Abstract
Cucurbitacin E (CuE) or α-elaterin is a natural compound previously shown to be an antifeedant as well as a potent chemopreventive agent against several types of cancer. The present study investigated the anticancer effects of CuE on colorectal cancer (CRC) using primary cell lines isolated from five CRC patients in Taiwan, Specifically, we explored the anti-proliferation and cell cycle G2/M arrest induced by CuE in CRC cells. MPM-2 flow cytometry tests show that CuE-treated cells accumulated in metaphase (CuE 2.5–7.5 μM). Results further indicate that CuE produced G2/M arrest as well as the downregulation of CDC2 and cyclin B1 expression and dissociation. Both effects increased proportionally with the dose of CuE; however, the inhibition of proliferation, arrest of mitosis, production of reactive oxygen species (ROS), and loss of mitochondrial membrane potential (ΔΨm) were found to be dependent on the quantity of CuE used to treat the cancer cells. In addition, cell cycle arrest in treated cells coincided with the activation of the gene GADD45(α, β, γ). Incubation with CuE resulted in the binding of GADD45γ to CDC2, which suggests that the delay in CuE-induced mitosis is regulated by the overexpression of GADD45γ. Our findings suggest that, in addition to the known effects on cancer prevention, CuE may have antitumor activities in established CRC.
Similar content being viewed by others
Main
Cucurbitacins are a group of tetracyclic triterpenes with medicinal properties derived from the climbing stem of Cucumic melo L.1 They have been used extensively in traditional folk medicines throughout Asia, providing selective biological activities against carcinogenesis.2 Interest in this herb has grown in recent years due to its putative beneficial pharmacological effects as an anti-inflammatory3, 4 and anticancer agent.5, 6 There have also been indications that cucurbitacins may help to prevent and treat oxidative damage as well as suppress specific inflammatory factors.3
Cucurbitacin E (CuE) is an active compound,7 which was previously shown to be a strong antifeedant with the ability to disrupt cell actin8 and cell adhesion.9 Recent studies have reported that CuE has an inhibitory effect on the proliferation of cancer cells, as well as on the polymerization and permeability of actin.10 However, it remains unclear whether CuE inhibits the growth of colorectal cancer (CRC) cells. Furthermore, the mechanism underlying the anticancer effects of CuE has yet to be identified.
This study investigated the effects of CuE on five established human primary cancer cell lines (CP1–CP5).12, 13 We also initiated an investigation into whether CuE contributes to anti-proliferation and cell cycle G2/M arrest in primary cancer cells isolated from CRC patients in Taiwan. We hope that findings from this work will help provide scientific basis and technical support for CRC therapy.
Results
CuE inhibited cell survival/proliferation of five primary CRC cell lines
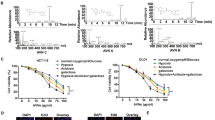
This study hypothesized that CuE could mediate the survival of primary CRC cell lines, thereby inhibiting proliferation. To explore the antitumor activity of CuE against CRC cells, we initiated an in vitro study in which each of the CRC cell lines was exposed to increasing doses of CuE (0, 2.5, 5, and 7.5 μM) over a period of 24 h. The proliferation of the CuE-treated cancer cells was then measured using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) method. The results, outlined in the upper portion of Figure 1a, indicate that cancer cell survival and proliferation rates decreased with the dosage of CuE throughout the 24-h treatment period. This suggests a dose-dependent reduction (y=−16.237x+108.12, R2=0.8547). The same results were not obtained in normal skin fibroblasts Hs-68 cells and normal lung fibroblasts MRC-5 cells as control (normal cell) group (data not shown). Moreover, CuE was shown to induce morphological changes in the primary colon cancer cells. Microscopic observation showed that following exposure to CuE (5 μM) between 6 and 24 h, the primary colon cancer cells underwent a remarkable change in morphology. In some cases, CuE induced cell death, resulting in the formation of a suspension in the medium (data not shown).
As shown in the upper panel, CuE was found to mediate the survival of five human primary CRC cell lines (five groups, n=6 per group), thereby inhibiting proliferation. An in vitro study was conducted by treating each of the cell lines with increasing doses of CuE (0, 2.5, 5, and 7.5 μM) for 24 h. The reversibility of the inhibitory effect CuE has on growth is presented in the middle and lower panels. To determine the extent to which the cancer cells recovered, following the first 24 h treatment period, the CuE was washed off the cells and the cultures were re-incubated for an additional 24–48 h. The survival of the CuE-treated CRC cells was then measured using MTT method. Results were expressed as a percentage of control recovery, which was considered to be 100%. All data were reported as the mean (±S.E.M.) of at least three separate experiments. Statistical analysis was performed using the t-test, with the significant difference set at a level of *P<0.05 versus the control group
Partial irreversibility of growth-inhibitory effects following CuE treatment
In order to determine whether the growth-inhibitory effects of CuE are reversible, we re-cultivated the primary colon cancer cells in fresh culture medium following exposure to CuE for 24 h. We then assessed the recovery of cell proliferation for an additional 24 (Figure 1a middle) to 48 h (Figure 1a lower), whereupon an MTT assay was performed. The results in Figure 1a suggest that the cell proliferation ability of the cancer cells remained substantially degraded (y=−22.822x+104.09, R2=0.6893) following CuE treatment for 48 and 72 h (y=−24.655x+102.47, R2=0.6489). These observations imply that the primary CRC cells underwent an irreversible change, such as apoptosis or cell cycle arrest, at least to a partial extent.
