Abstract
The disruption of DNA replication in cells triggers checkpoint responses that slow-down S-phase progression and protect replication fork integrity. These checkpoints are also determinants of cell fate and can help maintain cell viability or trigger cell death pathways. CHK1 has a pivotal role in such S-phase responses. It helps maintain fork integrity during replication stress and protects cells from several catastrophic fates including premature mitosis, premature chromosome condensation and apoptosis. Here we investigated the role of CHK1 in protecting cancer cells from premature mitosis and apoptosis. We show that premature mitosis (characterized by the induction of histone H3 phosphorylation, aberrant chromatin condensation, and persistent RPA foci in arrested S-phase cells) is induced in p53-deficient tumour cells depleted of CHK1 when DNA synthesis is disrupted. These events are accompanied by an activation of Aurora kinase B in S-phase cells that is essential for histone H3 Ser10 phosphorylation. Histone H3 phosphorylation precedes the induction of apoptosis in p53−/− tumour cell lines but does not appear to be required for this fate as an Aurora kinase inhibitor suppresses phosphorylation of both Aurora B and histone H3 but has little effect on cell death. In contrast, only a small fraction of p53+/+ tumour cells shows this premature mitotic response, although they undergo a more rapid and robust apoptotic response. Taken together, our results suggest a novel role for CHK1 in the control of Aurora B activation during DNA replication stress and support the idea that premature mitosis is a distinct cell fate triggered by the disruption of DNA replication when CHK1 function is suppressed.
Similar content being viewed by others
Main
To ensure genetic integrity, cells have elaborate mechanisms that maintain the accuracy of DNA replication and correct errors induced by DNA damage. Damage response pathways efficiently recognize disruptions of DNA replication or structure, arrest cell cycle progression and recruit appropriate repair complexes to preserve the accuracy of genetic information. However, if damage persists they may also commit cells to death. The PIK-like protein kinases Ataxia-telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) are major coordinators of the damage response.1 ATM delays DNA replication and the onset of mitosis primarily in response to DNA ends generated by double-strand breaks induced by DNA-damaging agents such as ionizing radiation through a complex signalling cascade including p53, CHK2 and the MRE11-NBS1-RAD50 complex as phosphorylation targets.2 This signalling cascade may also trigger apoptosis through p53-mediated transcriptional activation of pro-apoptotic genes such as BAX and PUMA.3, 4
ATR and its downstream phosphorylation target CHK1 are largely activated as a result of single-strand (ss) DNA formation that occurs during disruption of DNA replication. DNA lesions or disruptions of DNA precursor supply can lead to an uncoupling of the replication and helicase complexes to produce ssDNA.5 ssDNA is rapidly coated by the replication protein A (RPA) complex that, in turn, recruits ATR through the ATR-interacting protein (ATRIP).6 The formation of this complex drives the activation of CHK1 through phosphorylation at Ser345 and Ser317.7, 8 Activated CHK1 then plays pivotal roles in coordinating cellular responses to replication stress, slowing S-phase progression by suppressing inappropriate firing of replication origins, maintaining fork integrity, facilitating resolution of stalled forks and triggering G2/M arrest.9, 10, 11, 12
DNA damage response pathways are frequently mutated in individuals with inherited forms of cancer predisposition,1 and there is ample evidence that these changes accelerate the accumulation of genetic alterations that drive tumour-cell development. However, these adaptations may also serve to protect tumour cells from replication stress or DNA damage resulting from oncogene transformation.13, 14, 15, 16 In particular, work from a number of laboratories has identified the ATR-CHK1 pathway as a key mediator of cell death in response to DNA replication stress or some types of DNA damage.17, 18, 19 Inhibition or siRNA-mediated depletion of ATR or CHK1 triggers cell death in cells in response to DNA replication stress, particularly in tumour cells where oncogene activation may inappropriately drive DNA replication. As a result, there has been renewed interest in the use of CHK1 inhibitors for therapies that target tumour cells.20, 21, 22, 23 Loss of ATR or CHK1 function has also been shown to commit cells to other fates such as induction of premature chromosome condensation (PCC)24 or mitotic catastrophe.25 However, the molecular pathways that determine the fate of such cells and the relationships between these different outcomes are not well understood. Here we investigated the role of mitotic catastrophe in the induction of death in tumour cells depleted of CHK1. We show that premature mitosis is mainly induced in p53-deficient tumour cells depleted of CHK1 when DNA synthesis is inhibited through the premature induction of Aurora B kinase in S-phase cells. Although this induction precedes the onset of apoptosis, it is not required for this fate.
Results
Premature Ser10 phosphorylation of histone H3 and aberrant chromosome condensation in tumour cells depleted of CHK1
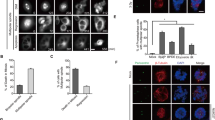
Previous studies have shown premature phosphorylation of histone H3 in cells defective in CHK1 function.10, 25, 26 To determine whether HCT116 cells showed a similar premature mitotic response when CHK1 function was compromised, control and CHK1-depleted cultures treated or not treated with thymidine for 24 h were collected and analysed for DNA content and histone H3 Ser10 phosphorylation by flow cytometry. In cultures treated with control or CHK1 siRNAs or thymidine alone, pSer10 histoneH3 was detected only in cells with a 4-N DNA content (Figures 1a and b). In contrast, phospho-histone H3 was clearly evident in cells with an S-phase DNA content as well as cells with a 4-N DNA content after CHK1 depletion and thymidine treatment. Although the frequency of S-phase cells showing histone H3 phosphorylation was very low in HCT116, it was significantly higher in the p53−/− derivative (Figures 1b and c). Similar results were obtained with another p53 defective colon cancer cell line SW480 (Supplementary Figure S1).
