Abstract
The insulin-like growth factor-2 mRNA-binding protein 3 (IGF2BP3) is a member of a highly conserved protein family that is expressed specifically in placenta, testis and various cancers, but is hardly detectable in normal adult tissues. IGF2BP3 has important roles in RNA stabilization and translation, especially during early stages of both human and mouse embryogenesis. Placenta is an indispensable organ in mammalian reproduction that connects developing fetus to the uterine wall, and is responsible for nutrient uptake, waste elimination and gas exchange. Fetus development in the maternal uterine cavity depends on the specialized functional trophoblast. Whether IGF2BP3 plays a role in trophoblast differentiation during placental development has never been examined. The data obtained in this study revealed that IGF2BP3 was highly expressed in human placental villi during early pregnancy, especially in cytotrophoblast cells (CTBs) and trophoblast column, but a much lower level of IGF2BP3 was detected in the third trimester placental villi. Furthermore, the expression level of IGF2BP3 in pre-eclamptic (PE) placentas was significantly lower than the gestational age-matched normal placentas. The role of IGF2BP3 in human trophoblast differentiation was shown by in vitro cell invasion and migration assays and an ex vivo explant culture model. Our data support a role of IGF2BP3 in promoting trophoblast invasion and suggest that abnormal expression of IGF2BP3 might be associated with the etiology of PE.
Similar content being viewed by others
Main
The placenta is a remarkable biological structure of eutherian mammals. Consisting of both zygote-derived and maternal cells, the placenta mediates interactions between the mother and the fetus, which are essential for fetal growth and survival. Although there are some data showing that unknown genetic etiologies of placenta are thought to be the cause of many types of pregnancy complications such as unexplained miscarriage, intrauterine growth retardation, pre-eclampsia (PE) and so on,1, 2, 3, 4 the molecular basis of human placentation remains mainly unclear.
Trophoblast is an extraembryonic tissue that plays a crucial role during embryo implantation and placentation.5 Proper invasion of trophoblast cells into maternal decidua is a prerequisite for a successful pregnancy. Trophoblast invasion shares many similarities with the invasion of tumor cells into host tissues. However, unlike the uncontrolled tumor invasion, invasion of trophoblast cells is strictly regulated by many endocrine factors and placenta-specific genes that either promote or inhibit invasion of these cells and ensure that trophoblast invasion in human placenta is temporally and spatially restricted to the first trimester, the whole endometrium and the upper third myometrium, respectively.6, 7, 8, 9 Many adhesion molecules or extracellular matrix proteins are involved in the invasion process. For example, cytotrophoblasts (CTBs) secrete matrix metalloproteinases (MMPs) and their tissue inhibitors (TIMPs) that are involved in maintaining the balance and tight control of the invasion of extravillous cytotrophoblast cells (EVTs) into maternal endomyometrium.
The insulin-like growth factor-2 mRNA-binding proteins (IGF2BPs) are known to bind to RNA and influence the fate of the transcript targets.10, 11, 12 IGF2BP family has three members, IGF2BP1, IGF2BP2 and IGF2BP3, of which only IGF2BP1 knockout mice are reported so far, with a phenotype of dwarfism of the fetus.13 IGF2BP3, also known as IMP3, or c-myc coding region determinant-binding protein 3 (CRD-BP3) or zipcode-binding protein (ZBP), is first identified as a novel KH-domain containing protein belonging to a highly conserved protein family (IGF2BPs) from a pancreatic cancer tumor screen termed KOC.14 IGF2BP3 is also associated with other cancers, including hepatocellular carcinoma, ovarian carcinoma, adenocarcinoma of the lung and so on.15, 16, 17, 18 Recent functional studies reported two validated target mRNAs for IGF2BP3, insulin-like growth factor-2 (IGF2) and epidermal growth factor receptor (EGFR). IGF2BP3 activates IGF2 at translational level and exerts its proproliferative effects through IGF2/phosphatidylinositide 3-kinase (PI3K) and IGF2/mitogen-activated protein kinase (MAPK) cascades in glioblastoma cells. On the other hand, in triple-negative breast cancer cells, IGF2BP3 was reported as an effector of EGFR-mediated migration and invasion.19, 20 These results indicated that IGF2BP3 might have various targets in different systems.
Furthermore, Vg1-RBP/Vera, the homologous protein of IGF2BP3 in Xenopus, regulates the localization of the mRNA for transforming growth factor-β-like factor Vg1 to the vegetative pole during stages III and IV of Xenopus oocyte development, and it is important for mesoderm induction and left–right axis formation. These data indicate an important role of IGF2BP3 in embryonic development.21, 22, 23 IGF2BP3 is thus regarded as an oncofetal protein highly expressed in fetal tissues and malignant tumors, but rarely found in adult benign tissues except placenta and testis.24
As a highly expressed gene in placenta, however, the role of IGF2BP3 in human placentation or implantation has not been reported. In the current study, expression of IGF2BP3 in human placental villi during early pregnancy was examined, and the expression level between normal pregnant and gestational age-matched PE placentas was compared. The role of IGF2BP3 in trophoblast invasion was further investigated. The results showed that IGF2BP3 protein is highly expressed in CTBs and trophoblast column (TC), but lower expressed in syncytiotrophoblast (STB) of the placental villi from the first trimester. It suggests that IGF2BP3 may be involved in the regulation of trophoblast invasion and migration. This was proved by our in vitro cell invasion and migration assays, followed by gelatinolytic zymography assays, and an ex vivo trophoblast explant culture model. Furthermore, we demonstrated that several predicted targets of IGF2BP3 were decreased by IGF2BP3 siRNA. In addition, protein kinase B (AKT) signaling pathway was shown to be involved in IGF2BP3-mediated trophoblast cell invasion and migration. The above evidences support a role of IGF2BP3 in promoting invasion of human trophoblast cells, and also suggest that dysregulation of IGF2BP3 expression may be associated with PE.