Non-CuE-induced apoptosis of primary CRC cells
In order to identify the role played by CuE in the apoptosis of primary CRC cells, this study used Annexin V-FITC and propidium iodide (PI) staining to reveal the formation of apoptotic cells following 4 h of exposure to CuE. The percentage of apoptotic cells was assessed using flow cytometric analysis (Supplementary Figure S1). The dot-plot of Annexin V-FITC fluorescence versus PI fluorescence indicates a non-significant increase in the percentage of apoptotic cells treated with CuE, compared with untreated cells. No significant increase was observed in the percentage of five CRC cell lines undergoing necrosis, apoptosis (Figure 2a), or caspase 3 activation at CuE concentrations of 2.5–7.5 μM (data not shown). Furthermore, the loss of mitochondrial membrane potential (MMP) is a hallmark of apoptosis and an early event that coincides with caspase activation. The percentages of apoptotic CRC cell lines in different CuE-treated groups were determined using flow cytometry. In non-apoptotic cells, JC-1 exists as a monomer in the cytosol (green) and aggregates in the mitochondria, where it appears red. In apoptotic and necrotic cells, JC-1 exists in a monomeric form and stains the cytosol green. CuE-free CRC cell lines with red fluorescing J-aggregates did not undergo apoptosis. Taken together, our observations indicate that treatment with CuE did not lead to a significant reduction in the MMP of CRC cell lines (data not shown). However, the results shown in Figure 1, Supplementary Figures S1 and 2A indicate that CuE may mediate the survival of primary CRC cells. Thus, we hypothesize that the proliferation of these cells was inhibited by pathways other than apoptosis.
(a) Total apoptosis and necrosis in CRC cells after 4 h of incubation with CuE. Results were expressed as a percentage of the total number of apoptotic cells (early and late apoptosis); (b) the level of intracellular ROS; and (c) the total ROS/Superoxide was detected by flow cytometry.(d) Arrest of cell cycle progression during the G2/M phase in response to CuE treatment. The cell cycle distribution of CRC cell lines was assessed by flow cytometry following staining with PI; (e) results were expressed as a percentage of G2/M; (f) MPM-2 (anti-phospho-Ser/Thr-Pro) expression in untreated and treated cancer cells. MPM-2 is an antibody capable of identifying proteins that are phosphorylated only in mitosis. Cells were dually stained using PI to analyze DNA content, and protein expression was quantified by flow cytometry. As a positive control, separate groups of cells were treated for 24 h with nocodazole (15 μg/ml), an anti-fungal agent known to induce metaphase arrest; (g) cell cycle analysis and the quantification of MPM-2 expression were performed by flow cytometry following treatment with CuE for 24 h. Asterisk (*) in each group of bars indicates that the difference resulting from treatment with CuE 0 μM is statistically significant at P<0.05
CuE-induced accumulation of reactive oxygen species (ROS) in CRC cells
Chemotherapy is known to induce cell death in a variety of tumor types, in part by promoting the production of intracellular ROS. To determine whether ROS production is associated with the CuE-induced cell cycle arrest of CRC cells, we assessed the state of ROS at multiple time points following the administration of various dosages of CuE (0, 2.5, 5, and 7.5 μM). Flow cytometry (Figure 2b) was performed using the Total ROS/Superoxide Detection Kit (Figure 2c) to examine the fluorescence intensity of DCHF-DA-incubated cells. Signals produced by peroxides, peroxynitrite, and hydroxyl radicals were detected in the FL1 channel. Superoxide production was detected in the FL2 channel.
The representative fluorescence patterns of the control and CuE treatment groups showed no significant differences in intracellular ROS levels. These findings suggest that CuE-induced cell cycle arrest in CRC cells occurs via a mechanism other than the accumulation of intracellular ROS.
CuE-induced accumulation of G2/M phase in CRC cells
The cell cycle distribution of CuE-treated cells was analyzed using flow cytometry. Cells were exposed to CuE for 24 h before processing and analysis. As shown in Figure 2d, exposure to CuE increased the number of G2/M phase cells, while simultaneously reducing the number of cells in the S and G1 phases (*P<0.05 versus CuE 0 μM). This suggests a dose-dependent induction (y=21.343x+3.8567, R2=0.9855) (Figure 2e), and may imply that the CRC cells underwent cell cycle arrest.
Effects of CuE on the mitotic index
To differentiate G2 arrest from mitotic arrest, we used an additional marker, MPM-2. This antibody is capable of recognizing proteins, the epitopes of which are exclusively phosphorylated during mitosis, specifically from early prophase to metaphase.14 MPM-2 is commonly used as an indicator of mitotic disturbance. To provide a positive control, we treated separate groups of primary CRC cells with nocodazole (15 μg/ml), an inducer of metaphase arrest.15
Treating the five CRC cell lines with nocodazole for 24 h synchronized the entire cell populations in the G2/M phase and increased MPM-2 labeling (Figure 2f). That is, in all CuE-treated cells, MPM-2 staining was elevated above the level of the negative controls (48–59% for five CRC cells, respectively; Figure 2g).