Induction of Ser10-histone H3 phosphorylation in S-phase CHK1-depleted HCT116 cells during DNA replication stress. (a) HCT116 cells pretreated with control or CHK1 siRNAs were exposed or not exposed to 2 mM thymidine (TdR) for 24 h before collection and analysis for pSer10 histone H3 (top panels, scatter plots) and DNA (bottom panels) content by flow cytometry. Cells with elevated pSer10-histone H3 are in the top quadrants of the scatter plots. S-phase cells showing phosphorylated histone H3 are in the top left quadrant those in G2/M are in the top right. Percentages of cells in each of the gated populations are indicated. (b) Mean values (± S.D.) for three independent experiments as presented above. The effects of nocodazole are also presented. (c) p53−/− HCT116 cells were treated with the CHK1 siRNA in the presence or absence of 2 mM thymidne for 24 h before collection and analysis of pSer10 Histone H3 and DNA content by flow cytometry. (d) Effects of nocodazole treatment on HCT116 or HCT116 p53−/− cells treated as in a, c
To determine whether mitotic arrest induced by nocodazole affected the frequency of cells showing premature phosphorylation of histone H3, control or CHK1 siRNA-transfected HCT116 or HCT116 p53−/− cells were treated with nocodazole for 16 h in the presence or absence of thymidine before collection and analysis of phosphorylated histone H3 and DNA content by flow cytometry as above. Nocodazole treatment clearly increased the frequency of cells with a G2/M (4N) DNA content exhibiting histone H3 phosphorylation. However, it did not further alter the frequency of S-phase HCT116 or HCT116 p53−/− cells containing pSer10 histone H3 (Figures 1b and d). Thus, DNA replication stress in CHK1-depleted HCT116 or p53−/− tumour cells triggers premature histone H3 phosphorylation in response to DNA replication stress, and this is not affected by agents that cause mitotic arrest. Our data also suggest that p53 helps protect these tumour cells from such premature mitosis.
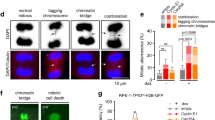
CHK1-depleted cells treated with thymidine were also fixed and analysed for pSer10 histone H3 and RPA2 by immunofluorescence. RPA2 forms foci at replication forks during DNA replication stress and this is greatly enhanced in CHK1-depleted cells.27 Therefore, cells showing both RPA2 foci and phospho-histone H3 staining indicate premature entry into mitosis before completion of replication. pSer10-histone H3-positive cells showing clear RPA foci were evident in CHK1-depleted HCT116 or HCT116 p53−/− cultures treated with thymidine (Figure 2a). These cells also showed aberrant patterns of chromosome condensation (Figure 2a, white arrow indicates a normal mitosis and yellow arrows, aberrant mitoses). In addition, these cultures presented other features of mitotic catastrophe such as the appearance of multinucleate cells28 (Figure 2b, green arrows). Aberrant mitoses are found in a high proportion of cells showing histone H3 phosphorylation in both HCT116 and HCT116 p53−/− CHK1-depleted cultures treated with thymidine (Figure 2c). However, the frequency of multinucleate cells is low and appears to depend upon p53 and CHK1 status rather than replication stress (Figure 2d).
Induction of aberrant mitoses and RPA foci in CHK1-depleted HCT116 and HCT116 p53−/− cells treated with thymidine. (a) Representative images of HCT116 p53−/− cells grown on coverslips were treated with CHK1 or control siRNAs in the presence or absence of thymidine (2 mM) for 24 h before fixation and staining for DAPI, pSer10 histone H3 and RPA2. These cells were then imaged by fluorescence microscopy. A typical mitosis in cells treated with control siRNA in the absence of thymidine is indicated by the white arrow in the top panel. Aberrant mitoses in CHK1 depleted cells treated with thymidine are indicated by yellow arrows. Several of the aberrant mitoses show clear RPA foci (red). (b) Representative multinucleate cells from the above cultures. Mean values for aberrant mitoses (c) or multinucleate cells (d) determined in three independent experiments ± S.D. values
Premature entry into mitosis in CHK1-depleted cells is not dependent on CDC45
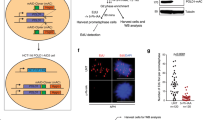
We previously showed that enhanced formation of RPA foci, RPA2 hyperphosphorylation, elevated levels of γH2AX and increased apoptosis in CHK1-depleted cells treated with replication inhibitors are dependent upon the replication helicase co-factor CDC45.27, 29 To determine whether premature entry into mitosis in CHK1-depleted cells showed a similar dependency on this protein, HCT116 or HCT116 p53−/− cultures transfected, with either control or CHK1 and/or CDC45 siRNAs were treated or not with thymidine for 24 h, collected and analysed for DNA and pSer10-histone H3 content by flow cytometry. Despite the general suppression of events triggered by CHK1 depletion during replication stress in CDC45-depleted cells, there was no decrease in the frequencies of pSer10-histone-H3-positive cells with a <4-N DNA content (Figures 3a–c).