Results
IGF2BP3 is highly expressed in the human placental trophoblast cells from the first trimester
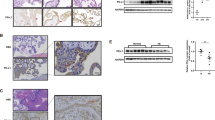
We first examined the expression of IGF2BP3 proteins in different types of trophoblast cells in human placental villi at different stages of pregnancy. Paraffin sections of placental tissues from the first and the third trimesters of normal pregnant women were immunostained with anti-IGF2BP3. The β-human chorionic gonadotrophin (β-hCG) immunostaining combined with H&E staining were used to distinguish villous CTBs (negative for β-hCG staining), TC (negative for β-hCG staining and located at the tip of the villus) and STB (positive for β-hCG staining) (Figure 1Ad and e). As shown in Figure 1, the IGF2BP3 protein was intensively and specifically stained in CTBs and TC during the first trimester, whereas less immunostaining was observed in the STB (Figure 1Aa). In the normal third trimester placentas, the IGF2BP3 protein level was lower than the first trimester villi (Figure 1Ab).
Expression of IGF2BP3 in human placental villi at different stages of pregnancy or from PE patients by IHC and western blotting. (A) IHC. (a, b, c) Immunostaining of IGF2BP3 (rabbit) in normal human placental villi from the first and third trimesters and the pre-eclamptic patients. (a) Placental villi of 7 weeks of pregnancy. (b) Placental villi from the third trimester of normal human. (c) Placental villi from pre-eclamptic patients. (d, e, f) Immunostaining of β-hCG, a marker for syncytiotrophoblast. (g, h, i) Negative controls in which serum IgG from rabbit was used in place of primary antibody. CTB, cytotrophoblast cells; STB, syncytiotrophoblast; TC, trophoblastic column; bar=50 μm. (B) Expression of IGF2BP3 proteins in the placental villi from the first trimester human placentas and the pre-eclamptic women or gestational age-matched normal pregnant women by western blotting. (C) Statistical analysis of the level of IGF2BP3 as representatively shown in B (n=8; **P<0.01), β-actin is a loading control (here and after)
IGF2BP3 is downregulated in PE placentas
To compare the expression level of IGF2BP3 under normal or pathological conditions such as PE, we collected gestationally matched normal human term placentas and late-onset PE placentas and performed immunohistochemistry (IHC) (Figure 1Ab and c). The results showed that IGF2BP3 was hardly detectable in the PE patients (Figure 1Ac). On the other hand, placental villi from the first trimester, the late-onset PE patients and normal pregnancy controls of close gestational stages were dissected for protein extraction and immunoblotting. Western blotting (Figure 1B) and the statistical analysis (Figure 1C) showed that the expression of IGF2BP3 in the normal human third trimester placentas was significantly lower than that in the first trimester placentas, and IGF2BP3 was significantly decreased in the placental villi from PE patients as compared with the normal pregnancy controls of close gestational stages (**P<0.01).
IGF2BP3 siRNA significantly inhibits invasion and migration capacities of HTR8/SVneo cells
As it is known, cell invasion and migration are important events in placenta. To determine the role of IGF2BP3 in trophoblast invasion and migration, matrigel cell invasion and transwell cell migration models were employed. HTR8/SVneo cells transfected with IGF2BP3 siRNA or scrambled siRNA for 24 h were plated onto matrigel or pure filter for invasion or migration assays, respectively. Transfected cells were parallelly cultured to examine the silencing efficiency. Transfection efficiency, as evaluated by the fluorescence signal in FITC siRNA-transfected HTR8/SVneo cells, was almost 90% (data not shown). Specific siRNA targeting IGF2BP3 significantly decreased the expression of IGF2BP3 protein, but not IGF2BP1 or IGF2BP2 in HTR8/SVneo cells (Figures 2a and b). Compared with the scrambled siRNA-transfected HTR8/SVneo cells, IGF2BP3 siRNA significantly decreased the percentage of cells that invaded (Figures 2c and d, **P<0.01) or migrated (Figures 2c and d, **P<0.01) to the other side of the filters.
IGF2BP3 siRNA inhibits migration and invasion capacity of HTR8/SVneo cells. (a) Confirmation of RNA interference of IGF2BP3 by western blotting. (b) Statistical bar graphs showing the knockdown efficiency of IGF2BP3 siRNA (n=4; **P<0.01). (c) Representative images of filters carrying invaded or migratory cells transfected with scrambled siRNA or IGF2BP3 siRNA in matrigel cell invasion assay and transwell cell migration assay, respectively. (d) Statistical bar graphs showing the effect of IGF2BP3 siRNA on invasion and migration of HTR8/SVneo cells (n=4; **P<0.01). (e) MTT assay to show proliferation of HTR8/SVneo cells under indicated treatment (n=5). (f) Ratio of apoptotic cells in HTR8/SVneo cells transfected with indicated siRNAs (n=3). (g) Western blotting of HTR8/SVneo cells transfected with indicated siRNA using indicated antibodies. GAPDH is a loading control (here and after). (h) Statistical analysis of the level of caspase 3 of the western blotting result as representatively shown in (g)
To determine whether the decrease in cell numbers on the opposite sides of the filters in the cell invasion and migration assays reflected a real decrease in cell invasion and migration capacities, or was simply a consequence of a decrease in cell proliferation or an increase in cell apoptosis, we examined proliferation and apoptosis of these cells after siRNA transfection. Proliferation of HTR8/SVneo cells was not significantly changed with IGF2BP3 siRNA treatment based on an MTT assay (Figure 2e, P>0.05). On the other hand, HTR8/SVneo cells transfected with IGF2BP3 siRNA or the scrambled siRNA were stained with Hoechst 33258 and apoptotic cells were counted under microscopy. Compared with the control cells, there were no more apoptotic cells in the IGF2BP3 siRNA-transfected group (Figure 2f, P>0.05). Further examination by western blotting showed that the expression of Caspase 3, an apoptotic marker, was not changed significantly after IGF2BP3 siRNA transfection in HTR8/SVneo cells (Figures 2g and h).
As reported, adhesion molecules or extracellular matrix proteins such as MMPs were involved in the invasion process. To examine gelatinolytic activities of MMP-2 and MMP-9 in HTR8/SVneo cells after IGF2BP3 siRNA transfection, gelatin zymography was performed. Results (Figure 3A) and the statistical analysis (Figure 3B) revealed that IGF2BP3 siRNA significantly decreased pro-MMP-9 but not pro-MMP-2 activities in the supernatants of HTR8/SVneo cells as compared with the scrambled siRNA (*P<0.05).