G2/M phase arrest by CuE in CRC cells via cell division cycle gene 2 (CDC2) downregulation and dissociation of the cyclin B1/CDC2 complex
Figure 3 illustrates the immunoblotting results for cellular proteins from CRC cells treated with CuE. In this test, CDC2 protein expression was quantified by measuring relative intensities. We found that CDC2 decreased following incubation with CuE (upper portion of Figure 3a and Supplementary Figure S2), and CDC2 levels were significantly lower in cells incubated with CuE concentrations of 2.5, 5, and 7.5 μM (Figure 3b) (y=−6.974x+103.12, R2=0.8300). We also quantified the activity of the cyclin B1/CDC2 complex (important for G2–M transition during the cell cycle) by performing co-immunoprecipitation (Co-IP; Figure 3a lower) and measuring relative band intensities. Through this test, we found that the activity of the cyclin B1/CDC2 complex was significantly suppressed in cells incubated with CuE (Figure 3c) (y=−6.6879x+104.76, R2=0.9453). These findings indicate that the increase in G2/M phase CRC cells resulted from non-CuE-induced apoptosis of primary CRC cells, the downregulation of CDC2, and the dissociation of the cyclin B1/CDC2 complex following incubation with CuE.
Cell cycle arrest by CuE in CRC cells via inhibition of CDC2. The CRC Cells (CP3 and CP4) were treated with CuE followed by (a) western blotting analysis (upper) and Co-IP (lower); (b) quantification of intensities by Li-COR near infrared imaging system; (c) delay in mitosis in CuE-treated CRC cells via the inhibition of Cdc2-cyclin B1 complex dissociation. Significant differences were set at a level of *P<0.05 versus the 0 μM CuE control group
Gene expression profile of CRC cells following exposure to CuE
Next, we used human genome SurePrint G3 arrays to study the genome-wide gene expression profiles of CRC cells exposed for 4 h to the vehicle (dimethyl sulfoxide (DMSO)) or CuE (5 μM). Three sets of the experiments were performed independently to enable the comparative analysis between the CP3, CP4, and CP5 cell lines.
Principal component analysis showed that the microarray data derived from CuE-treated cells and DMSO-treated cells constituted two spatially separated planes. This suggests that treatment with CuE had a far greater impact on the gene expression profile than could be reasonably attributed to technical errors. Therefore, as calculated by the expression levels in the CuE-treated cells divided by those in the vehicle-treated cells, we considered changes of>2-fold as substantial upregulation and changes of<0.5-fold as downregulation. To identify biologically relevant molecular networks of these genes, we used distinct pathway analysis tools of bioinformatics, endowed with the comprehensive knowledgebase of the Kyoto Encyclopedia of Genes and Genomes (KEGG) (www.kegg.jp). In so doing, we identified the KEGG pathway (Supplementary Table S3: CP3; Supplementary Table S4: CP4; Supplementary Table S5: CP5) as well as a battery of downregulated (Supplementary Table S6: CP3; Supplementary Table S7: CP4; Supplementary Table S8: CP5) and upregulated genes (Supplementary Table S9: CP3; Supplementary Table S10: CP4; Supplementary Table S11: CP5). Specifically, CuE treatment reduced the expression of cyclin B1 and CDC2 genes and also elevated the levels of growth arrest and DNA-damage-inducible protein 45 (GADD45)-α, -β, and -γ genes (Figure 4a). These findings suggest that common molecular pathways are involved in the induction of cell cycle G2/M arrest.16 The RT-PCR (Figure 4b and Supplementary Figure S3) and qPCR analysis further validated microarray analysis findings, which showed substantial cyclin B1 (y=−16.699x+109.01, R2=0.8743) and CDC2 (y=−11.385x+109.72, R2=0.9841) downregulation, as well as notable upregulation of GADD45 −α (y=37.573x+76.187, R2=0.8958), −β (y=78.13x+33.907, R2=0.9821), and −γ(y=571.91x−717.67, R2=0.8028) in CRC cell lines following exposure to CuE (Figures 4c and d).
Genes downregulated or upregulated in CRC cells following exposure to CuE: (a) The gene expression profiles of cyclin B1, CDC2, as well as GADD45-α, -β, and -γ were studied in CRC cells exposed for 4 h to the vehicle (DMSO) or to 5 μM of CuE; (b) The RT-PCR results of cyclin B1, CDC2, and GADD45-α, -β, and -γ mRNAs in CRC cells following exposure to CuE. (c and d) Quantitative RT-PCR (qPCR) analysis of mRNA expression in cyclin B1 and CDC2, as well as in GADD45-α, -β, and -γ. Expression levels for these proteins were standardized against the levels of GAPDH in CRC cells exposed for 4 h to DMSO (CuE 0 μM control) or CuE. Asterisk (*) in each group of bars indicates that the difference resulting from treatment with CuE 0 μM is statistically significant at P<0.05
G2/M arrest by CuE in CRC cells via integration of GADD45 γ with CDC2
Figure 4d illustrates the gene expression in five CuE-treated CRC cell lines, revealing an increase in GADD45γ. Moreover, the activity of the GADD45γ /CDC2 complex (important for the blockade of G2–M transition during the cell cycle) was determined by Co-IP (Figure 5a) and quantified by measuring the relative band intensities. Our results indicated that the activity of GADD45γ/CDC2 complex was significantly suppressed in cells incubated at a CuE concentration of 7.5 μM (Figure 5b) (y=7.8819x+86.851, R2=0.7695). These findings indicate that the increase in G2/M phase cells resulted from the downregulation of CDC2 and cyclin B1 as well as the dissociation of the cyclin B1/CDC2 complex by GADD45γ following incubation with CuE.
