues. The cluster analysis revealed 476 out of these 753 genes showed a consistent and marked repression in tumors compared with the normal controls, pointing to a clinical relevance of this set of genes beyond just the initial cell lines studied (Figure 1a and Supplementary Table S1C). Among this 476 gene list are many genes previously reported to be silenced by DNA hypermethylation in colon cancer (e.g., SFRP1, CDH2, UCHL1 and ELOVL4) but it also included many genes previously not reported to be DNA hypermethylated.
Genes silenced by DNA hypermethylation in colon cancer. (a) Strategy used to determine genes repression associated with DNA methylation in colon cancer by using microarray. Left, 1275 genes show the upregulation ≥3-fold change in either DKO cells or HCT116 cells treated with 5-Aza-dC compared with parental HCT116 cells (left); expression profiles of 1275 genes in normal colon tissue and multiple colon cancer cell lines, and 753 genes show consistent downregulation in multiple colon cancer cell compared with normal colon tissue (middle); 476 out of 753 genes show consistent downregulation in human primary colon tumor tissues compared with the normal controls (right). (b) Genome-wide location analysis of H3K4me3 was done in HCT116 and DKO cells by using ChIP-seq and Solexa Genome Analyzer. Venn diagram depicts the overlapping of genomic regions marked by H3K4me3 surrounding the transcriptional start site (<1.0 kb) from the two cell lines. A total of 2507 promoters were identified to carry high H3K4me3 in DKO cells but not in HCT116 cells. (c) Venn diagram depicting an overlap of 203 genes that were repressed by DNA hypermethylation with no detectable H3K4me3 in HCT116 cells, thus defined as genes silenced by DNA methylation. (d) Representative genes showing the differential gene expression, methylation status and H3K27me3 in HCT116 and DKO cells. RT-PCR shows that SFRP1 and CDH2 were silenced in HCT116 and re-expressed in DKO cells (left); MSP shows that SFRP1 and CDH2 promoters were methylated in HCT116 cells but demethylated in DKO cells (middle); ChIP-seq result shows that H3K4me3 was detected at SFRP1 and CDH2 promoters in DKO cells but not in HCT116 cells
Histone H3 lysine 4 trimethylation (H3K4me3) is associated with active gene promoter,10, 19 with the majority of these marks located within 1.0 kb of transcription start sites (TSS). Moreover, H3K4me3 and DNA hypermethylation are mutually exclusive: DNMTs only recognize sites with unmethylated H3K420, 21 and loss or gain of H3K4 methylation is a strong predictor of promoter DNA methylation status.12 As such, measuring the H3K4me3 status in our original HCT116 cells and the corresponding DNMT double knock out line would provide an independent measure of promoter activity (and DNA methylation status) to our expression Beadchips. Thus, we next generated genome-wide maps of H3K4me3 in both HCT116 and DKO cells by chromatin immunoprecipitation combined with high-throughput sequencing (ChIP-seq). A total of 11 747 and 13 074 high confidence H3K4me3 sites were identified in HCT116 and DKO cells, respectively, within 1.0 kb of a TSS (using a false discovery rate (FDR) of 1%). Comparing the two data sets revealed 2507 gene promoters that lacked H3K4me3 in HCT116 cells but gained this mark in the DKO cells (Figure 1b and Supplementary Table S2).
To distinguish the most robust gene set that appeared to be silenced in colon cancer cells by DNA methylation, we next identified the overlapped genes between the 476 silenced genes identified by expression Beadchips and the 2075 genes bound by H3K4me3 ChIP-seq. These H3K4me3-negative and expression-negative genes in parental HCT116 cells consisted of 203 genes (Figure 1c and Supplementary Table S3) and they are re-expressed in DKO cells with a gain of H3K4me3. For convenience, we define these genes as colon cancer methylation silenced (CMS) genes for the remainder of the paper. Figure 1d illustrates two CMS genes (SFRP1 and CDH2) to exemplify such changes in DNA methylation and H3K4me3 in HCT116 and DKO cells. We also randomly validated an additional set of CMS genes by RT-PCR and methylation-specific PCR (MSP) (see Supplementary Figures S1A and S1B). In each case, the gene was verified to be promoter methylated and silenced in HCT116 cells but became demethylated and re-expressed in DKO cells, validating the accuracy of this genomic approach.