Possible mechanisms involved in IGF2BP3-mediated invasion and migration of HTR8/SVneo cells. (A) Gelatinolytic analysis of MMP-2 and MMP-9 after IGF2BP3 siRNA transfection. (B) Statistical analysis of the zymographic results as representatively shown in (A) (n=4; *P<0.05). (C) Immunostaing of F-actin organization and IGF2BP3 knockdown monitored by confocal microscopy upon phalloidin labeling in HTR8/SVneo cells transfected with indicated siRNAs for 72 h. Bar=10 μm. (D) IGF2, CD164 and CD44 mRNA levels in HTR8/SVneo cells transfected with indicated siRNA. (E) The protein levels of p-AKT (S473), p-AKT (T308) and PAN-AKT in IGF2BP3 siRNA-transfected HTR8/SVneo cells by western blotting. (F) Statistical analysis of the levels of p-AKT (S473) and p-AKT (T308) of the western blotting result as representatively shown in (E)
Cytoskeleton dynamics play an important role in cell motility. To determine the cytoskeleton dynamics in IGF2BP3-mediated trophoblast invasion and migration, immunofluorescence (IF) was performed. IGF2BP3 knockdown inhibited the linear polymer microfilament (F-actin) formation in HTR8/SVneo cells (Figure 3C). Compared with the scrambled siRNA-transfected group the needle-like F-actin structures in HTR8/SVneo cells transfected with IGF2BP3 siRNA were significantly increased, and the stress fibers were severely lost at the same time (Supplementary Figure S1).
To find out the possible targets of IGF2BP3 during trophoblast invasion and migration, reverse transcription and real-time quantitative polymerase chain reaction (qPCR) were employed. The mRNA levels of IGF-2 and cluster of differentiation 44 (CD44) were significantly decreased in IGF2BP3 siRNA-treated HTR/SVneo cells as compared with the control group (Figure 3D).
Kinases are often involved in the cell invasion and migration. To investigate the underlying signaling pathways involved in IGF2BP3-mediated trophoblast invasion and migration, western blotting was performed to detect the phosphorylation of AKT. As expected, the levels of phospho-AKT (Ser473 and Thr308) were lower in the IGF2BP3 siRNA-transfected HTR/SVneo cells than the scrambled siRNA-transfected cells (Figures 3E and F).
Knockdown of IGF2BP3 inhibits the invasion and migration ability of EVTs from the first trimester human placenta villi
To further confirm the role of IGF2BP3 in trophoblast invasion and migration in vivo, explants were freshly obtained from one placenta and separated into two groups. One group was treated with scrambled siRNA and the other group was treated with IGF2BP3 siRNA. Scrambled siRNA labeled with Alexa Fluor 488 exhibited green fluorescence in almost all the cells in the outgrowth EVTs (Figure 4A). The whole mount IF staining showed the silencing efficiency of IGF2BP3 (Figure 4B). Bright field images (Figures 4Ba, f and k) showed the explants. Dark field images showed the IF staining (Figures 4Bb–e, g–j, and i–o). Cytokeratin 7 (CK7) is a marker for cytotrophoblast cells, and was used to distinguish trophoblast cells of the outgrowth area from the villous tip (Figure 4Bb and g). Compared with the group treated by scrambled siRNA, the IGF2BP3 level was significantly decreased in EVTs treated by its specific siRNA (Figure 4Bc and h).
As shown in Figures 4C–E, the average outgrowth length in the control explants was 0.7–1.8 mm. However, in the IGF2BP3 siRNA-transfected group, the average outgrowth length was ∼0.2–0.8 mm (**P<0.01). Ratio of area of outgrowth compared with the total area of the explant in the Scrambled siRNA group was ∼70–90%; the ratio was ∼30–50% in the IGF2BP3 siRNA-transfected group (**P<0.01).
Discussion
A number of placenta-specific genes have been identified. However, only a few of them are strictly specifically expressed in the placenta,25 such as mouse trophoblast-specific protein (Tpbp) a and b, that may act as inhibitors for cathepsin proteases.26 Placental-specific protein 1 (PLAC1) has traditionally been considered to be placenta specific, but recent investigations showed that, besides being expressed in the placenta, it may also be transcribed in the testis in humans, and elevated in various cancers.27 The cellular function of PLAC1 was thought to be membrane associated and has been linked to trophoblast differentiation28 and fibroblast growth factor 7 (FGF7) signaling.29 Similar to PLAC1, IGF2BP3 is also highly expressed in the placenta and testis and is associated with various cancers. In mammals, IGF2BPs have extremely similar order and spacing of domains. Especially, IGF2BP1 and 3 show an identity of 73% amino acid sequence with each other. These similarities suggest that the proteins may share biochemical functions. A large body of in vitro evidences indicated that IGF2BP1 promotes cancer metastasis. Similar functions were found in IGF2BP3. In contrast to IGF2BP1 and IGF2BP3, IGF2BP2 has been suggested as a candidate involved in type 2 diabetes (T2D). Although many investigators have suggested the relevance between IGF2BP3 and cancers, the role of IGF2BP3 in trophoblast cells is still a puzzle.
In this study, IGF2BP3 was found to be specifically highly expressed in human placental cytotrophoblast cells and TC. Furthermore, the expression level of IGF2BP3 in placentas from the first trimester was much higher than the third trimester. Trophoblast cells from placentas of the first trimester exhibited much higher proficiency of migration and invasion as compared with those from the third trimester.30 We further showed that IGF2BP3 siRNA inhibited invasion and migration of trophoblast HTR8/SVneo cells. In line with this, the F-actin of HTR/SVneo cells transfected with IGF2BP3 siRNA was seriously altered as compared with the control cells. More interestingly, silencing of IGF2BP3 significantly inhibited the outgrowth capacity of first trimester human placental villi by an explant culture model. Above all, these data strongly supported a role of IGF2BP3 in invasion and migration of trophoblast cells.