Delay in mitosis in CRC cells by CuE via the combined effects of CDC2 and GADD45γ. Cells were treated with CuE followed by (a) Co-IP and western blotting analysis and (b) quantification of intensities by Li-COR near infrared imaging system. Delay in mitosis in CuE-treated CRC cells was caused by Cdc2–GADD45γ complex combination. Significant differences were set at a level of *P<0.05 versus the 0 μM control group
Discussion
Cucurbitacins are a group of naturally tetracyclic triterpenes that have been shown to act as a potent cytotoxic agent in cancer cells.17 In our previous study, treating human oral squamous cell carcinoma cells of the SAS cell line with CuE led to growth arrest and apoptosis.18 The most common mode of cell death following treatment with cucurbitacins appears to be apoptosis.19 Two major apoptotic pathways exist: the death receptor pathway and the mitochondrial pathway.20 Multiple apoptotic stimuli trigger the activation of proteases called caspases, which in turn initiate and execute the apoptotic program.21 In a previous study, CuE was shown to activate the caspase-dependent pathway, coinciding with the activation of the mitochondrial pathway in bladder cancer cells.22 Throughout Asia, CuE has been used in traditional medicine for cancer therapy, and many studies have attempted to elucidate the mechanism underlying its antitumor activity.23
However, the results summarized in Figures 1 and 2 indicate that CuE may mediate the survival of CRC cells. Thus, the proliferation of these cells was inhibited by a pathway other than apoptosis. Recent studies have shown that CuE can inhibit the growth of tumor precursors24 and enhance VEGFR2-mediated Jak2–STAT3 pathways.25 These effects lead to cell cycle arrest, apoptosis, and anti-angiogenesis.26
In this study, CuE demonstrated anticancer activity as well as the ability to induce cell cycle G2/M arrest. The results collected through our series of tests provide experimental evidence to support the contention that CuE may irreversibly arrest the growth of primary CRC cells. The results of mechanistic analysis further led to the conclusion that both the inhibition of proliferation and the induction of cell cycle G2/M arrest are highly dependent upon the accumulation of CuE in cancer cells. The role of CuE in the inhibition of tumor growth was highlighted by the delay of mitosis through the downregulation of cyclin B1 as well as CDC2 gene expression and dissociation.
The arrested GADD45 encodes three highly conserved nuclear proteins, which contribute to cellular homeostasis by responding to stress.27 Evidence suggests that the GADD45 family fulfills similar functions in survival, cell cycle control, apoptosis, and the repair of DNA damage.16, 28
Gadd45γ has been shown to interact with several key cellular regulators, including cyclin B1 and p21. These interactions result in the proliferation of cell nuclear antigens and mitogen-activated protein kinase.29, 30, 31, 32 The cellular function of Gadd45γ is dependent on the partner with which it interacts. Notably, Gadd45γ is able to suppress G2–M progression in response to stress through its ability to interact with and suppress the kinase activity of the cyclin B1/CDC complex.33, 34 Accordingly, the RNA silencing of Gadd45 expression impairs G2–M checkpoint activity. Whether interactions between Gadd45 and p21 have a role in G1 arrest has yet to be determined.35 Additionally, the downregulation of Gadd45 is closely associated with the degree of malignancy in cancers. Thus, the Gadd45 gene family may have an important role in carcinogenesis.
Unlike the G2 arrest mediated by radiation, the effects of CuE in CRC cells appears to be independent of DNA damage in the Chk1-cdc2-mediated pathway. Rather, these effects predominantly appear to result from metaphase arrest.36 Interestingly, our findings suggest that cell cycle G2/M arrests occurred primarily at higher CuE doses in the five CRC cell lines (7.5 μM), whereas apoptosis and caspase activation were more likely at lower doses (5 μM) in SAS18 and other cancer cells.37, 38 The results of mechanical analysis led us to conclude that both the inhibition of proliferation and the induction of apoptosis,18 cell cycle arrest and autophagy5, 39, 40, 41 are dependent upon the type of cancer cell. However, upon further investigation, our data suggested the existence of a more complex mechanism involving cell cycle deregulation and apoptosis. This mechanisms appears to reflect differences in the degree of CuE-induced toxicity between cancer cell lines.
In conclusion, we have demonstrated for the first time that CuE inhibits tumor growth by arresting the cell cycle in the G2/M phase via GADD45γ gene expression and the blockage of cyclin B1/CDC2 complex in primary CRC cells (Supplementary Figure S4). The role of CuE in the inhibition of tumor growth was highlighted by a delay in mitosis through the upregulation of the GADD45 gene family. These findings suggest the applicability of CuE as an antitumor agent.
Materials and Methods
Materials
CuE, DMSO and MTT were obtained from Sigma (St. Louis, MO, USA). Cell culture medium (DMEM), fetal bovine serum, antibiotics, sodium pyruvate, trypsin, and phosphate-buffered saline (PBS) were purchased from Gibco, BRL (Grand Island, NY, USA). Polyvinylidene fluoride (PVDF) membrane was purchased from Merck Millipore (Darmstadt, Germany), and molecular weight markers were purchased from Bio-Rad (Berkeley, CA, USA). All other reagents and compounds were of analytical grades.