CDC45 depletion does not affect induction of pSer10-histone H3 in S-phase HCT116 or HCT116 p53−/− cells depleted of CHK1. (a) Indicated cells pretreated with CHK1 or CHK1 and CDC45 siRNAs were exposed or not exposed to 2 mM thymidine (TdR) for 24 h before collection and analysis for pSer10-histone H3 (top panels, scatter plots) and DNA content (bottom panels) by flow cytometry. (b) Western blot analysis of CDC45 and CHK1 depletion in a representative experiment. β-Actin levels are presented as loading controls. (c) Mean values (± S.D.) for three independent experiments as presented above
CHK1 depletion does not lead to aberrant activation of cyclin B-CDK1 in HCT116 cells
In human cells, mitosis is initiated through localized activation of CDK1 and its binding to the regulatory partner Cyclin B, whose level rises in late S phase and peaks in mitosis.30 Previous work showed that Chk1 deletion in mouse ES cells led to activation of the cyclinB-Cdk1 complex (through loss of inhibitory phosphorylation at Tyr15 in Cdk1) followed by premature mitosis and cell death.25 To determine whether a similar mechanism drove premature mitosis and death in colon cancer cells, we analysed the levels of proteins regulating the S-G2 transition in CHK1-depleted HCT116 or HCT116 p53−/− cells in the presence or absence of thymidine. p53 is stabilized after a 24-h treatment with thymidine and is further upregulated in CHK1-depleted HCT116 cells (Figure 4). As previously reported,18 the downstream effector CDKN1A/p21 is not significantly affected after 24-h treatment with thymidine but is upregulated in CHK1-depleted cells treated with thymidine. Phosphorylation of CHK2 at Thr68 also increased, indicating activation of the ATM-CHK2 signalling cascade. CHK1 is phosphorylated at Ser345 in thymidine-treated HCT116 and HCT116 p53−/− cells, but CHK1 depletion largely eliminates this response. Cyclin B1 levels remain elevated in CHK1-depleted cells after thymidine treatment. Importantly, the inhibitory phosphorylation of CDK1 at Tyr15 triggered by the S-phase checkpoint in thymidine-treated cells is still evident after CHK1 depletion, thus limiting progression of such cells into G2/M. Similar results were obtained with SW480 cells (Supplementary Figure S2). Although CHK2 depletion alone caused a detectable reduction in pTyr15 CDK1 and cyclin B stability, co-depletion of CHK1 and 2 or treatment with an inhibitor of both checkpoint kinases (AZD7762) did not affect pTyr15 CDK1 or cyclin B levels (Supplementary Figure S3). Yet, despite the inhibitory phosphorylation of CDK1 when CHK1 is depleted, histone H3 phosphorylation increases in both HCT116 parental and p53−/− cells (Figure 4).
CDK1 phosphorylation is maintained in CHK1-depleted HCT116 cells during DNA replication stress. Western blot analysis of indicated S-phase regulatory proteins from extracts of HCT116 or HCT116 p53−/− cells pretreated with CHK1 or control siRNAs and exposed or not exposed to thymidine (2 mM, 24 h). β-Actin levels are presented as loading controls
Autophosphorylation of Aurora kinases in S-phase cells depleted of CHK1
In mitotic cells, histone H3 is phosphorylated on Ser10 by Aurora Kinase B following its activation by phosphorylation at a number of sites including the activation of autophosphorylation site at Thr232.31 To determine whether Ser10 phosphorylation of histone H3 in S-phase cells was accompanied by autophosphorylation of Aurora kinase B, CHK1-depleted cells treated with thymidine for 24 h were collected for western blot analysis. HCT116 p53−/− or SW480 cultures treated with thymidine alone showed lower levels of histone H3 or Aurora B phosphorylation than control (Figure 5a, Supplementary Figure S2), most likely due to the arrest of such cells in S-phase. However, when CHK1-depleted cells were treated with thymidine, phosphorylation of both histone H3 and Aurora B was enhanced relative to cells depleted of CHK1 alone. Interestingly, Aurora A was also phosphorylated at Thr288 during this treatment; however, Aurora A is not required for Ser10 histone H3 phosphorylation due to its differential targeting on the centrosome, distinct from Aurora B on the centromere.32 The specific Aurora B inhibitors ZM447439 and AZD1152 substantially reduced phosphorylated forms of Aurora B and histone H3 in CHK1-depleted cells, whereas the Aurora A-specific inhibitor MLN8237 did not reduce Aurora kinase B phosphorylation or histone H3 phosphorylation (Figure 5b). Taken together, these data prove that histone H3 phosphorylation in CHK1-depleted tumour cells treated with thymidine was the result of Aurora B activity. Consistently, these inhibitors did not affect the levels of cyclin B, pTyr15 CDK1 or phosphorylated forms of RPA2.