Identification of CMS genes associated with ES cell differentiation
It has been reported that genes identified as having methylated promoters in cancer, in ES cells, contain repressive histone marks.16, 17, 18 The expression of these PRC2 target genes is repressed in ES cells and only activated upon induction of differentiation.15, 22 This link prompted us to investigate the possibility that our CMS genes may represent a set of genes involved in a differentiation program that is subsequently lost/repressed in human cancer. To determine if any of our CMS genes are repressed in human ES cells and activated upon differentiation, we examined gene expression in undifferentiated human ES cells to human ESCs induced to differentiate along the trophoblast lineage. In this differentiation system, hESC pluripotency regulators Oct4, Sox2 and Nanog are significantly downregulated within 96 h (Figure 2a and Supplementary Table S4) and markers of the trophoblast lineage (e.g., CDX2, GATA3, GATA2 and TFAP2A) are greatly upregulated (data not shown). Of 203 CMS genes, 40 were absent or detectable at very low levels in the undifferentiated human ES cells but were upregulated upon differentiation (Figure 2a). We next compared these gene lists with earlier studies including ours characterizing the bivalently marked genes (H3-K4me3 and H3-K27me3) in undifferentiated hESCs.10, 22, 23 Of the 40 CMS genes identified to be upregulated upon differentiation of hESCs, 21 of these also contained the bivalent marks in undifferentatiated hESCs (Figure 2b). Among these 21 genes associated with ES cell differentiation, HAND1, a transcription factor involved in the regulation of development, showed the highest level of induction during differentiation (Figure 2a). Thus, of our initial 203 colon cancer DNA methylation repressed genes, 21 of these contain repressive histone marks in undifferentiated human ESCs and are subsequently activated upon differentiation. Of particular interest is HAND1, one of only two transcription factors in this 21 gene list and the most upregulated in the hESC differentiation system.
CMS genes associated with ES cell differentiation. (a) Gene clustering showing the gene expression profiles of CMS genes during retinotic acid (RA)-induced differentiation in human ES cells. Only genes with twofold or greater change in at least one time point during differentiation are shown. Self-renewing factors, OCT4, SOX2 and NANOG2, were included in this analysis as experimental control. The genes were rank ordered in terms of fold-changes. In total, 40 CMS genes were found to be associated with differentiation and 68 genes were found to be associated with self-renewal. Hand1 was ranked ordered as top CMS gene associated with differentiation. (b) Listed are 21 PRC2 target genes previously identified in ES cells
Removal of DNA methylation promotes EZH2 recruitment to a small set of CMS genes, including HAND1
We next wished to determine whether differentiation-associated CMS genes are also PcG targets in adult colon cancer cells. It has been previously shown that PRC2 protein EZH2 can directly control DNA methylation for gene silencing at certain gene promoters.24 However, a recent genome-wide analysis of chromatin patterns in human breast and prostate cancer cells does not seem to support a mechanistic link between the two silencing events.9 Moreover, a recent study in colon cancer cells suggests that removal of DNA methylation does in fact promote a gain of bivalency in methylated gene promoters.11 Thus, the observed bivalency may be associated with enhanced EZH2 recruitment upon loss of DNA methylation in colon cancer cells.
To test this hypothesis, we generated genome-wide maps of EZH2 binding in both HCT116 and DKO cells. Of the 203 CMS genes identified above, only 20 genes (< 10%) were found to be bound by EZH2 in HCT116 cells (FDR 5%, within in 2.0 kb of TSS site) (data not shown). This finding is consistent with the recent genome-scale analysis showing DNA hypermethylated promoters are largely not marked by H3K27me3,9 further confirming these two silencing events are mechanistically separable in colon cancer. In DKO cells, however, we detected 39 out of 203 CMS genes displaying a marked increase in EZH2 binding and 6 of these, including HAND1, were associated with differentiation in ES cells (Figure 3a). We confirmed the EZH2 and H3K4me3 binding profiles in four of these genes (in addition to two control genes) by ChIP-PCR in both HCT116 and DKO cells. This showed a marked increase in enrichments of both EZH2 and H3K4me3 in DKO cells compared with HCT116 cells (Figure 3b). As a control, CDH2 and SFRP1 were only weakly bound by EZH2 in both cell lines (Figure 3b). These data indicate that, by enhanced recruitment of EZH2, a subset of CMS genes remain repressed in the DNA methylation-defective DKO cells (Figure 3c), suggesting an additional mechanism to prevent their re-expression. By contrast, CMS genes that do not show an enhanced EZH2 binding in DKO cells (such as CDH1 and SFRP1) appear to be sufficiently re-expressed with an open chromatin carrying mainly H3K4me3 (Figure 3b, right panel; and Figure 3c).