Cell culture
The five primary cell lines of colon cancer cells were derived, as a gift, from the cell bank maintained in the MedicoGenomics Research Center at KMU. The cells were grown at 37 °C in Dulbecco’s Modified Eagle Medium (Gibco, BRL) supplemented with 10% (v/v) fetal bovine serum (HyClone, South Logan, UT, USA) and a combination of antibiotics (penicillin, 200 unit/ml, and streptomycin, 200 g/ml) (HyClone) under an atmosphere of CO2/air (5%) for this series of studies.
Cell proliferation assay
The cells were seeded into 96-well culture plates at 5000 cells/well. The cells were treated with 0, 2.5, 5, and 7.5 μM CuE for 1–3 days. MTT dye (1 mg/ml) was added to each well for at least 4 h of treatment. The reaction was stopped by the addition of DMSO, and optical density was measured at 540 nm on a multi-well plate reader. Background absorbance of the medium in the absence of cells was subtracted. All samples were assayed in triplicate, and the mean for each experiment was calculated. Results were expressed as a percentage of control, which was considered as 100%. Each assay was carried out in triplicate, and the results were expressed as the mean (±S.E.M.).
Evaluation of apoptosis
The apoptosis was assessed by the ApopNexin FITC apoptosis detection kit (Chemicon, Millipore, Billerica, MA, USA). The cells were treated with 0, 2.5, 5, and 7.5 μM CuE for 6 h, and the apoptotic cells were detected by ApopNexin FITC apoptosis detection kit and flow cytometry (FACSCalibur, BD, Franklin Lakes, NJ, USA), and data were analyzed by WinMDI 2.8 free software (BD).
Cell cycle analysis
The method for the cell cycle analysis was by using PI, that is, using the fluorescent nucleic acid dye PI to identify the proportion of cells that are in one of the three interphase stages of the cell cycle. The cells were treated with 0, 2.5, 5, and 7.5 μM CuE for 24 h. Cells were harvested and fixed in 1 ml cold 70% ethanol at least 8 h at −20 °C. DNA was stained in PI/RNaseA solution and the cell cycle (at least 10 000 single cells) was detected by flow cytometry (FACSCalibur, BD). Data were analyzed by the WinMDI 2.8 free software (BD).
Evaluation of MMP (ΔΨm)
The cells were first seeded in 24-well plates (Orange Scientific, Braine, Alleud, Belgium). Following the treatment with CuE for 6 h, JC-1 (10 μg/ml, Sigma) was added to the culture medium, 50 μl per well, and then incubated (at 37 °C for 20 min) for mitochondria staining. After washing twice with warm PBS, the cells were fixed with 2% paraformaldehyde, inspected by Flow cytometry (FACSCalibur, BD) and data were analyzed by the WinMDI 2.8 free software (BD).
Measurement of intracellular ROS
Intracellular ROS generation was measured using a DCFHDA fluorescent dye (Molecular Probes/Invitrogen, Thermo Fisher Scientific, Grand Island, NY, USA) and a Total ROS/Superoxide Detection Kit (ENZO life Sciences, Plymouth Meeting, PA, USA). The CPs cells were cultured in six-well plates at a density of 1 × 104/well. Following treatment with an appropriate concentration of CuE, the cells were incubated with 10 μM DCFH-DA at 37 °C for 30 min and then washed twice with PBS. ROS/superoxide concentration was assessed by staining ROS and superoxide detection reagent according to the protocol provided by the manufacturer (ENZ-51010, ENZO life Sciences). For each experiment, the cells were analyzed for fluorescence using flow cytometry. Data were analyzed using the WinMDI 2.8 free software.
Mitotic index analysis
The mitotic index was assessed by MPM-2 (anti-phospho-Ser/Thr-Pro) expression. After 24 h of treatment with CuE, cells were harvested and fixed in 70% ethanol overnight. Cells were then washed and suspended in 100 μl of IFA-Tx buffer (4% FCS, 150 nM NaCl, 10 nM HEPES, 0.1% sodium azide, 0.1% Triton X-100) with a primary MPM-2 antibody (1 μg/ml; Upstate Cell Signaling Solutions, Millipore, Watford, UK) at room temperature for 1 h. Cells were washed and resuspended in IFA-Tx buffer with a rabbit anti-mouse FITC-conjugated secondary antibody (1: 50; Serotec, Oxford, UK) for 1 h at room temperature in darkness. Finally, cells were washed and resuspended in 500 μl of PBS with 20 μg/ml of PI (Sigma) for 30 min in the dark. MPM-2 expression was analyzed using flow cytometry (FACSCalibur, BD). Data were analyzed using the WinMDI 2.8 free software (BD).