Ser10-histone H3 and Aurora kinase B phosphorylation in CHK1-depleted HCT116 p53−/− cells during replication stress are sensitive to Aurora B inhibitors. Western blot analysis of indicated S-phase regulatory and mitotic proteins from extracts of HCT116 p53−/− cells pretreated or not with CHK1 or control siRNAs and exposed or not exposed to thymidine (2 mM, 24 h). Effects of co-treatment with the Aurora A inhibitor MLN8237 (10 nM) or the Aurora B inhibitors AZD1152 (50 nM) and ZM447439 (2 μM) are also presented. Inhibitors were added 1 h before cell collection. β-Actin levels are presented as loading controls
To determine whether Aurora B autophosphorylation occurred in S-phase cells, cultures treated with CHK1 siRNA in the presence or absence of thymidine as above were collected and analysed for DNA content, pSer10 histone H3 and pThr232 Aurora B by flow cytometry. These analyses showed clear induction of both phosphorylated proteins in cells with an S-phase DNA content after CHK1 depletion and thymidine treatment (Figures 6a and b). Consistently, phosphorylation of both Aurora B and histone H3 was reduced in cells treated with the aurora kinase inhibitor ZM447439.33 To determine whether the cells showing phosphorylation of Aurora B also showed phosphorylation of histone H3, CHK1-depleted HCT116 p53−/− cells treated with thymidine for 24 h were fixed and stained for pSer10-histone H3 and pThr232 Aurora B content using mouse anti-pSer10 histone H3 and rabbit anti-phospho Aurora kinase antibodies. Alexa Fluor 647—R-Phycoerythrin goat anti-mouse IgG and FITC goat anti-rabbit IgG were then used to visualize the proteins by flow cytometry. Cells gated for phospho-Aurora kinase were then analysed for phospho-histone H3, whereas cells gated for phospho-histone H3 were subsequently analysed for phospho-Aurora kinase (Figure 6c). This analysis revealed that the cells containing elevated phospho-Aurora kinase also contained elevated phospho-histone H3. Similarly, cells first gated for phospho-histone H3 content had elevated phospho-Aurora kinase. CHK1-depleted SW480 cells gave similar results (Supplementary Figure S4). Taken together, our results show that CHK1 depletion triggers autophosphorylation of Aurora B in S-phase cells during DNA replication stress and that this activation of Aurora B is necessary for histone H3 phosphorylation on Ser10.
Aurora kinase autophosphorylation is required for Ser10-histone H3 phosphorylation in S-phase CHK1-depleted p53−/−HCT116 cells but is not required for cell death. (a) Control or CHK1-depleted HCT116 p53−/− cells were exposed or not exposed to 2 mM thymidine (TdR) for 24 h before collection and analysis of pSer10-histone H3, phospho-Aurora kinase and DNA content by flow cytometry. Cells showing elevated levels of the indicated phospho-proteins are gated and the % of cells presenting elevated levels is indicated. Effects of the Aurora B inhibitor (ZM447439, 2 μM, 24 h) treatment are also presented. It is important to note that Aurora B inhibition has been reported to induce polyploidy;34 however, when the cells are treated with ZM447439 and thymidine, they fail to traverse mitosis. (b) Mean values (± S.D.) for three independent experiments as presented above. (c) CHK1-depleted HCT116 p53−/− cells exposed to thymidine for 24 h were fixed and stained for pSer10-histone H3 and phospho-Aurora kinase content using mouse anti-pSer10-histone H3 and rabbit anti-phospho Aurora kinase. Alexa Fluor 647—R-Phycoerythrin goat anti-mouse IgG and FITC goat anti-rabbit IgG were then used to visualize the proteins by flow cytometry. Cells gated for phospho-Aurora kinase were then analysed for pSer10-histone H3. Similarly, cells gated for pSer10-histone H3 were subsequently analysed for phospho-Aurora kinase. Analyses show that gated cell populations co-stain for both proteins. (d) CHK1-depleted HCT116 p53−/− cells exposed to 2 mM thymidine for the indicated times in the presence or absence of 2 μM ZM447439 were collected and analysed for DNA and pSer10-histone H3 content by flow cytometry. The percentages of cells with a subG1 DNA content or showing histone H3 phosphorylation presented are mean values from three independent experiments ±S.D.
To determine whether activated Aurora kinase B could be detected at the site of stalled replication forks, CHK1-depleted HCT116 cells treated with thymidine were fixed and co-stained for phosphorylated forms of Aurora kinase and RPA foci (Supplementary Figure S5). While it is important to note that the antibody employed detects the phosphorylated (active) forms of Aurora kinases A, B and C, there was no indication of co-localization of any of the active Aurora kinases with RPA foci. The dispersed focal pattern of staining of the phosphorylated Aurora kinases was typically found in cells with aberrant chromosome condensation after such treatment and was not found in control cells.
Phosphorylation of histone H3 is not necessary for the induction of apoptosis in CHK1-depleted HCT116 cells treated with thymidine
To investigate the relationship between histone H3 phosphorylation in S-phase and apoptosis, we followed accumulation of these cells together with the onset of apoptosis. CHK1-depleted HCT116 p53−/− cells were treated with thymidine and then collected at various times up to 48 h. Cells were fixed and the levels of pSer10-histone H3 and DNA content were measured by flow cytometry. Cells with a <4-N DNA content showing histone H3 Ser10 phosphorylation increased by 18 h after thymidine treatment reaching a peak at 24 h (Figure 6d). After that the fraction of S-phase cells containing pSer10 histone H3 declined. Control cells or cells treated with thymidine alone or CHK1 siRNA alone showed little change in the level of pSer10 histone H3 (Supplementary Figure S6). pSer10-histone H3-containing cells with a 4-N DNA content depleted of CHK1 decreased after thymidine treatment (reflecting the slower transition of cells through S-phase) but then increased at 18–24 h before falling at later times Supplementary Figure S7). However, the frequency of these cells was lower than those showing pSer10-histone H3 in S-phase. The appearance of p53−/− cells with a subG1 DNA content lagged behind, initially increasing at 24 h and reaching a maximum at 40–48 h post thymidine treatment (Figure 6d). Similar results were obtained with SW480 cells (Supplementary Figure S4D). Thus, the accumulation of subG1 cells distinctly follows the induction of histoneH3 phosphorylation.