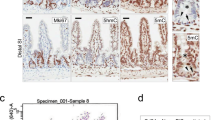
A subset of CMS genes are enriched with EZH2 in the absence of DNA methylation in colon cancer cells. (a) Box plot showing the enhanced EZH2 enrichment (P<0.003) in six differentiation-associated CMS genes in DKO cells compared with HCT116 cells based on EZH2 ChIP-seq data. (b) ChIP-PCR validation of four CMS genes showing the EZH2 and H3K4me3 enrichments in DKO cells compared with HCT116 cells. SFRP1 and CDH2 were included as control showing no different EZH2 enrichments between the two cell lines. (c) Differential expression levels of six CMS genes in HCT116 and DKO cells. Although CDH2 and SFRP1 were fully re-expressed in DKO cells, HAND1, EPHB1, KCNK12 and COLEC12 remained repressed in DKO cells. (d) Schematic representation of HAND1 locus. The locations of two MSP regions analyzed in HCT116 and DKO cells are indicated. (e) Left panel: MSP showing hypermethylated HAND1 promoter in HCT116 cells but demethylated in DKO cells. M1 and M2 indicate the distal and the immediate HAND1 promoter regions, respectively. Right panel: two indicated genomic regions of HAND1 promoter from HCT116 and DKO cells analyzed using bisulfite genomic sequencing (BGS). (f) RT-PCR analysis of HAND1 expression in normal colon tissue and a panel of colorectal cancer cell lines. GADPH was used as PCR control. (g) MSP analysis of HAND1 promoter in a series of colon cancer cell lines. M1 and M2 as indicated in (E). (h) ChIP-PCR detection of H3K4me3 and H3K27me3 at HAND1 locus in HCT116 and DKO cells. Location of ChIP-qPCR fragment covering the −4.0 to +4.0-kb genomic region surrounding the TSS of HAND1 were indicated with numbers
To further illustrate the epigenetic events in detail, we chose to focus on HAND1. Both MSP and bisulfite sequencing analysis detected the hypermethylated DNA in the distal and immediate promoter regions of HAND1 in HCT116 but not in DKO cells (Figure 3d and e), as well as in other colon cancer cell lines as validated by RT-PCR (Figure 3f) and MSP (Figure 3g). Furthermore, in striking contrast to HCT116 cells, HAND1 in DKO cells exhibited marked enrichments of both H3K4me3 and H3K27me3, clearly indicating a bivalent chromatin surrounding the TSS (Figure 3h). Thus, the gain of this bivalency of HAND1 in DKO cells may be responsible for its remained repression in these cells.
Pharmacological reactivation of HAND1 in colon cancer cells
We have previously shown that histone methylation inhibitor Deazaneplanocin A (DZNep) is able to inhibit histone methylations, such as H3K27me3 and H4K20me3, and reverses EZH2-mediated gene silencing in cancer cells.25 Moreover, genes silenced by bivalent histone modifications, such as DACT3 or CDKN1C, but not those silenced by DNA hypermethylation, are strongly reactivated by DZNep in combination with histone deacetylase inhibitor Trichostatin A (TSA).6, 26 Accordingly, HAND1, whose expression was only slightly increased in DKO cells, was strongly induced by combined treatment of DZNep and TSA and this was not seen in the parental HCT116 cells (Figure 4a, left panel). By contrast, the expression of SFRP1, which was not an EZH2 target and was already strongly re-expressed in DKO cells, was not further induced by such a combination treatment (Figure 4a, right panel). ChIP analysis indicated a decreased recruitment of both EZH2 and H3K27me3 to HAND1 and a concomitant further increase in H3K4me3 in DKO cells treated with the DZNep/TSA combination but not in HCT116 cells (Figure 4b). These findings suggest that HAND1 promoter demethylation in DKO cells is insufficient to give rise to a full HAND1 re-expression due to a gain of bivalent chromatin modifications but make it render to the DZNep/TSA treatment that is able to target bivalency. Consistently, pre-treatment of HCT116 cells with the DNA methylation inhibitor 5-Aza-dC, followed by DZNep/TSA treatment resulted in a much greater level of HAND1 induction, compared with treatment with 5-Aza-dC or DZNep/TSA alone (Figure 4c). A similar result was also obtained in another colon cancer cell line DLD1 in which the HAND1 promoter was also found to be hypermethylated (Figures 4d and 3g).
Pharmacological reactivation of HAND1 in colon cancer cells. (a) qRT-PCR analysis showing the expression of HAND1 and SFRP1 mRNA in HCT116 and DKO cells treated with 5 μM DZNep, 100 nM TSA or their combination. (b) ChIP-PCR showing the changes of EZH2, H3K27me3 and H3K4me3 in HCT116 and DKO cells with or without DZNep plus TSA treatment. (c and d) qRT-PCR analysis of HAND1 mRNA expression in HCT116 and DLD1 cells following the indicated drug treatments. (e) qRT-PCR analysis of HAND1 mRNA expression in SW480 cells following the indicated drug treatments. (f) ChIP-PCR analysis indicated the changes of H3K4me3 and H3K27me3 in SW480 cells with or without DZNep plus TSA treatment. Error bars indicate±S.D.
Unlike HCT116 and DLD1 cells, HAND1 in SW480 cells was found to be hypermethylated in a distal M1 region, but not in the immediate promoter region (M2) (Figure 3g). In SW480 cells, HAND1 expression was strongly induced by DZNep/TSA treatment (Figure 4e). Accordingly, HAND1 in these cells was found to be bivalent and carries both H3K4me3 and H3K27me3, which was disrupted by DZNep/TSA (Figure 4f), a result similar to DKO cells. Thus, HAND1 in colon cancer is silenced by either DNA methylation or Polycomb-mediated histone methylation or both, depending on the cell type.