Western blotting assay
A total of 30–50μg proteins were separated by SDS-PAGE (10–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis), and transferred to PVDF membranes (Merck Millipore) in a tank blotter (in 25 mM Tris/0.192 M glycine, pH 8.3/20% methanol) at 30 V overnight. The membranes were blocked with 5% non-fat milk (in 10 mM Tris-HCl, pH 8.0/150 mM NaCl/0.05% tween-20) overnight and incubated with anti-β-actin (AC-15 Sigma), anti-CDC 2 (SC-747, Santa Cruz BioTechnology, Dallas, TX, USA) antibody for 1.5∼2 h. The blots were washed with Tris-HCl (pH 8.0/150 mM NaCl/0.05% Tween-20) for 3 × 10 min and incubated with second antibody (anti-rabbit or anti-mouse immunoglobulins) (IRDye Li-COR, Dallas, TX, USA) at 1/200 dilution for 1 h. The antigen was then visualized and analyzed by Odyssey infrared imaging system (Odyssey LI-COR, Lincoln, NE, USA).
Co-IP
Co-IP is an effective means of quantifying protein–protein interaction in cells. Briefly, 500 μg of cellular proteins were labeled using anti-cyclin B1 (SC-752, Santa Cruz BioTechnology) and GADD45γ (TA505437, OriGene Technologies, Rockville, MD, USA) following overnight incubation at room temperature. The protein-antibody immunoprecipitates were collected by protein A/G plus-agarose (SC-2003, Santa Cruz BioTechnology). Following the final wash, the samples were boiled and centrifuged to pellet the agarose beads. Western blotting analysis of the CDC2 protein in the supernatant was then conducted. Antigens were visualized using a near infrared imaging system (Odyssey LI-COR), and data were analyzed using the Odyssey 2.1 software (Odyssey LI-COR).
Gene expression profiling (GEP)
Briefly, the cells untreated or treated with CuE for 4 h were harvested, and total RNA was isolated utilizing an RNasey kit (Qiagen, Hilden, Germany) as described by the manufacturer. Total RNA was sent to Welgene Company for whole human genome SurePrint G3 arrays GEP analysis (Agilent Technologies, Santa Clara, CA, USA).
RT-PCR
A reverse transcriptase system (Promega, Southampon, UK) was used to synthesize complementary DNA (cDNA) from 1 μg of total RNA. Between 2 and 4 μl of cDNA were used for PCR analysis. PCR (50 μl) reactions were performed using 100 ng of each primer (Supplementary Table S1) and 1 unit of Dynazyme II (Flowgen, Lichfield, UK). Thermal cycling was conducted for 35 cycles at the following temperature/durations: 98 °C for 10 s, 66 °C for 30 s, and 72 °C for 1 min using a Progene thermal cycler (Cambridge, UK). A final extension of 72 °C was performed for 10 min at the end of 35 cycles. The primers used for amplification of the target genes were checked against all other gene sequences for specificity. PCR reactions were analyzed on 1.5% agarose/TAE minigels and stained using 0.5μg/ml ethidium bromide. Gels were visualized using an Apligene UV CCD camera system (Oncer & Apligene, Shanghai, China).
Quantitative real-time PCR (qRT-PCR)
Real-time PCR was conducted using the SYBR Green PCR MasterMix (Applied Biosystems, Thermo Fisher Scientific, Grand Island, NY, USA) according to the manufacturer’s instructions. qRT-PCR was performed using approximately 200 ng of SYBR Green PCR MasterMix and primers (Supplementary Table S2) in an ABI 7300 system (Applied Biosystems, Foster City, CA, USA). PCR conditions were 95 °C for 120 s, 60 °C for 30 s, and 72 °C for 30 s for 40 cycles. Sample cells from three plates were run in duplicate, using the threshold suggested by the software for the instrument to calculate Ct. To normalize readings, we used Ct values from 18 s as internal controls for each run, obtaining a delta Ct value for each gene.
Statistical analysis
All data were reported as the means (±S.E.M.) of at least three separate experiments. Statistical analysis was performed by using t-test, with the significant differences determined at the level of P<0.05.
Abbreviations
- CRC:
-
colorectal cancer
- CuE:
-
cucurbitacin E
- GADD45G:
-
growth arrest and DNA-damage-inducible protein gamma
- cDNA:
-
complementary DNA
- qRT-PCR:
-
quantitative real-time PCR
- PI:
-
propidium iodide
- DMSO:
-
dimethyl sulfoxide
- CDC2:
-
cell division cycle gene 2
- Co-IP:
-
co-immunoprecipitation
- MMP:
-
mitochondrial membrane potential
- PBS:
-
phosphate-buffered saline
- PVDF:
-
polyvinylidene fluoride
- MTT:
-
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- GEP:
-
gene expression profiling
References
Ríos JL, Andújar I, Escandell JM, Giner RM, Recio MC . Cucurbitacins as inducers of cell death and a rich source of potential anticancer compounds. Curr Pharm Des 2012; 18: 1663–1676.
Huang WW, Yang JS, Lin MW, Chen PY, Chiou SM, Chueh FS et al. Cucurbitacin E induces G(2)/M phase arrest through STAT3/p53/p21 signaling and provokes apoptosis via Fas/CD95 and mitochondria-dependent pathways in human bladder cancer T24 Cells. Evid Based Complement Alternat Med 2012; 2012: 952762.
Chen X, Bao J, Guo J, Ding Q, Lu J, Huang M et al. Biological activities and potential molecular targets of cucurbitacins: a focus on cancer. Anticancer Drugs 2012; 23: 777–787.