In CHK1-depleted HCT116 (p53+/+) cultures treated with thymidine, the level of cells with an S-phase DNA content showing pSer10-histoneH3 is highest at 24 h, but the frequency of these is significantly lower than that in the p53−/− cultures (<4% of the cell population). The accumulation of cells with a subG1 DNA content begins earlier in p53+/+ HCT116 and reaches much higher levels (Supplementary Figure S8).
Despite the sequential onset of Ser10-histone H3 phosphorylation and subG1 cell appearance, inhibition of Ser10-histone H3 phosphorylation in p53−/− HCT116 or SW480 cells by the Aurora B inhibitor ZM447439 had only a small effect on cell death (Figure 6d, Supplementary Figure S4D). CHK1-depleted HCT116 p53−/− or SW480 cells treated with thymidine show a pronounced decrease in the level of pSer10-histone H3 after treatment with ZM447439 but only a small decrease in the level of cells with a subG1 DNA content. Consistent with another report,34 ZM447439 alone had little effect on apoptosis over the time period used in these experiments (Supplementary Figure S9). Induction of polyploidy was observed following treatment of our cells with ZM447439 alone, consistent with Aurora B inhibition and the prevention of cytokinesis (see Figure 6a), but this phenotype was not seen in cells treated with both ZM447439 and thymidine as such cells fail to traverse mitosis. Taken together, our data suggest that pSer10-histone H3 induction precedes the onset of cell death but is not required for its execution.
Discussion
The ATR-CHK1 signalling cascade has a pivotal role in the response of tumour cells to DNA replication stress. In addition to slowing S-phase progression when DNA synthesis is inhibited by arresting inappropriate origin firing and aiding the resolution of stalled replication forks, it protects tumour cells from apoptosis. If the function of this signalling cascade is compromised, a number of catastrophic outcomes have been reported, including the induction of premature mitosis,10, 25, 26 PCC24 and apoptosis.17, 18, 19 The objective of this work was to determine how these different cell fates are related, particularly the relationship between premature mitosis and cell death. We find that premature mitosis (characterized by Aurora B activation, phosphorylation of its substrate histone H3 on Ser10 and aberrant chromosome condensation in cells arrested in S-phase) is only weakly induced in the parental p53+/+ colon cancer cell line HCT116 despite a robust apoptotic response. In contrast, p53-deficient colon cancer cell lines show significantly higher levels of premature mitosis but a delayed and lower level of apoptosis. In p53-deficient cells, the increased level of pSer10 histone H3 is accompanied by marked S-phase activation of Aurora B. Furthermore, Aurora B, but not Aurora A, inhibitors prevent Ser10-histone H3 phosphorylation in S-phase cells, suggesting that an inappropriate activation of Aurora B underlies histone H3 phosphorylation. However, the inhibition of Ser10-histone H3 phosphorylation does not significantly affect the onset of apoptosis in these cells. Thus, although Ser10-histone H3 phosphorylation precedes the induction of apoptosis, it is not required for this cell fate. Consistent with this conclusion, CDC45 depletion suppresses the apoptotic response in HCT116 cells;27 however, it does not affect premature Ser10-histone H3 phosphorylation and thus these cell fates are uncoupled.
Previous work with mouse embryonic stem cells showed that premature mitotic entry induced by targeted knockout of Chk1 was the result of an inappropriate activation of the cyclin B-Cdk1 complex.25 This activation is thought to occur through the dephosphorylation of Cdk1 at Tyr15 by Cdc25A, a dual-specificity phosphatase. CDC25A is normally targeted for proteasomal degradation by CHK1-mediated phosphorylation following the inhibition of DNA replication.35 Thus, in CHK1-depleted cells dephosphorylation of CDK1 would be predicted to lead to premature mitotic entry. However, in our tumour cell lines, CDK1 was not dephosphorylated during replication stress after CHK1 depletion. Instead, we found that Aurora B kinase was inappropriately activated in CHK1-depleted tumour cells during S-phase arrest and this in turn triggered phosphorylation of its substrate, histone H3. Taken together, these findings suggest that there are two pathways that prevent premature mitosis in tumour cells during DNA replication stress (Figure 7). The first is through the inhibition of pTyr15 CDK1 dephosphorylation, while the second is through a CHK1-mediated suppression of Aurora B phosphorylation. How CHK1 controls Aurora B activation during replication stress is not clear. Phosphatases have been implicated in the regulation of Aurora B,36, 37, 38 and we speculate that CHK1 may control the activity of a subset of these phosphatases as well as CDC25A. However, chronic transcriptional alterations resulting from CHK1 depletion39 may also have a role in this process.