The family of insulin-like growth factor-2 mRNA-binding proteins was identified based on their ability to bind to IGF-2 mRNA and thereby regulate its stability and translation,14, 16, 31 which was verified in our study as well. The mRNA level of IGF-2 was found to be decreased by IGF2BP3 siRNA in HTR8/SVneo cells when the IGF-2 level in scrambled siRNA or IGF2BP3 siRNA-transfected HTR8/SVneo cells was examined by qPCR (Figure 3D).
Several putative mRNA targets of IGF2BP3, which contribute to cell motility and invasion, are recently identified based on photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP).32 These targets include cluster of differentiation 164 (CD164, endolyn), Ras homolog gene family, member A (RhoA), Cyclin-G1 (CCNG1) and G1/S-specific cyclin-D2 (CCND2) that are known to have a role in carcinogenesis.33, 34, 35, 36 Interestingly, CD164 mRNA expression was reported to be significantly decreased in IGF2BP3-depleted MDA-MB-231 cells. We therefore explored CD164 mRNA level by qPCR in scrambled or IGF2BP3 siRNA-transfected HTR8/SVneo cells, but no significant change was observed (Figure 3D). We also detected the hyaluronan receptor CD44 expression by qPCR because it is known to contribute to tumor progression. The mRNA level of CD44 in HTR8/SVneo cells transfected with IGF2BP3 siRNA is decreased as compared with scrambled siRNA-transfected cells (Figure 3D). It is shown that IGF-2 activates PI3K/AKT pathway and stimulates migration of ovine trophectoderm cells.37, 38 The phosphorylation of AKT is reported to enhance the metastasis of several tumor cells.38, 39 According to our data, the AKT signaling pathway may also has function in IGF2BP3-mediated trophoblast cell invasion and migration on the maternal–fetal interface (Figures 3E and F), but the direct targets of IGF2BP3 that were involved in the AKT pathway remain to be further investigated.
PE is a critical pregnancy-related disease complicated by hypertension and proteinuria, and is a major cause of maternal mortality, morbidities, perinatal deaths, preterm birth and intrauterine growth restriction.40, 41 Of the worldwide pregnancies, 3–5% are affected by this disease.42 The mechanisms involved in PE remain was poorly known, although our understanding of this pathological condition has been improved.43 Exploration of the roles of differentially expressed placenta-specific genes in PE patients will enrich our understanding of the pathogenesis of this disease and contribute to its diagnosis and management. Impaired trophoblast invasion/migration that lead to poor spiral arterial remodeling and consequently inadequate placental perfusion was believed to induce PE.44, 45 In this study, we found that the level of IGF2BP3 in the placentas from PE patients is significantly lower than their age-matched normal placentas (Figure 1), that is consistent with the results that the knockdown of IGF2BP3 inhibits EVT invasion and migration. Nevertheless, the molecular basis remains to be further investigated in order to clarify the mechanism of the downregulation of IGF2BP3 in PE placentas and the relationship between IGF2BP3 and the pathogenesis of PE.
In summary, our data provide the first insight into the function of IGF2BP3 in human placenta trophoblast invasion and migration, and suggest a possible pathological mechanism involved in PE.
Materials and Methods
Tissues collection
First trimester human placental tissues (5–8 weeks) were obtained from healthy pregnant women undergoing legal abortion. Term placentas (PE and gestational age-matched controls) were collected after caesarean birth. Informed consent for placenta donation was obtained from each woman who donated her placenta. The standard to define PE placentas has been as described previously.46 Two of the eight PE patients were complicated with intrauterine growth restriction (IUGR). Ethical approval was granted by the Ethic Committee of the Beijing Obstetrics and Gynecology Hospital. This study and the use of samples were under standard experimental protocols approved by the Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences. All the placental tissues were collected and stored in ice-cold DMEM (HyClone Laboratories, Inc., Logan, UT, USA), and transported to the laboratory within 1 h after surgery. The placentas were separated into small pieces. For western blotting and IHC, the placentas were cut into 5–10 mm, and for explant culture, the placentas were cut into 2–5 mm. For western blotting, the samples were washed with ice-cold phosphate-buffered saline (PBS) and quickly frozen in liquid nitrogen forprotein extraction (first trimester: n=8, normal third trimester: n=8, PE third trimester: n=8). For explant culture, the tissues were cultured immediately after washing (first trimester: n=13; see explant culture method for details). For IHC, the tissues were washed with ice-cold PBS and fixed with 4% neutral PFA (paraformaldehyde; Sigma-Aldrich Inc., St. Louis, MO, USA) at 4 °C, followed with gradient ethanoldehydration and embedding in paraffin (first trimester: n=3, normal third trimester: n=3, PE third trimester: n=3).
IHC and IF
IHC was performed using a biotin-streptavidin-peroxidase (SP) kit (SP-9001, SP-9002, Zhongshan Golden Bridge Co., LTD, Beijing, China) and a diaminobenzidine kit (DAB, Zhongshan Golden Bridge Co., LTD) as previously described.47 Sections (5 μm) were deparaffinized and rehydrated in xylene and gradients of ethanol. Slides were boiled in citrate buffer (10 mM sodium citrate, 10 mM citric acid, pH 6.0) in a microwave oven at 92–98 °C for 15 min to retrieve antigen. The sections were then incubated with 3% H2O2 in methanol for 10 min to quench endogenous peroxidase and blocked with normal goat serum for 20 min. Sections were incubated with primary antibodies against IGF2BP3 (1 : 400; 14642-1-AP, Proteintech, Chicago, IL, USA) or β-hCG (1 : 200; ab9582, Abcam, Cambridge, MA, USA) at 4°C overnight. The IGF2BP3-treated sections were then incubated with biotinylated goat-anti-rabbit IgG (SP-9001) secondary antibody, and the β-hCG-treated sections were then incubated with biotinylated goat-anti-mouse IgG (SP-9002) secondary antibody. After incubations, the sections were stained with DAB working reagent (freshly mixed according to the manufacturer's instructions) for 30–60 s. Then counterstained with hematoxylin. Finally, sections were mounted with histomount (Zhongshan Golden Bridge Co., Ltd) on the slides. Negative control was performed by normal rabbit serum IgG (011-000-003, whole molecule, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) instead of the primary antibody. Images were acquired with a microscope (Nikon Eclipse 80i, Nikon, Tokyo, Japan).