Differentiation regulator HAND1 is silenced in multiple human malignancies
Among 40 CMS genes associated with ES differentiation, HAND1 ranks as the most downregulated in primary colon tumor samples compared with the normal controls in 90% of the primary colon tumors analyzed (Figure 5a). This striking downregulation of HAND1 in colon cancer was further confirmed by RT-PCR analysis using randomly selected colon tumor samples and matched normal tissues (Figure 5b), which was consistent with the hypermethylation of HAND1 promoter, as determined by MSP and bisulfite sequencing, in the primary colon tumors but not in the normal controls (Figures 5c and d). In addition, Oncomine analysis revealed the downregulation of HAND1 in a variety of other human tumors, including bladder, brain and cervix (Figure 5e), indicating that a loss of HAND1 expression may be involved in tumorigenesis of many cancers.
HAND1 is silenced in multiple human cancers. (a) Expression profiles of 40 differentiation-related CMS genes in human colorectal tumors and matched normal mucosa. Genes are rank ordered in terms of fold expression between tumor and normal samples. (b) RT-PCR analysis of HAND1 expression from eight randomly selected pairs of human colorectal tumor (T) and matched mucosa (N). β-Actin was used as PCR control. (c) MSP analysis of HAND1 promoter in eight selected tumors. (d) Bisulfite sequencing of HAND1 promoter from normal colon epithelium and tumor. (e) HAND1 is significantly suppressed in multiple-type of cancer. Representative data are shown across multiple independently published microarray studies as indicated. P-values are shown below
Restoration of HAND1 expression in colon cancer induces terminal differentiation, inhibits cell proliferation and prevents xenograft tumor formation
To investigate the function of HAND1 in colon cancer, we ectopically expressed HAND1 in colon cancer cell lines HCT116 and DLD1 (Supplementary Figure S2A). Overexpression of HAND1 resulted in marked inhibition of colony formation (Figure 6a) and anchorage-independent growth in soft agar (Figure 6b). Ectopic HAND1 was also able to abolish the transformation of mouse embryonic fibroblast cells induced by Ras/E1A oncoproteins (Figure 6c). These results supported a role of HAND1 in suppressing cancer cell growth and cellular transformation.
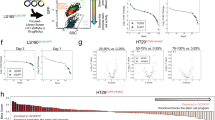
Restoration of HAND1 disrupts cancer phenotypes. (a) HCT116 and DLD1 colony formation at day 12. Cell colonies were stained with 0.5% gentian violet. (b) Anchorage-independent growth assessed by soft agar assay. (c) HAND1 inhibits RAS/E1A induced transformation in MEFs. (d) DLD1 growth curve. Dox treatment resulted in growth inhibition of HAND1-expressing DLD1 cell (lower panel) but not the control cells (upper panel). (e) Dox treatment resulted in anchorage-independent growth in DLD1 cells expressing Hand1. (f) Morphologies of above cells upon exposure to Dox for 9 days. (g) FACS showing the decreased BrdU staining and accumulation of tetroploid cells at day 9 following inducible Hand1 expression. (h) FACS analysis showing the induced HAND1 expression resulted in caspase 3 activation at day 12. FSC-H means forward scattered height. (i) DLD1 cells infected with pMN-GFP-HAND1 retrovirus were unable to develop tumors in nude mice, as shown by representative mice and tumors through GFP imaging (left). Cells infected with retrovirus pMN-GFP (vector control) and pMN-GFP-Hand1 (Hand1) were implanted into right and left flank, respectively, and measure the tumor growth for indicated times (n=8, error bars indicate±S.D.) (right panel)
To further characterize HAND1 as a tumor suppressor, we generated a stable line of DLD1 expressing a doxycycline (Dox)-inducible HAND1 (Supplementary Figure S2B). Induction of HAND1 expression resulted in marked inhibition of both proliferation and anchorage-independent growth (Figures 6d and 6e). Notably, HAND1-expressing DLD1 cells maintained in Dox progressively displayed morphological changes characteristic of differentiation, including giant cell phenotype, flat and elongated morphology and multinucleate formation, which was not seen in the control (Figure 6f). In contract, another tumor-suppressor candidate THY1, whose expression was also silenced by DNA methylation in colon cancer cells (Supplementary Table S1), did not induce such morphological changes, despite the proliferation inhibition (Supplementary Figures S2C–F). Moreover, DLD1 cells expressing HAND1 lost proliferation over time, by measuring the proportion of cells in S phase with incorporation of bromodeoxyuridine (BrdU), which was concomitant with marked accumulation of tetraploid cells (Figure 6g) and multinucleate formation observed under microscopy. Of note, the above phenotype induced by HAND1 in colon cancer cells resembles its established role in promoting mouse trophoblast stem cell differentiation toward giant cells undergoing endoreduplication.27, 28, 29 Ultimately, prolonged exposure of HAND1-expressing DLD1 cells to Dox for over 2 weeks led to massive apoptosis upon detachment from matrix, as indicated by marked activation of caspase-3 (Figure 6h).
Next, we investigated the effect of HAND1 in vivo. DLD1 cells expressing GFP-GFP-HAND1 exhibited a similar morphological change of terminal differentiation (Supplementary Figure S3). Although the control cells expressing GFP only developed subcutaneous tumors progressively (8 out of 8 inoculations), cells expressing GFP-HAND1 did not (0 out of 8 inoculations) (Figure 6i). Thus, HAND1 functions as a tumor suppressor with a remarkable ability to disrupt cancer phenotypes and loss of HAND1 may be a crucial epigenetic event contributing to tumorigenesis.