Model for control of premature mitosis during DNA replication stress. Mitosis is triggered when activated CDK1 binds to its regulatory partner cyclinB. During DNA replication stress, activation of ATR elicits phosphorylation of CHK1 that, in turn, phosphorylates CDC25A, targeting it for ubiquitin-mediated degradation. In the absence of CDC25A, pTyr15 CDK1 is not dephosphorylated to active CDK1 thus preventing the onset of mitosis. In CHK1-depleted HCT116 cells, CDK1 phosphorylation at Tyr15 is unexpectedly sustained, which should suppress the initiation of mitosis. However, our work indicates that activated CHK1 also suppresses phosphorylation of Aurora B to prevent premature entry into mitosis in cells arrested in S-phase (left). When CHK1 is depleted, Aurora B autophosphorylation is no longer suppressed leading to its activation, histone H3 phosphorylation and premature chromosome condensation (right)
Interestingly, CHK1 is thought to contribute to the full activation of Aurora B by phosphorylation at Ser311 during an unperturbed prometaphase.40 Thus, the suppression of Aurora B activation by CHK1 (direct or indirect) seems counter-intuitive. However, this putative suppressive role is only found in cells after CHK1 activation during DNA replication stress. We found no evidence for Aurora B activation during an unperturbed S-phase after CHK1 depletion. To our knowledge, this is also the first report of Aurora B activation in S-phase cells. In support of this finding, Aurora B has been reported to have a role in G1/S transition through its regulation of CDKN1A/p21 expression.41 In T lymphocytes, it can also form a complex with mTOR to promote the G1/S transition.42
CHK1-depleted HCT116 cells (p53+/+) do not exhibit high levels of premature mitosis despite having a robust apoptotic response to replication stress. As loss of p53 delays and reduces the intensity of the death response, it seems likely that apoptosis in the p53+/+ cells is triggered by the p53-mediated death pathway.
Given the interest in the use of CHK1 inhibitors in therapy, it is important to understand the mechanism underlying the response of tumours to such agents to improve our ability to predict which tumours will respond most favourably. One of the attractions of CHK1 inhibitors is their ability to kill p53−/− tumour cells. Further work should make it possible to understand how further exploitation of these pathways can enhance the induction of death in such tumour cells.
Materials and Methods
Cell lines and cultures
HCT116 and SW480 human colon cancer cell lines were obtained from ATCC. The HCT116 p53 −/− derivative was generously provided by Dr. Bert Vogelstein (Johns Hopkins University, Baltimore, MD). Cells were maintained in DMEM supplemented with 10% foetal bovine serum (FBS). For experiments using thymidine, dialysed FBS was used to remove deoxynucleosides that might alter the cellular response to this agent. The Aurora kinase B inhibitors (ZM447439 and AZD1152) and the Aurora A inhibitor MLN823743 were all obtained from Selleckchem, Houston, TX). They were added at the indicated concentrations at the same time as thymidine treatment in the flow cytometry experiments or 1 h before cell harvest for western blot analysis. The CHK1/CHK2 inhibitor AZD7762 obtained from Selleckchem and was added 1 h before thymidine treatment. Nocodazole (0.2 μg/ml) was added 16 h before cell collection to arrest cells in pro-metaphase.
SiRNA transfection
The sequences for the CHK1 and CHK2 siRNAs used in these experiments were 5′-GAAGCAGUCGCAGUGAAGA-3′ and 5′-AACCUGAGGACCAAGAA-3′, respectively, as described previously.18, 29 CDC45 (M-003232-02) siRNAs were from Dharmacon (Lafayette, CO, USA) and the negative control low GC content siRNA was from Eurofins MWG Operon (London, UK). siRNA duplexes were transfected into cells using DharmaFECT1 (Thermo Scientific, Loughborough, UK) according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were treated.
Cell cycle analysis
Cell cycle analysis of floating and adherent cells was carried out as previously described.18
Cell extraction and western blotting
Whole-cell extracts were prepared as described previously.44 The cell lysis buffer was supplemented with 500 nM okadaic acid (Merck Chemicals, VWR International, Beeston, UK) when samples were used to analyse Aurora kinase sites of phosphorylation. Proteins were separated by SDS-PAGE and blotted onto nitrocellulose (Schleicher & Schuell). Proteins were detected using the enhanced chemiluminiscence detection system (ECL, Amersham) according to the manufacturer’s recommendations using anti-CHK1 (#2345), pAurora: Phospho-Aurora A (Thr288)/Aurora B (Thr232)/Aurora C (Thr198) (#2914), p21 (#2947), pSer345 CHK1 (#2348), CHK1 (#2360), pThr68 CHK2 (#2661), CHK2 (#2662), pTyr15 CDK1 (#9111) from Cell Signaling, Hitchin, UK; anti-CDC45 (sc-20685), CDK1 (sc-54), Cyclin B1 (sc-245), pSer10 H3 (sc-8656), p53 (sc-126) from Santa Cruz Biotechnology (Wembley, UK) and anti-RPA34 (NA19L) from Calbiochem (Feltham, UK); or anti-β-actin (A-5060) from Sigma-Aldrich (Poole, UK).