To prepare the IF samples, the cells were fixed on the coverslip and were blocked with 5% donkey serum for 1 h at room temperature. Cells were incubated with primary antibodies against IGF2BP3 (1 : 400, Proteintech) at 4 °C overnight. After removing the primary antibodies, cells were incubated with the fluorescent secondary antibody for 1 h at room temperature. Phalloidin (1 : 10 000; P5282, Sigma-Aldrich Inc.) was incubated for 20 min after the incubation with the fluorescent secondary antibody, DAPI (4',6-diamidino-2-phenylindole, 1 : 500; D1306, Invitrogen, Carlsbad, CA, USA) was used to stain the nuclei. Carl Zeiss LSM 780 confocal laser-scanning microscope (Carl Zeiss MicroImaging GmbH, Jena, Germany) was used to capture the images.
Western blotting
The cells or tissues were lysed in whole-cell lysis buffer (50 mM HEPES, 150 mM NaCl, 1 mM EGTA, 10 mM sodium pyrophosphatate, 1.5 mM MgCl2, 100 mM NaF, 10% glycerol and 1% Triton X-100, pH 7.2) containing an inhibitor cocktail (1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin and 1 mM sodium orthovanadate) to extract total protein. Protein concentrations were determined using a standard bicinchoninic acid (BCA) assay (23225, Thermo Fisher Scientific Inc., Waltham, MA, USA), and 50 μg of total protein was subjected to 10% SDS-PAGE followed by electrotransfer onto nitrocellulose membranes. The membranes were blocked in 5% skim milk, and then incubated overnight at 4 °C with primary antibodies against human IGF2BP3 (1 : 4000, Proteintech), IGF2BP2 (1 : 10 000; 11601-1-AP, Proteintech), IGF2BP1 (1 : 10 000; ab82968, Abcam), GAPDH (1 : 20 000; ab37187, Abcam), β-hCG (1 : 500; ab9582, Abcam ), β-actin (1 : 20 000; TA-09, Zhongshan Golden Bridge Co., LTD), Caspase 3 (1 : 200; SC-7272, Santa Cruz Biotechnology, Santa Cruz, CA, USA), Phospho-AKT (Thr308) (1 : 500; 2965, Cell Signaling Technology, Beverly, MA, USA), Phospho-AKT (Ser 473) (1 : 500; 4060, Cell Signaling Technology) or AKT (pan) (1 : 500; 4691, Cell Signaling Technology). Membranes were then washed with TBST for 3 × 10 min. This was followed by incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature in 5% skim milk and washed with TBST for 3 × 10 min. Immunoreactive signals were detected using enhanced chemiluminescence (Pierce, Rockford, IL, USA). Three independent experiments were performed.
Cell culture and RNA interference
The HTR8/SVneo cell line, which was derived from the invasive extravillous trophoblast with a stable transfection of simian virus 40 large T antigen48 and was widely used as a model for the first trimester EVT invasion and migration, is a kind gift from Dr. Benjamin K Tsang (Department of Obstetrics & Gynecology and Cellular & Molecular Medicine, University of Ottawa, Ottawa, ON, Canada; Chronic Disease Program, Ottawa Hospital Research Institute, Ottawa, ON, Canada). Cells were cultured in RPMI-1640 medium (HyClone Laboratories, Inc.) with 10% fetal bovine serum (FBS, Gibco BRL, Carlsbad, CA, USA) at 37 °C in an incubator with 5% CO2. For RNA interference, cells were transfected with 100 nM IGF2BP3 siRNA (5′-UCACGAUAUCUCCAUUGCAGGAAUU-3′, Invitrogen (Frederick, MD, USA), Genbank ID for IGF2BP3: NM_006547.2) or scrambled siRNA (a universal negative control; Invitrogen, Frederick, MD, USA) with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) kit according to the manufacturer’s instructions.
Matrigel cell invasion and transwell cell migration assay
For Matrigel cell invasion assay, transwell inserts (6.5 mm, Costar, Cambridge, MA, USA) containing polycarbonate filters with 8 μm pores were precoated with 50 μl of 1 mg/ml Matrigel matrix (Becton Dickinson, Bedford, MA, USA). For cell migration assay, the inserts were not precoated with matrigel. 1.0 × 105 ofHTR8/SVneo cells in serum-free medium were plated in the upper chamber, whereas medium with 10% FBS was added to the lower chamber. After incubating for 24 h, the cells on the Matrigel side of the inserts were removed by cotton swab. The inserts were fixed in methanol and stained with hematoxylin and eosin (Zhongshan Golden Bridge Co., LTD). The number of invaded or migrated cells attached to the other side of the insert was counted under a light microscope (Olympus IX51, Japan) in eight random fields at a magnification of × 100. Three independent experiments were performed. Numbers of invasive or migrated cells under different treatments were normalized to the control and expressed as a means of invasion or migration percentage (%)±S.D. Conditional culture media were collected for gelatinolytic activity assay.
Explant culture and whole mount immunofluorescent
The first trimester (5–8 weeks) placental villi were dissected into the explants of 2–5 mm in diameter, and explanted in Millicell-CM culture dish inserts (0.4 mm pore size, Millipore, Carrigtwohill, Co., Cork, Ireland). The inserts were precoated with phenol red-free matrigel substrate (Becton Dickinson), and placed into 24-well culture plates (Costar). The explants were cultured at 3% O2 and 5% CO2 as previously described.49 The serum-free DMEM/F12 medium was added into the inserts, and the DMEM/F12 medium with 10% FBS was added into the 24-well culture plates. The explants, which were successfully anchored in the matrigel and initiated to grow, were used for the consequent experiments and were recorded every 24 h by using an inverted microscope system (Nikon Eclipse Ti). IGF2BP3 siRNA (300 nM) or an equal concentration of the scrambled siRNA was introduced into the two wells of explants that derived from the same placenta. Evaluation of EVT outgrowth was conducted as previously described.50, 51 EVT migration distance at individual sites was measured from the villous tip to the distal edge of the outgrowth sheet, as shown in Figure 4C. For each cytotrophoblast explant, four villous tips were randomly chosen, and at least two distances were measured for each tip. EVT migration distances of scrambled siRNA-treated and IGF2BP3 siRNA-treated groups were then subjected to statistical analysis. The distances from the cell column base to the tip of the outgrowth were measured with SPOT software (SPOT Imaging Solutions, a division of diagnostic instruments, Inc., Sterling Heights, MI, USA). EVT outgrowths were quantified as the area 72 h after plating divided by the initial area of the explant at 0 h using the Adobe Illustrator software (Adobe Systems Inc., San Jose, CA, USA). Outgrowth areas of IGF2BP3 siRNA-treated group were expressed as relative units to the scrambled siRNA-treated group, which was set to 100%.52
Samples from 13 placentas were used. Six explants from each placenta was collected and were divided into two groups, 3 for siRNA treatment and 3 for control treatment.