To investigate the molecular mechanism by which HAND1 induces growth inhibition and differentiation, we compared the gene expression profiles of DLD1 GFP and DLD1 GFP-HAND1, and identified 145 genes (using threefold cutoff) as induced by HAND1 (Supplementary Table S5). As expected, these genes were also found to be downregulated in colon tumors, as shown through the Gene set enrichment analysis (GSEA) (Figure 7a).
HAND1-associated gene signature in vitro and in vivo. (a) GSEA of gene expression in human colon tumor versus normal control using the 145 HAND1-associated gene set. Left: GSEA enrichment plot. Right: the top 50 genes showing decreased expression in human colon tumors as compared with the normal controls. (b) Gene clustering showing the upregulation of 145 genes in HAND1-overxpressing DLD1 cells as compared with the vector (PMN) control cells. DHRS9 was found to be most increased. Right, bar graph showing a 256-fold induction of DHRS9 in HAND1-expressing cells. (c) Downregulation of DHRS9 in human colon tumors as compared with the normal tissues. (d) Schematic representation of the human DHRS9 promoter. The promoter constructs used in this study are indicated (upper panel). HCT116 cells were transfected with PGL3-basic, and HAND1 (100 ng), together with a luciferase reporter construct containing the DHRS9 promoter (–1989/–950) or (−1061/+451). Relative luciferase activities were measured 48 h after transfection. Results are depicted as fold induction, after normalization to Renilla luciferase activity (bottom panel). (e) DLD1 colony formation with or without DHRS9 expression. Cell colonies were stained with 0.5% gentian violet. (f) Soft agar assay of DLD1 cells expressing pMN or pMN-DHRS9
Among HAND1-associated genes, we identified DHRS9 as the most induced gene (210-fold induction; Figure 7b). DHRS9 encodes a retinol-metabolizing enzyme (retinol dehydrogenase) functionally implicated in retinoid-induced colon epithelial cell differentiation,30, 31 and is downregulated in less differentiated colon carcinomas.30, 32 Indeed, we found that DHRS9 was consistently downregulated in colon tumors (Figure 7c). In addition, ectopic HAND1 was able to activate the DHRS9 promoter activity (Figure 7d), indicating DHRS9 as a transcriptional target of HAND1. Like HAND1, DHRS9 overexpression in DLD1 cells was able to reduce both the anchorage-dependent and -independent growth of these cells (Figures 7e and f). These findings suggest that tumor suppression function of HAND1 may be at least partially mediated through DHRS9.
Discussion
Although PRC2 is implicated in ES cell maintenance by repressing differentiation-associated genes22, 33 and some of these genes are targeted by DNA hypermethylation for gene silencing during oncogenesis,16, 17, 18 a functional link between genes important in (ES) cell differentiation and tumorigenesis yet to be established. In this study, we identify a Polycomb-targeted differentiation program transcriptionally inactivated in human colorectal cancer and show that restoration of a key component of this program, HAND1, is able to induce colon cancer cell terminal differentiation and apoptosis. Thus, we propose that a HAND1-associated differentiation program acts to limit cancer development and thus by extension, provides a functional link between gene silencing, maintenance of proliferation, repressed differentiation by PRC2, in the context of tumorigenesis. Our study provides the first detailed description of temporal, bivalent DNA methylation and Polycomb silencing of a known (ES cell) differentiation gene in the context of tumor differentiation and progression.
A previous study suggests a similarity between the transcriptional signatures of poorly differentiated tumors to that of ES cells and this similarity extends to PRC2 target genes.34 Although DNA methylation events in cancer have recently been suggested to be linked to a Polycomb-impeded differentiation program, a fundamental issue that remains unresolved is the mechanism by which the developmental program is inactivated during cancer initiation and development and whether it has a tumor-suppressor role. Our work here suggests that the mechanism involves epigenetic regulation of specific developmental regulatory genes, genes that are switched off and associated with repressive histone marks in ES cells, activated upon differentiation, but inactivated in adult cancer cells. More precisely, we found that the bivalency of a Polycomb target gene identified in ES cells, HAND1, is lost during cancer progression through the acquisition of promoter DNA hypermethylation resulting in gene silencing. This observation is consistent with the recent proposal that cancers arise from tumor-initiating stem cells that may share similar epigenetic properties with ES cells.34, 35, 36
Although genes silenced by DNA hypermethylation in colon cancer have been well documented previously by several labs,37, 38, 39, 40 the main aim of our study is to functionally investigate its relationship with EZH2-mediated H3K27me3 in the context of cell differentiation and cancer development. Our data suggest a mutually exclusive nature of EZH2 binding with DNA methylation in at least some of the DNA methylated genes. This observation is in agreement with several recent reports indicating the two silencing pathways are largely independent,6, 9, 25 although this contrasts with a previous report showing that EZH2 directly controls DNA methylation.24 It is also consistent with the previous observation that inhibition of DNA methylation facilitates the formation of bivalency and hence increasing the efficiency of reprogramming to a pluripotent state.12
This information is critical in designing epigenetic therapy as distinct pharmacologic approaches are required to target these two separate pathways.6, 25 At the mechanistic level, we show that EZH2 and H3K27me3 are minimally enriched in the hypermethylated HAND1 promoter, but their enrichments are increased when the promoter becomes demethylated. Similarly, HAND1 is also a Polycomb and H3K27me target in human ES cells where its promoter is unmethylated.10 This reversibility may suggest a model in which the two silencing events may participate in HAND1 inactivation in different stages of colon tumorigenesis. Although a previous study shows that HAND1 promoter in colon cancer carries both PRC2 and DNA methylation,41 it did not address the role of PRC2 and H3K27me3 in regulating HAND1 expression both before and after promoter demethylation. In fact, our result shows that HAND1 promoter demethylation is insufficient to fully reactivate HAND1 expression because of enhanced engagement of PRC2, resulting in a bivalent histone methylations, which, however, is sensitive to the combination treatment of histone modifying compounds DZNep and TSA. We speculate that many cancer genes carrying bivalent epigenetic configuration such as HAND1 may be susceptible to the pharmacologic perturbation with combinatorial epigenetic treatment as described herein.