Immunofluorescence
Cells were cultured on glass coverslips and treated as indicated. Cells were then fixed with 3% buffered paraformaldehyde for 15 min at RT, incubated in PBS containing 30 mM NH4Cl for 5 min and permeabilized in PBS containing 0.5% Triton X-100 for 8 min at RT. For RPA and pS10 Histone H3, cells were incubated with the mouse anti-RPA34 (Calbiochem, NA19L) antibody diluted to 2 μg/ml and rabbit anti-pSer10-Histone H3 (Upstate Biotechnology, Feltham, UK; #06-570) antibody diluted 1:100 for 45 min at RT and detected with a Alexa-594-conjugated goat anti-mouse IgG (A11032; Molecular Probes, Invitrogen, Paisley, UK) and Alexa 488-conjugated goat anti-rabbit IgG (A11008; Molecular Probes, Invitrogen) diluted 1:1000. Antibody dilutions and washes after incubations were performed in PBS containing 0.5% BSA and 0.05% Tween 20. Coverslips were finally mounted in Vectashield mounting medium with DAPI (H-1500; Vector Laboratories, Burlingame, CA, USA). For RPA and phospho Aurora Kinase, a rabbit anti-phospho-Aurora A (Thr288)/Aurora B (Thr232)/Aurora C (Thr198) (Cell Signaling, #2914) diluted 1:50 were used. Mitotic cells were distinguishable from apoptotic cells in which areas of condensed chromatin were totally fragmented and lost the nuclear envelope. Resulting fluorescence was visualized using a Nikon Eclipse T200 microscope equipped with a Hamamatsu Orca ER camera and the Volocity 3.6.1 (Improvision, Cambridge, UK) software or an Olympus FV1000 confocal BX61 upright microscope equipped with a X 60(1.42 NA) objective lens and the Fluoview 3.1 software (Olympus, (Southend-on-Sea, UK). The pictures were further processed by using the Image J software (http://rsbweb.nih.gov/ij/).
Detection of pSer10 histone H3 and phospho-Aurora kinases by flow cytometry
Cells were collected after the treatment, fixed with 70% ice-cold ethanol and stored at −20 °C. After fixation, cells were washed twice with PBS and incubated for 15 min in PBS-T (PBS/0.1% BSA/0.25% Triton X-100). After centrifugation, the cell pellet was suspended in the same buffer plus an antibody specifically recognizing pSer10 histone H3 (ab-14955, Abcam, Cambridge, UK) diluted 1:500 and/or phospho-Aurora A (Thr288)/Aurora B (Thr232)/Aurora C (Thr198) (#2914, Cell Signaling) diluted 1:50 and incubated for 3 h at room temperature. The cells were then rinsed with PBS containing 0.25% Triton X-100 and incubated with the FITC-conjugated goat anti-rabbit immunoglobulins antibody (sc-2012, Santa Cruz Biotechnology) diluted at a ratio of 1:30 in PBS containing 1% BSA and/or Alexa Fluor 647—R-Phycoerythrin goat anti-mouse IgG (H+L) (A20990, Life Technologies, Paisley, UK). After a 30-min incubation at room temperature in the dark, the cells were stained with PI solution (PBS with 5 μg/ml PI and 100 μg/ml RNAse A), and cellular fluorescence was measured by flow cytometry (LSRII, Becton Dickinson, Oxford, UK) and analysed using FlowJo software.
Abbreviations
- CHK1:
-
checkpoint kinase 1
- CHK2:
-
checkpoint kinase 2
- ATM:
-
ataxia-telangiectasia mutated
- ATR:
-
ATM and Rad3 related
- RPA:
-
replication protein A
- ssDNA:
-
single-strand DNA
- PCC:
-
premature chromosome condensation
References
Ciccia A, Elledge SJ . The DNA damage reponse: making it safe to play with knives. Mol Cell 2010; 40: 179–204.
Shiloh Y, Ziv Y . The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 2013; 14: 197–210.
Nakano K, Vousden KH . PUMA a novel proapoptotic gene, is induced by p53. Mol Cell 2001; 7: 683–694.
Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B . PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell 2001; 7: 673–682.
Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA . Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Gene Dev 2005; 19: 1040–1052.
Zou L, Elledge SJ . Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300: 1542–1548.
Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Gene Dev 2000; 14: 1448–1459.
Zhao H, Piwnica-Worms H . ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol 2001; 21: 4129–4139.
Zachos G, Rainey MD, Gillespie DA . Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J 2003; 22: 713–723.
Zachos G, Rainey MD, Gillespie DA, Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Mol Cell Biol 2005; 25: 563–574.
Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA . Chk1 regulates the density of active replication origins during the vertabrate S phase. EMBO J 2007; 26: 2719–2731.
Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 2005; 7: 195–201.
Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM et al. C-myc can induce DNA damage, Increase reactive oxygen species, and mitigate p53 function:a mechanism for oncogene-induced genetic instability. Mol Cell 2002; 9: 1031–1044.
Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005; 434: 864–870.
Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006; 444: 638–642.
Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M et al. Nucleotide deficiency promotes genetic instability in early stages of cancer development. Cell 2011; 145: 435–446.
Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR . and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet 2009; 5: e1000324.
Rodriguez R, Meuth M . Chk1 and p21 cooperate to prevent apoptosis during DNA replication fork stress. Mol Biol Cell 2006; 17: 402–412.
Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell 2008; 133: 2110–2118.
Parsels LA, Qian Y, Tanska DM, Gross M, Zhao L, Hassan MC et al. Assessment of chk1 phosphorylation as a pharmacodynamic biomarker of chk1 inhibition. Clin Cancer Res 2011; 17: 3706–3715.
Carrassa L, Damia G . Unleashing Chk1 in cancer therapy. Cell Cycle 2011; 10: 2121–2128.
Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci USA 2011; 108: 3336–3341.
Blackwood E, Epler J, Yen I, Flagella M, O'Brien T, Evangelista M et al. Combination drug scheduling defines a ‘window of opportunity’ for chemopotentiation of gemcitabine by an orally bioavailable, selective CHK1 inhibitor, GNE-900. Mol Cancer Ther 2013; 12: 1968–1980.
Nghiem P, Park PK, Kim Y, Vaziri C, Schreiber SL . ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc Natl Acad Sci USA 2001; 98: 9092–9097.