Whole mount immunofluorescent staining was performed to confirm the silencing efficiency of the siRNA. The explants cultured for 72 h together with matrigel were fixed by 4% neutral PFA at room temperature for 30 min, and then washed 3 × 30 min in PBS with 1% Triton X-100. The explanted villi were blocked with blocking buffer (PBS with 1% Triton X-100, 10% FBS and 0.2% sodium azide) for 1 h, and then incubated with primary antibodies against CK7 (1 : 100; Dako) or IGF2BP3 (1 : 400; Proteintech) at 4 °C for 2 days. The explants were washed 3 × 1 h in PBS with 1% Triton X-100 and 10% FBS, and 3 × 10 min in PBS with 1% Triton X-100. Explants were then incubated with fluorescent secondary antibody for 1 day and DAPI for 1 h sequentially. Finally, the fluorescent signals were photographed using an inverted microscope system (Nikon Eclipse Ti).
Cell proliferation assay
HTR8/SVneo cells transfected with IGF2BP3 siRNA or the scrambled siRNA for 24 h were seeded at a density of 1.0 × 104 cells per well in a 24-well plate. The culture medium was removed after 20, 44 and 68 h of culture, and then 500 μl MTT reagent (3-[4,5–dimethylthiazol–2–yl]-2,5-diphenyltetrazolium bromide; Apllygen Co., Beijing, China) was added. The MTT reagent was gently removed 4 h later and 500 μl dimethyl sulfoxide (DMSO) was added to each well. The optical density of each well was measured at 570 nm wavelength (Beckman DU530, Fullerton, CA, USA). Five independent experiments were performed.
Hoechst 33258 staining
Hoechst staining of HTR8/SVneo cells was performed to evaluate cell apoptosis after treatments of scrambled siRNA or IGF2BP3 siRNA.53 At the end of the culture, cells attached to the growth surface were trypsinized. Attached and detached cells were pooled, pelleted and resuspended in neutral-buffered formalin (10%) containing Hoechst 33258 dye (12.5 ng/ml, Sigma-Aldrich Inc.). Cells were spotted onto slides and assessed for typical apoptotic nuclear morphology (nuclear shrinkage, condensation and fragmentation) with Hoechst dye under a fluorescence microscope with the appropriate filter combination. At least 200 cells per treatment group were counted and assessed in randomly selected fields of blinded slides to avoid experimental bias. Three independent experiments were performed.
Gelatin zymography
Gelatin zymography assay was performed as previously described.54 Briefly, conditioned medium was diluted in 4 × sample buffer (8% SDS (w:v), 0.04% bromophenol blue (w:v), 0.25 M Tris) and incubated at 37 °C for 30 min. Then, 10 μl total protein was electrophoresed through a 10% polyacrylamide gels containing 0.5 mg/ml gelatin (Difco Laboratories, Detroit, MI, USA). After electrophoresis, the gel was washed in 2.5% Triton X-100 and 50 mM Tris-HCl (pH 7.5) for 2 × 30 min, followed by incubation in calcium assay buffer (50 mM Tris, 10 mM CaCl2, 1 mM ZnCl2, 1% Triton X-100, pH 7.5) for 24 h at 37 °C. Coomassie Brilliant Blue R250 in 50% methanol and 10% acetic acid was used to stain the gel for 1 h, and the gel was destained in 10% acetic acid for 2 × 30 min. Clear bands can be visualized in the areas where the gelatin was degraded. Four independent experiments were performed.
Reverse transcription and real-time PCR
Total RNA was extracted and purified from HTR8/SVneo cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The concentration of each RNA sample was determined by using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc.). Then, 2 μg of total RNA was used as template to reversely transcribe the RNA to cDNA with Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Quantitative real-time PCR was performed with SYBR Premix Ex Taq kit (Takara, Dalian, China) according to the manufacturer’s instructions using real-time PCR System (ABI PRISM 7500 Real-time PCR System; Applied Biosystems, Foster City, CA, USA). The samples were subjected to an initial stage of 3 min at 95 °C. The conditions for cDNA amplification were as follows: 40 cycles at 95 °C for 5 s, and 60 °C for 30 s. The experiment was performed in triplicates.
Specific primers used for amplification were as follows:
IGF2BP3 forward: 5′-GTCAAGTGCAGAAGTTGTTGTC-3′,
IGF2BP3 reverse: 5′-GCAATCTGTCTTTGGTTTGGC-3′,
IGF2 forward: 5′-CCGAAACAGGCTACTCTCCT-3′,
IGF2 reverse: 5′-AGGGTGTTTAAAGCCAATCG-3′,
CD44 forward: 5′-GCCCTTCCATAGCCTAATCC-3′,
CD44 reverse: 5′-CTTTGGTGTCTCCCAGAAGC-3′,
CD164 forward: 5′-GAGTGCTGTAGGATTAATTGGAAAAT-3′,
CD164 reverse: 5'-GGGAGGAATGGAATTCTGC-3',
GAPDH forward: 5'-GTCGCCAGCCGAGCCACATC-3',
GAPDH reverse: 5'-CCAGGCGCCCAATACGACCA-3'.