Materials and Methods
Tissue samples, cell lines and drug treatment
Twenty-four pairs of colon tumor and matched normal colon tissue samples were obtained from Singapore Tissue Network using protocols approved by Institutional Review Board of National University of Singapore. Study samples were collected with informed patient consent. All patients had surgery as primary treatment and all samples were kept in liquid nitrogen until RNA extraction. The colorectal cancer cell lines DLD1, RKO, HT29, SW480 and HT15 were purchased from the American Type Culture Collection (Manassas, VA, USA). HCT116 cells with genetic disruption of DNMT1 and DNMT3B (HCT116 DKO) were kindly provided by Dr. Bert Vogelstein (Johns Hopkins University, Baltimore, MD, USA). Cells were treated with 5 μM 3-DZNep (obtained from National Cancer Institute, Bethesda, MD, USA) or 5 μM 5-Aza-dC (Sigma, St. Louis, MO, USA) for 72 h and TSA (Cell Signaling, Beverly, MA, USA) at 100 nM for 24 h. For 5-Aza-dC treatment, the medium was replaced with freshly added 5-Aza-dC for every 24 h. For co-treatment of cells with DZNep and TSA, DZNep was added for 24 h followed by TSA for additional 24 h.
The hESC cell line WA01 (WiCell H1) was maintained in feeder-free conditions. Media containing 80% DMEM/F12 (Gibco, Grand Island, NY, USA) supplemented with 20% KNOCKED OUT serum replacement (Gibco), 1 mM L-glutamine (Gibco), 0.1 mM nonessential amino acids (Gibco) and 0.1 mM β-mercaptoethanol (Sigma) were conditioned on inactivated mouse embryonic fibroblasts and subsequently supplemented with 8 ng/ml FGF2 (Invitrogen, Grand Island, NY, USA) before use in hESC culture. hESC were induced to differentiate along the trophoblast lineage by a combined treatment of 100 ng/ml BMP4 (R&D Systems, Minneapolis, MN, USA) and 20 μM SU5402 (Calbiochem, San Diego, CA, USA).
Expression vectors and stable cell lines
HAND1 ORF was PCR amplified from normal colon tissue cDNA and was first subcloned into p3 × FLAG-CMV10 and then into pcDNA4/myc-hisB (Invitrogen) for transient transfection and pcDNA4/TO/Flag-HAND1 for inducible expression. Retroviral-mediated gene transfer was performed using pMN-GFP/IRES retrovirus vector-expressing Hand1. Infected DLD1 cells were sorted by GFP signals and expanded for in vitro and in vivo studies. To generate the Hand1-inducible cell line, DLD1 stable cell line expressing pcDNA6-TR (Invitrogen) and selected with Blasticidin (10 μg/ml) was first isolated according to the protocols of T-Rex system kit (Invitrogen), and then transfected with the pcDNA4/TO/Flag-HAND1 and selected for Zeocin (100 μg/ml). The generated clones were screened for Dox-inducible expression of Flag-HAND1. For Dox treatment, inducible cells were treated with Dox to a final concentration of 1 μg/ml.
Microarray gene expression analysis, semiquantitative RT-PCR and Taqman assay
Total RNA was isolated using Trizol (Invitrogen) and purified with the RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany). Reverse transcription was performed using an RNA Amplification kit (Ambion, Grand Island, NY, USA). The microarray hybridization was performed using the Illumina Gene Expression Sentrix BeadChip HumanRef-8_V2, and data analysis was performed using GeneSpring software from Agilent Technologies (Santa Clara, CA, USA) as described.6 For RT-PCR, total RNA was reverse-transcribed using oligo(dT)12-18 primer with Superscript II reverse transcriptase (Invitrogen). One hundred nanograms of cDNA was used for PCR and the primer sequences are shown in Supplementary Table 6. Quantitative real-time PCR was performed on a PRISM 7900 Sequence Detection System (Applied Biosystems, Grand Island, NY, USA) using TaqMan probes (Applied Biosystems). Samples were normalized to the levels of GAPDH mRNA. GSEA of gene expression levels were analyzed with the GSEA2.0 software package (http://broad.mit.edu/tools/software.html).