Qiao J, Xu LH, He J, Ouyang DY, He XH . Cucurbitacin E exhibits anti-inflammatory effect in RAW 264.7 cells via suppression of NF-κB nuclear translocation. Inflamm Res 2013; 62: 461–469.
He J, Wang Y, Xu LH, Qiao J, Ouyang DY, He XH . Cucurbitacin IIa induces caspase-3-dependent apoptosis and enhances autophagy in lipopolysaccharide-stimulated RAW 264.7 macrophages. Int Immunopharmacol 2013; 16: 27–34.
Aribi A, Gery S, Lee DH, Thoennissen NH, Thoennissen GB, Alvarez R et al. The triterpenoid cucurbitacin B augments the antiproliferative activity of chemotherapy in human breast cancer. Int J Cancer 2013; 132: 2730–2737.
Lang KL, Deagosto E, Zimmermann LA, Machado VR, Campos Bernardes LS, Schenkel EP et al. Chemical modification produces species-specific changes in cucurbitacin antifeedant effect. J Agric Food Chem 2013; 61: 5534–5539.
Sörensen PM, Iacob RE, Fritzsche M, Engen JR, Brieher WM, Charras G et al. The natural product cucurbitacin E inhibits depolymerization of actin filaments. ACS Chem Biol 2012; 7: 1502–1508.
Musza LL, Speight P, McElhiney S, Barrow CJ, Gillum AM, Cooper R et al. Cucurbitacins, cell adhesion inhibitors from Conobea scoparioides. J Nat Prod. 1994; 57: 1498–1502.
Gabrielsen M, Schuldt M, Munro J, Borucka D, Cameron J, Baugh M et al. Cucurbitacin covalent bonding to cysteine thiols: the filamentous-actin severing protein Cofilin1 as an exemplary target. Cell Commun Signal 2013; 11: 58.
Zhang T, Li J, Dong Y, Zhai D, Lai L, Dai F et al. Cucurbitacin E inhibits breast tumor metastasis by suppressing cell migration and invasion. Breast Cancer Res Treat 2012; 135: 445–458.
Chen MJ, Tang WY, Hsu CW, Tsai YT, Wu JF, Lin CW et al. Apoptosis induction in primary human colorectal cancer cell lines and retarded tumor growth in SCID mice by sulforaphane. Evid Based Complement Alternat Med 2012; 2012: 415231.
Hsu YC, Weng HC, Lin S, Chien YW . Curcuminoids-cellular uptake by human primary colon cancer cells as quantitated by a sensitive HPLC assay and its relation with the inhibition of proliferation and apoptosis. J Agric Food Chem 2007; 55: 8213–8222.
Escargueil AE, Larsen AK . Mitosis-specific MPM-2 phosphorylation of DNA topoisomerase IIalpha is regulated directly by protein phosphatase 2A. Biochem J 2007; 403: 235–242.
Coldwell MJ, Cowan JL, Vlasak M, Mead A, Willett M, Perry LS et al. Phosphorylation of eIF4GII and 4E-BP1 in response to nocodazole treatment: a reappraisal of translation initiation during mitosis. Cell Cycle 2013; 12: 3615–3628.
Salvador JM, Brown-Clay JD, Fornace AJ Jr . Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv Exp Med Biol 2013; 793: 1–19.
Alghasham AA . Cucurbitacins—a promising target for cancer therapy. Int J Health Sci (Qassim) 2013; 7: 77–89.
Hung CM, Chang CC, Lin CW, Ko SY, Hsu YC . Cucurbitacin E as inducer of cell death and apoptosis in human oral squamous cell carcinoma cell line SAS. Int J Mol Sci 2013; 14: 17147–17156.
Habib L, Khreich N, Jraij A, Abbas S, Magdalou J, Charcosset C et al. Preparation and characterization of liposomes incorporating cucurbitacin E, a natural cytotoxic triterpene. Int J Pharm 2013; 448: 313–319.
Bincoletto C, Bechara A, Pereira GJ, Santos CP, Antunes F, Peixoto da-Silva J et al. Interplay between apoptosis and autophagy, a challenging puzzle: new perspectives on antitumor chemotherapies. Chem Biol Interact 2013; 206: 279–288.
Hensley P, Mishra M, Kyprianou N . Targeting caspases in cancer therapeutics. Biol Chem 2013; 394: 831–843.
Hsu YC, Chen MJ, Huang TY . Inducement of mitosis delay by cucurbitacin E, a Novel tetracyclic triterpenes from climbing stem of Cucumic melo L, through GADD45γ in human brain malignant glioma (GBM) 8401 Cells. Cell Death Dis 2014; 5: e1087.
Li Y, Wang R, Ma E, Deng Y, Wang X, Xiao J et al. The induction of G2/M cell-cycle arrest and apoptosis by cucurbitacin E is associated with increased phosphorylation of eIF2alpha in leukemia cells. Anticancer Drugs 2010; 21: 389–400.
Abdelwahab SI, Hassan LE, Sirat HM, Yagi SM, Koko WS, Mohan S et al. Anti-inflammatory activities of cucurbitacin E isolated from Citrullus lanatus var. citroides: role of reactive nitrogen species and cyclooxygenase enzyme inhibition. Fitoterapia 2011; 82: 1190–1197.