Statistical analysis
The bands from western blotting and gelatin zymography were quantified by MetaView Image Analyzing System (Version 4.50; Universal Imaging Co., Downingtown, PA, USA). Each experiment was performed in triplicates. Results were presented as means±S.D. The t-test was performed using the Statistical Package for Social Science (SPSS for Windows package release 10.0; SPSS Inc., Chicago, IL, USA) and indicated in Results and Figure legends. P<0.05 was considered as statistically significant. *P<0.05; **P<0.01.
Abbreviations
- IGF2BP3:
-
insulin-like growth factor-2 mRNA-binding protein 3
- CTB:
-
cytotrophoblast
- STB:
-
syncytiotrophoblast
- TC:
-
trophoblast column
- CK7:
-
cytokeratin 7
- hCG:
-
human chorionic gonadotropin
- EVT:
-
extravillous cytotrophoblast cell
- MMP:
-
matrix metalloproteinase
- PE:
-
pre-eclampsia
- IUGR:
-
intrauterine growth restriction
- F-actin:
-
linear polymer microfilament
- CD44:
-
cluster of differentiation 44
- IGF2:
-
insulin-like growth factor-2
References
Risteli J, Foidart JM, Risteli L, Boniver J, Goffinet G . The basement membrane proteins laminin and type IV collagen in isolated villi in pre-eclampsia. Placenta 1984; 5: 541–550.
Khong TY, De Wolf F, Robertson WB, Brosens I . Inadequate maternal vascular response to placentation in pregnancies complicated by pre-eclampsia and by small-for-gestational age infants. Br J Obstet Gynaecol 1986; 93: 1049–1059.
Hustin J, Jauniaux E, Schaaps JP . Histological study of the materno-embryonic interface in spontaneous abortion. Placenta 1990; 11: 477–486.
al-Okail MS, al-Attas OS . Histological changes in placental syncytiotrophoblasts of poorly controlled gestational diabetic patients. Endocr J 1994; 41: 355–360.
Lunghi L, Ferretti ME, Medici S, Biondi C, Vesce F . Control of human trophoblast function. Reprod Biol Endocrinol 2007; 5: 6.
Staun-Ram E, Shalev E . Human trophoblast function during the implantation process. Reprod Biol Endocrinol 2005; 3: 56.
Strickland S, Richards WG . Invasion of the trophoblasts. Cell 1992; 71: 355–357.
Karmakar S, Dhar R, Das C . Inhibition of cytotrophoblastic (JEG-3) cell invasion by interleukin 12 involves an interferon gamma-mediated pathway. J Biol Chem 2004; 279: 55297–55307.
Cohen M, Bischof P . Factors regulating trophoblast invasion. Gynecol Obstet Invest 2007; 64: 126–130.
Nielsen FC, Nielsen J, Christiansen J . A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand J Clin Lab Invest Suppl 2001; 234: 93–99.
Yaniv K, Yisraeli JK . The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene 2002; 287 49–54.
Yisraeli JK . VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell 2005; 97: 87–96.
Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM et al. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol 2004; 24: 4448–4464.
Müeller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H et al. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene 1997; 14: 2729–2733.
Jiang Z, Chu PG, Woda BA, Rock KL, Liu Q, Hsieh CC et al. Analysis of RNA-binding protein IMP3 to predict metastasis and prognosis of renal-cell carcinoma: a retrospective study. Lancet Oncol 2006; 7: 556–564.
Kapoor S . IMP3: a new and important biomarker of systemic malignancies. Clin Cancer Res 2008; 14: 5640 author reply 5640-1.
Findeis-Hosey JJ, Xu H . The use of insulin like-growth factor II messenger RNA binding protein-3 in diagnostic pathology. Hum Pathol 2011; 42: 303–314.
Findeis-Hosey JJ, Xu H . Insulin-like growth factor II-messenger RNA-binding protein-3 and lung cancer. Biotech Histochem 2012; 87: 24–29.
Samanta S, Sharma VM, Khan A, Mercurio AM . Regulation of IMP3 by EGFR signaling and repression by ERβ: implications for triple-negative breast cancer. Oncogene 2012; 31: 4689–4697.
Suvasini R, Shruti B, Thota B, Shinde SV, Friedmann-Morvinski D, Nawaz Z et al. Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. J Biol Chem 2011; 286: 25882–25890.
Deshler JO, Highett MI, Abramson T, Schnapp BJ . A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr Biol 1998; 8: 489–496.
Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP et al. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev 1998; 12: 1593–1598.
Zhang JY, Chan EK, Peng XX, Tan EM . A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J Exp Med 1999; 189: 1101–1110.
Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC . A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 1999; 19: 1262–1270.
Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG et al. Genes, development and evolution of the placenta. Placenta 2003; 24: 123–130.
Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y et al. Functional annotation of a full-length mouse cDNA collection. Nature 2001; 409: 685–690.
Silva WA Jr, Gnjatic S, Ritter E, Chua R, Cohen T, Hsu M et al. PLAC1, a trophoblast-specific cell surface protein, is expressed in a range of human tumors and elicits spontaneous antibody responses. Cancer Immun 2007; 7: 18.
Fant M, Barerra-Saldana H, Dubinsky W, Poindexter B, Bick R . The PLAC1 protein localizes to membranous compartments in the apical region of the syncytiotrophoblast. Mol Reprod Dev 2007; 74: 922–929.
Massabbal E, Parveen S, Weisoly DL, Nelson DM, Smith SD, Fant M . PLAC1 expression increases during trophoblast differentiation: evidence for regulatory interactions with the fibroblast growth factor-7 (FGF-7) axis. Mol Reprod Dev 2005; 71: 299–304.
Hohn HP, Denker HW . Experimental modulation of cell-cell adhesion, invasiveness and differentiation in trophoblast cells. Cells Tissues Organs 2002; 172: 218–236.
Monk D, Bentley L, Beechey C, Hitchins M, Peters J, Preece MA et al. Characterisation of the growth regulating gene IMP3, a candidate for Silver-Russell syndrome. J Med Genet 2002; 39: 575–581.
Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010; 141: 129–141.