DNA methylation analysis
The CpG island DNA methylation status was determined by PCR analysis after bisulfite modification of genomic DNA and followed by MSP and bisulfite genomic sequencing as previously described.6 Primer sequences and conditions for MSP and bisulfite sequencing PCR are shown in Supplementary Table 7.
ChIP assays
ChIP assays were performed as described previously.6 The immunoprecipitated DNA was quantitated by real-time quantitative PCR using Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems). We used the following antibodies in the ChIP study: anti-H3K27m3 (Upstate, Lake Placid, NY, USA), anti-H3K4me3 (Upstate) and anti-EZH2 (Active-motif, Carlsbad, CA, USA). The enrichments of these histone marks at the examined regions were normalized to a genome control region.6, 42 The sequences of the primer sets are shown in Supplementary Table 8.
Luciferase reporter assay
Genomic DNA containing the human DHRS9 promoter regions −1989/−950 and −1061/+451 were cloned into pGL3-luciferase construct (Promega, Madison, WI, USA). Luciferase assays were performed using the Dual Luciferase system (Promega). HCT116 cells were plated at a density of 3 × 105 cells per well of a 12-well plate and transfected with a mixture containing luciferase reporter plasmid, HAND1 expression vector. Forty-eight hours after transfection, the luciferase activities were analyzed using the Dual Luciferase system.
ChIP-seq
Purified immunoprecipitated DNA was prepared for sequencing according to a modified version of the solexa Genomic DNA protocol. Fragmented DNA was end paired and subjected to 15 cycle of linker-mediated-PCR using Oligos purchased from Illumina (San Diego, CA, USA). Amplified fragments between 200 and 300 bp were isolated by agarose gel electrophoresis and purified. The fragment DNA size was confirmed by Agilent DNA chip analysis. Ten nanograms of linker-ligated DNA was applied to the flow cell using the Solexa Cluster Station fluidics device (Illumina). Samples were then subjected to 36 bases of sequencing according to Illumina’s standard protocols. The data generated from Solexa Genome Analyzer instrument underwent ChIP-seq pipeline, managed by the Research Computing group.
Cell proliferation assay, apoptosis and flow cytometric analysis
Cell growth was measured with CellTiter-Glo Luminescent Cell Viability Assay (Promega). In brief, 1000 cells were seeded in 96-well plate and the cell number was measured following the manufacturer’s instructions. To measure caspase-3 activity, cells were harvested and fixed with Cytofix/Cytoperm solution (BD Biosciences, Franklin Lakes, NJ, USA) as instructed after treatment with Dox for 9 days and then stained with fluorescein isothiocyanate-conjugated rabbit anti-active caspase-3 monoclonal antibody (BD Biosciences). Quantification of cells positive for the caspase-3 was performed by flow cytometry.
For BrdU incorporation assay, cells were treated as above and incubated with BrdU reagent for 6 h before harvesting cells. Cells were fixed and stained with the BrdU antibody and 7-AAD according to the manufacturer’s protocol (BD Biosciences). Stained cells were analyzed by FACScalibur (BD Biosciences) and quantified by using CellQuest software (BD Biosciences).
Colony formation and soft-agar assay
Cells were plated at 3.0 × 105 per well using six-well plates, and transfected with either pcDNA4-HAND1 or empty vector (2.0 μg) using Fugen 6 (Roche, Indianapolis, IN, USA). The cells were replated in triplicates and cultured for 10–15 days in complete DMEM medium containing Zeocin (100 μg/ml) and stained with Gentian Violet after methanol fixation. For anchorage-independent growth, cells were transfected as above, and suspended in DMEM containing 0.3% agar, 10% fetal bovine serum, and layered on DMEM containing 0.6% agar, 10% FBS and Zeocin (100 μg/ml) in six-well plate at 48 h post-transfection. Colony formation was assessed at 4 weeks post-transfection.
Tumors and animal models
Eight female athymic BALB/c nude mice (5- to 8-week-old) were housed in the Biological Resource Centre. Mice were implanted subcutaneously in both sides of flank with 5 × 105 of human colon cancer DLD1 cells infected with either pMN-GFP-HAND1 construct or empty pMN-GFP retroviral vector, respectively. Tumor progression was monitored by whole-body GFP imaging by using Olympus OV100 Small Animal Imaging System (Shinjuku-ku, Tokyo, Japan). All mouse procedures were pre-approved by the Institutional Animal Care and Use Committee (Singapore) and performed according to institutional policies.