Dong Y, Lu B, Zhang X, Zhang J, Lai L, Li D et al. Cucurbitacin E, a tetracyclic triterpenes compound from Chinese medicine, inhibits tumor angiogenesis through VEGFR2-mediated Jak2-STAT3 signaling pathway. Carcinogenesis 2010; 31: 2097–2104.
Patlolla JM, Rao CV . Triterpenoids for cancer prevention and treatment: current status and future prospects. Curr Pharm Biotechnol 2012; 13: 147–155.
Geifman-Holtzman O, Xiong Y, Holtzman EJ . Gadd45 stress sensors in preeclampsia. Adv Exp Med Biol 2013; 793: 121–129.
Schäfer A . Gadd45 proteins: key players of repair-mediated DNA demethylation. Adv Exp Med Biol 2013; 793: 35–50.
Shih RS, Wong SH, Schoene NW, Zhang JJ, Lei KY . Enhanced Gadd45 expression and delayed G2/M progression are p53-dependent in zinc-supplemented human bronchial epithelial cells. Exp Biol Med (Maywood) 2010; 235: 932–940.
Caratozzolo MF, Micale L, Turturo MG, Cornacchia S, Fusco C, Marzano F et al. TRIM8 modulates p53 activity to dictate cell cycle arrest. Cell Cycle 2012; 11: 511–523.
Notas G, Alexaki VI, Kampa M, Pelekanou V, Charalampopoulos I, Sabour-Alaoui S et al. APRIL binding to BCMA activates a JNK2-FOXO3-GADD45 pathway and induces a G2/M cell growth arrest in liver cells. J Immunol 2012; 189: 4748–4758.
Yang Z, Song L, Huang C . Gadd45 proteins as critical signal transducers linking NF-kappaB to MAPK cascades. Curr Cancer Drug Targets 2009; 9: 915–930.
Zhu N, Shao Y, Xu L, Yu L, Sun L . Gadd45-alpha and Gadd45-gamma utilize p38 and JNK signaling pathways to induce cell cycle G2/M arrest in Hep-G2 hepatoma cells. Mol Biol Rep 2009; 36: 2075–2085.
Vairapandi M, Balliet AG, Hoffman B, Liebermann DA . GADD45b and GADD45g are cdc2/cyclinB1 kinase inhibitors with a role in S and G2/M cell cycle checkpoints induced by genotoxic stress. J Cell Physiol 2002; 192: 327–338.
Million K, Horvilleur E, Goldschneider D, Agina M, Raguénez G, Tournier F et al. Differential regulation of p73 variants in response to cisplatin treatment in SH-SY5Y neuroblastoma cells. Int J Oncol 2006; 29: 147–154.
Taylor WR, Stark GR . Regulation of the G2/M transition by p53. Oncogene 2001; 20: 1803–1815.
Shan XL, Zhou XY, Yang J, Wang YL, Deng YH, Zhang MX . Inhibitory effect of cucurbitacin E on the proliferation of ovarian cancer cells and its mechanism. Chin J Cancer 2010; 29: 20–24.
Duncan KL, Duncan MD, Alley MC, Sausville EA . Cucurbitacin E-induced disruption of the actin and vimentin cytoskeleton in prostate carcinoma cells. Biochem Pharmacol 1996; 52: 1553–1560.
Zhang T, Li Y, Park KA, Byun HS, Won M, Jeon J et al. Cucurbitacin induces autophagy through mitochondrial ROS production which counteracts to limit caspase-dependent apoptosis. Autophagy 2012; 8: 559–576.
Zhu JS, Ouyang DY, Shi ZJ, Xu LH, Zhang YT, He XH . Cucurbitacin B induces cell cycle arrest, apoptosis and autophagy associated with G actin reduction and persistent activation of cofilin in Jurkat cells. Pharmacology 2012; 89: 348–356.
Guo J, Wu G, Bao J, Hao W, Lu J, Chen X et al. Induced ATM-mediated DNA damage causes G2/M cell cycle arrest in a ROS-dependent manner. PLoS One 2014; 9: e88140.
Acknowledgements
The authors appreciate the funding support provided by the National Science Council (Taiwan) (grant NSC100-2313-B-309-001) and (NSC101-2313-B-309-001). The authors are also grateful to the technical assistance provided by Ms Ai-Jie Han of Innovative Research Center of Medicine.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Finazzi-Agrò
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Hsu, YC., Huang, TY. & Chen, MJ. Therapeutic ROS targeting of GADD45γ in the induction of G2/M arrest in primary human colorectal cancer cell lines by cucurbitacin E. Cell Death Dis 5, e1198 (2014). https://doi.org/10.1038/cddis.2014.151
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.151
Keywords
This article is cited by
-
Cucurbitacin C as an effective anti-cancer agent: unveiling its potential role against cholangiocarcinoma and mechanistic insights
Journal of Cancer Research and Clinical Oncology (2023)
-
Use of cucurbitacins for lung cancer research and therapy
Cancer Chemotherapy and Pharmacology (2021)
-
Naturally occurring anti-cancer compounds: shining from Chinese herbal medicine
Chinese Medicine (2019)
-
Inducement of apoptosis by cucurbitacin E, a tetracyclic triterpenes, through death receptor 5 in human cervical cancer cell lines
Cell Death Discovery (2017)
-
GADD45γ induces G2/M arrest in human pharynx and nasopharyngeal carcinoma cells by cucurbitacin E
Scientific Reports (2014)