Niida H, Tsuge S, Katsuno Y, Konishi A, Takeda N, Nakanishi M . Depletion of Chk1 leads to premature activation of Cdc2-cyclinB and mitotic catastrophe. J Biol Chem 2005; 280: 39246–39252.
Morgan MA, Parsels LA, Parsels JD, Mesiwala AK, Maybaum J, Lawrence TS . Role of Checkpoint Kinase 1 in preventing premature mitosis in response to gemcitabine. Cancer Res 2005; 65: 6835–6842.
Rodriguez R, Gagou ME, Meuth M . Apoptosis induced by replication inhibitors in Chk1-depleted cells is dependent upon the helicase cofactor Cdc45. Cell Death Differ 2008; 15: 889–898.
Vitale I, Galluzzi L, Castedo M, Kroemer G . Mitotic catastrophe: a mechanism for avoiding genetic instability. Nat Rev Mol Cell Biol 2011; 12: 385–392.
Gagou ME, Zuazua-Villar P, Meuth M . Enhanced H2AX phosphorylation, DNA replication fork arrest, and cell death in the absence of Chk1. Mol Biol Cell 2010; 21: 739–752.
Morgan DO . Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 1997; 13: 261–291.
Murnion ME, Adams RR, Callister DM, Allis CD, Earnshaw WC, Swedlow JR . Chromatin-assocoated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J Biol Chem 2001; 276: 26656–26665.
Cowley DO, Rivera-Perez JA, Schliekelman M, He YJ, Oliver TG, Lu L et al. Aurora-A kinase is essential for bipolar spindle formation and early development. Mol Cell Biol 2009; 29: 1059–1071.
Tyler RK, Shpiro N, Marquez R, Eyers PA . VX-680 inhibits Aurora A and Aurora B kinase activity in human cells. Cell Cycle 2007; 6: 2846–2854.
Li M, Jung A, Ganswindt U, Marini P, Friedl A, Daniel PT et al. Aurora kinase inhibitor ZM447439 induces apoptosis via mitochondrial pathways. Biochem Pharmacol 2010; 79: 122–129.
Mailand N, Falck J, Lukas C, Syljuâsen RG, Welcker M, Bartek J et al. Rapid destruction of human Cdc25A in response to DNA damage. Science 2000; 288: 1425–1429.
Posch M, Khoudoli GA, Swift S, King EM, Deluca JG, Swedlow JR . Sds22 regulates aurora B activity and microtubule-kineyochore interactions at mitosis. J Cell Biol 2010; 191: 61–74.
Foley EA, Maldonado M, Kapoor TM . Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol 2011; 13: 1265–1271.
Lesage B, Qian J, Bollen M . Spindle checkpoint silencing: PP1 tips the balance. Curr Biol 2011; 21: R898–R903.
Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H et al. Chk1 is a histone H3 Threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 2008; 132: 221–232.
Petsalaki E, Akoumianaki T, Black EJ, Gillespie DAF, Zachos G . Phosphorylation at serine 311 is required for Aurora B activation. J Cell Biol 2011; 195: 449–466.
Trakala M, Fernandez-Miranda G, Perez de Castro I, Heeschen C, Malumbres M . Aurora B prevents delayed DNA replication and premature mitotic exit by repressing p21 CIP. Cell Cycle 2013; 12: 1030–1041.
Song J, Salek-Ardakani S, So T, Croft M . The kinases Aurora B and mTOR regulate the G1-S cell cycle progression of T lymphocytes. Nat Immunol 2007; 8: 64–73.
Sloane DA, Trikic MZ, Chu ML, Lamers MB, Mason CS, Mueller I et al. Drug resistant Aurora A mutants for cellular target validation of the small molecule kinase inhibitors MLN8054 and MLN8237. ACS Chem Biol 2010; 5: 563–576.
Bolderson E, Scorah J, Helleday T, Smythe C, Meuth M . ATM is required for the cellular response to thymidine induced replication fork stress. Hum Mol Genet 2004; 13: 2937–2945.
Acknowledgements
This work was supported by a Programme grant S305PA from Yorkshire Cancer Research to MM and project grant S306 to PAE and by the Instituto de Salud Carlos III/Fondos FEDER (Miguel Servet program (CP11/00024) to RR and RTICC (RD12/0036/0015)); Grupo Español de Investigación en Sarcomas (beca JM Buesa-2012 to RR); Obra Social Cajastur-IUOPA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Raschellà
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Zuazua-Villar, P., Rodriguez, R., Gagou, M. et al. DNA replication stress in CHK1-depleted tumour cells triggers premature (S-phase) mitosis through inappropriate activation of Aurora kinase B. Cell Death Dis 5, e1253 (2014). https://doi.org/10.1038/cddis.2014.231
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.231
Keywords
This article is cited by
-
Development of targeted therapy therapeutics to sensitize triple-negative breast cancer chemosensitivity utilizing bacteriophage phi29 derived packaging RNA
Journal of Nanobiotechnology (2021)
-
Targeting TOPK sensitises tumour cells to radiation-induced damage by enhancing replication stress
Cell Death & Differentiation (2021)
-
Mithramycin delivery systems to develop effective therapies in sarcomas
Journal of Nanobiotechnology (2021)
-
Gold nanoparticles-induced cytotoxicity in triple negative breast cancer involves different epigenetic alterations depending upon the surface charge
Scientific Reports (2018)
-
11q deletion in neuroblastoma: a review of biological and clinical implications
Molecular Cancer (2017)