Havens AM, Jung Y, Sun YX, Wang J, Shah RB, Bühring HJ et al. The role of sialomucin CD164 (MGC-24v or endolyn) in prostate cancer metastasis. BMC Cancer 2006; 6: 195.
Ellenbroek SI, Collard JG . Rho GTPases: functions and association with cancer. Clin Exp Metastasis 2007; 24: 657–672.
Reimer CL, Borras AM, Kurdistani SK, Garreau JR, Chung M, Aaronson SA et al. Altered regulation of cyclin G in human breast cancer and its specific localization at replication foci in response to DNA damage in p53+/+ cells. J Biol Chem 1999; 274: 11022–11029.
Sicinski P, Donaher JL, Geng Y, Parker SB, Gardner H, Park MY et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature 1996; 384: 470–474.
Kim J, Song G, Gao H, Farmer JL, Satterfield MC, Burghardt RC et al. Insulin-like growth factor II activates phosphatidylinositol 3-kinase-protooncogenic protein kinase 1 and mitogen-activated protein kinase cell Signaling pathways, and stimulates migration of ovine trophectoderm cells. Endocrinology 2008; 149: 3085–3094.
Ye Q, Cai W, Zheng Y, Evers BM, She QB . ERK and AKT signaling cooperate to translationally regulate survivin expression for metastatic progression of colorectal cancer. Oncogene 2013 e-pub ahead of print 29 April 2013 doi:10.1038/onc.2013.122.
Wang FP, Li L, Li J, Wang JY, Wang LY, Jiang W . High mobility group box-1 promotes the proliferation and migration of hepatic stellate cells via TLR4-dependent signal pathways of PI3K/Akt and JNK. PLoS One 2013; 8: e64373.
Gauster M, Siwetz M, Orendi K, Moser G, Desoye G, Huppertz B . Caspases rather than calpains mediate remodelling of the fodrin skeleton during human placental trophoblast fusion. Cell Death Differ 2010; 17: 336–345.
Huppertz B . Placental origins of preeclampsia: challenging the current hypothesis. Hypertension 2008; 51: 970–975.
Hogberg U . The World Health Report 2005: “make every mother and child count” - including Africans. Scand J Public Health 2005; 33: 409–411.
Levine RJ, Lam C, Qian C, Yu KF, Maynard SE, Sachs BP et al. Soluble endoglin and other circulating antiangiogenic factors in preeclampsia. N Engl J Med 2006; 355: 992–1005.
Roberts JM, Lain KY . Recent insights into the pathogenesis of pre-eclampsia. Placenta 2002; 23: 359–372.
George EM, Granger JP . Recent insights into the pathophysiology of preeclampsia. Expert Rev Obstet Gynecol 2010; 5: 557–566.
Zhang Q, Chen Q, Lu X, Zhou Z, Zhang H, Lin HY et al. CUL1 promotes trophoblast cell invasion at the maternal-fetal interface. Cell Death Dis 2013; 4: e502.
Fu JJ, Lin P, Lv XY, Yan XJ, Wang HX, Zhu C et al. Low molecular mass polypeptide-2 in human trophoblast: over-expression in hydatidiform moles and possible role in trophoblast cell invasion. Placenta 2009; 30: 305–312.
Graham CH, Hawley TS, Hawley RG, MacDougall JR, Kerbel RS, Khoo N et al. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res 1993; 206: 204–211.
Seeho SK, Park JH, Rowe J, Morris JM, Gallery ED . Villous explant culture using early gestation tissue from ongoing pregnancies with known normal outcomes: the effect of oxygen on trophoblast outgrowth and migration. Hum Reprod 2008; 23: 1170–1179.
Hu Y, Tan R, MacCalman CD, Eastabrook G, Park SH, Dutz JP et al. IFN-gamma-mediated extravillous trophoblast outgrowth inhibition in first trimester explant culture: a role for insulin-like growth factors. Mol Hum Reprod 2008; 14: 281–289.
Kuang H, Chen Q, Zhang Y, Zhang L, Peng H, Ning L et al. The cytokine gene CXCL14 restricts human trophoblast cell invasion by suppressing gelatinase activity. Endocrinology 2009; 150: 5596–5605.
Hromatka BS, Drake PM, Kapidzic M, Stolp H, Goldfien GA, Shih IeM et al. Polysialic acid enhances the migration and invasion of human cytotrophoblasts. Glycobiology 2013; 23: 593–602.
Sasaki H, Sheng Y, Kotsuji F, Tsang BK . Down-regulation of X-linked inhibitor of apoptosis protein induces apoptosis in chemoresistant human ovarian cancer cells. Cancer Res 2000; 60: 5659–5666.
Wang HX, Wang HM, Lin HY, Yang Q, Zhang H, Tsang BK et al. Proteasome subunit LMP2 is required for matrix metalloproteinase-2 and -9 expression and activities in human invasive extravillous trophoblast cell line. J Cell Physiol 2006; 206: 616–623.
Acknowledgements
This study was supported by the National Key Basic Research Program of China (2011CB944400), the Fundamental Research Funds for the Central Universities (105564GK) and grants from the Natural Science Foundation of China (81225004, 31171438 and 31070750).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by Y Shi
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Li, W., Liu, D., Chang, W. et al. Role of IGF2BP3 in trophoblast cell invasion and migration. Cell Death Dis 5, e1025 (2014). https://doi.org/10.1038/cddis.2013.545
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.545
Keywords
This article is cited by
-
GPC3 and PEG10 peptides associated with placental gp96 elicit specific T cell immunity against hepatocellular carcinoma
Cancer Immunology, Immunotherapy (2023)
-
N6‐methyladenosine (m6A) Modification in Preeclampsia
Reproductive Sciences (2023)
-
The role of insulin-like growth factor 2 mRNA binding proteins in female reproductive pathophysiology
Reproductive Biology and Endocrinology (2022)
-
Fragile X-Related Protein 1 (FXR1) Promotes Trophoblast Migration at Early Pregnancy via Downregulation of GDF-15 Expression
Reproductive Sciences (2022)
-
Dysfunction of Shh signaling activates autophagy to inhibit trophoblast motility in recurrent miscarriage
Experimental & Molecular Medicine (2021)