Abbreviations
- DNMT:
-
DNA methyltransfease
- DZNep:
-
Deazaneplanocin A
- ES:
-
embryonic stem
- PRC2:
-
Polycomb repressor complex 2
References
Cao R, Zhang Y . The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 2004; 14: 155–164.
Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–629.
Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 2003; 100: 11606–11611.
Kirmizis A, Bartley SM, Farnham PJ . Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol Cancer Ther 2003; 2: 113–121.
Yu J, Cao Q, Mehra R, Laxman B, Tomlins SA, Creighton CJ et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell 2007; 12: 419–431.
Jiang X, Tan J, Li J, Kivimae S, Yang X, Zhuang L et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 2008; 13: 529–541.
Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell 2007; 11: 513–525.
Chen H, Tu SW, Hsieh JT . Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem 2005; 280: 22437–22444.
Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 2008; 40: 741–750.
Zhao X, Han X, Chew J, Liu J, Chiu K, Choo A et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 2007; 3: 286.
McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W et al. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res 2008; 68: 5753–5759.
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454: 766–770.
Tenen DG . Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer 2003; 3: 89–101.
Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125: 315–326.
Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006; 441: 349–353.
Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 2007; 39: 237–242.
Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007; 39: 232–236.
Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C et al. Epigenetic stem cell signature in cancer. Nat Genet 2007; 39: 157–158.
Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007; 448: 553–560.
Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007; 39: 457–466.
Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007; 448: 714–717.
Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006; 125: 301–313.
Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 2007; 1: 299.
Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006; 439: 871–874.
Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 2007; 21: 1050–1063.
Yang X, Karuturi RK, Sun F, Aau M, Yu K, Shao R et al. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS One 2009; 4: e5011.
Martindill DM, Risebro CA, Smart N, Franco-Viseras Mdel M, Rosario CO, Swallow CJ et al. Nucleolar release of Hand1 acts as a molecular switch to determine cell fate. Nat Cell Biol 2007; 9: 1131–1141.
Cross JC, Flannery ML, Blanar MA, Steingrimsson E, Jenkins NA, Copeland NG et al. Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development 1995; 121: 2513–2523.
Scott IC, Anson-Cartwright L, Riley P, Reda D, Cross JC . The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol Cell Biol 2000; 20: 530–541.
Jette C, Peterson PW, Sandoval IT, Manos EJ, Hadley E, Ireland CM et al. The tumor suppressor adenomatous polyposis coli and caudal related homeodomain protein regulate expression of retinol dehydrogenase L. J Biol Chem 2004; 279: 34397–34405.
Jones RJ, Dickerson S, Bhende PM, Delecluse HJ, Kenney SC . Epstein-Barr virus lytic infection induces retinoic acid-responsive genes through induction of a retinol-metabolizing enzyme, DHRS9. J Biol Chem 2007; 282: 8317–8324.
Duester G . Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem 2000; 267: 4315–4324.
Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K . The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 2007; 27: 3769–3779.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507.
Reya T, Morrison SJ, Clarke MF, Weissman IL . Stem cells, cancer, and cancer stem cells. Nature 2001; 414: 105–111.
Stingl J, Caldas C . Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer 2007; 7: 791–799.
Chan TA, Glockner S, Yi JM, Chen W, Van Neste L, Cope L et al. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med 2008; 5: e114.
Gius D, Cui H, Bradbury CM, Cook J, Smart DK, Zhao S et al. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell 2004; 6: 361–371.
Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 2007; 3: 1709–1723.
Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet 2001; 10: 3001–3007.
Jin B, Yao B, Li JL, Fields CR, Delmas AL, Liu C et al. DNMT1 and DNMT3B modulate distinct polycomb-mediated histone modifications in colon cancer. Cancer Res 2009; 69: 7412–7421.
Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133: 1106–1117.
Acknowledgements
We thank Dr. Bert Vogelstein for the HCT116 DKO cells. We thank Zhuang Li for performing the microarray hybridization. We thank the Singapore Tissue Network for providing the human samples. This work was supported by the Agency for Science, Technology and Research of Singapore.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by D Heery
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Tan, J., Yang, X., Jiang, X. et al. Integrative epigenome analysis identifies a Polycomb-targeted differentiation program as a tumor-suppressor event epigenetically inactivated in colorectal cancer. Cell Death Dis 5, e1324 (2014). https://doi.org/10.1038/cddis.2014.283
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.283
This article is cited by
-
Game-theoretic link relevance indexing on genome-wide expression dataset identifies putative salient genes with potential etiological and diapeutics role in colorectal cancer
Scientific Reports (2022)
-
Transcriptome profiling unveils GAP43 regulates ABC transporters and EIF2 signaling in colorectal cancer cells
BMC Cancer (2021)
-
An embedded method for gene identification problems involving unwanted data heterogeneity
Human Genomics (2019)
-
Prognostic value of histone marks H3K27me3 and H3K9me3 and modifying enzymes EZH2, SETDB1 and LSD-1 in colorectal cancer
Journal of Cancer Research and Clinical Oncology (2018)
-
The role of EZH2 in overall survival of colorectal cancer: a meta-analysis
Scientific Reports (2017)