Abstract
Yme1L is an AAA protease that is embedded in the mitochondrial inner membrane with its catalytic domain facing the mitochondrial inner-membrane space. However, how Yme1L regulates mammalian mitochondrial function is still obscure. We find that endogenous Yme1L locates at punctate structures of mitochondria, and that loss of Yme1L in mouse embryonic fibroblast (MEF) cells results in mitochondrial fragmentation and leads to significant increased ‘kiss-and-run’ type of mitochondrial fusion; however, Yme1L knockdown (shYme1L (short hairpin-mediated RNA interference of Yme1L)) cells still remain normal mitochondrial fusion although shYme1L mitochondria have a little bit less fusion and fission rates, and the shYme1L-induced fragmentation is due to a little bit more mitochondrial fission than fusion in cells. Furthermore, shYme1L-induced mitochondrial fragmentation is independent on optic atrophy 1 (OPA1) S1 or S2 processing, and shYme1L results in the stabilization of OPA1 long form (L-OPA1); in addition, the exogenous expression of OPA1 or L-OPA1 facilitates the shYme1L-induced mitochondrial fragmentation, thus this fragmentation induced by shYme1L appears to be associated with L-OPA1’s stability. ShYme1L also causes a slight increase of mitochondrial dynamics proteins of 49 kDa and mitochondrial fission factor (Mff), which recruit mitochondrial key fission factor dynamin-related protein 1 (Drp1) into mitochondria in MEF cells, and loss of Drp1 or Mff inhibits the shYme1L-induced mitochondrial fragmentation. In addition, there is interaction between SLP-2 with Yme1L and shYme1L cells retain stress-induced mitochondrial hyperfusion. Taken together, our results clarify how Yme1L regulates mitochondrial morphology.
Similar content being viewed by others
Main
Mitochondria are dynamic double-membraned organelles whose shapes are determined by the equilibrium between organelle fusion and fission.1 These two events are essential for maintenance of a normal mitochondrial network and cellular function. Mitochondrial fission relies on the dynamin-related protein 1 (Drp1), mitochondrial fission factor (Mff), mitochondrial dynamics proteins of 49 kDa and 51 kDa (Mid49 and Mid51) recruit Drp1 to mitochondria from the cytosol in mammals.2 In humans, disruption of Drp1 is associated with neonatal lethality, microcephaly, abnormal brain development, hypoplasia and optic atrophy.3 Optic atrophy 1 (OPA1), Mfn1 and Mfn2 have been shown to be central for the fusion of mammalian mitochondria.4 The long form of OPA1, Mfn1 and SLP-2 are also required for the stress-induced mitochondrial hyperfusion (SIMH).5 Mitochondria fusions proceed with two modes: complete fusion and transient fusion events (‘kiss-and-run’ type), and the protein level of OPA1 governs the transition of these two fusion events.6 In humans, mutations in OPA1cause dominant optic atrophy,7 and mutations in Mfn2 cause Charcot-Marie-Tooth type 2A, an inherited peripheral neuropathy.8
The functions of OPA1 are regulated by mRNA splicing and proteolysis. OPA1 is proteolytic processed at S1 and S2 sites by the mitochondrial proteases OMA1 and Yme1L to generate short forms, uncleaved OPA1 forms are considered as OPA1 long forms, and both long and short forms of OPA1 are required for mitochondrial fusion.9, 10, 11, 12 Human Yme1L, identified as the ortholog of the Yme1p subunit of yeast mitochondrial i-AAA protease, is an ATP-dependent proteolytic complex in the mitochondrial inner membrane.13, 14 Yme1L belongs to the highly conserved family of AAA proteases, which contain the AAA domain and the M41 metallopeptidase domain harboring the consensus metal-binding site HEXXH.15 In yeast, Yme1 has chaperone-like activity in vitro,16 and Yme1 also assists in protein folding in the mitochondrial intermembrane space and affects mt-DNA escaping from mitochondria to nucleus.17, 18 Importantly, yeast Yme1 is responsible for mitochondrial protein quality control and mediates the degradation or proteolytic processing of its substrates.19, 20 In mammals, Yme1L regulates the processing of OPA1 at S2 site.11 The depletion of Yme1L leads to mitochondrial fragmentation,21, 22 but the mechanism for Yme1L in regulation of mitochondrial morphology is still obscure. Moreover, the roles of Yme1L in mammalian mitochondria are largely unknown. This present study reveals a crucial role for Yme1L in regulation of mitochondrial morphology.
Results
Yme1L regulates mitochondrial morphology depending on its protease activity
In yeast, Yme1 is a mitochondrial i-AAA protease with its catalytic domain located in the mitochondrial inner-membrane space.23 To further determine the localization of Yme1L in mammalian cells, we used immunofluorescence assay with an anti-Yme1L antibody to detect endogenous Yme1L in mouse embryonic fibroblasts (MEFs) expressing matrix-targeted GFP (mito-GFP). As shown in Figure 1A, red flourescences indicate the localization of Yme1L, they colocalize with mito-GFP green flourescences; in addition, the red flourescences display puncti form, suggesting that endogenous Yme1L is localized to punctate structures on mitochondria in MEF cells.
The knockdown of Yme1L results in mitochondrial fragmentation independent of OPA1 S2 processing. (A) Yme1L locates in punta on mitochondria. Matrix-tagged GFP-expressed MEF cells were fixed and were assessed by immunostainning with anti-Yme1L antibody and Cy3-conjugated second antibody, the localization of Yme1L was visualized by confocal microscope. (B) Effect of Yme1L knockdown on mitochondrial morphology. Mitochondrial morphology in control and Yme1L knockdown MEF cells were visualized by expression of mito-DsRed (a). Lysates from control cells and cells expressing shRNA against Yme1L were analyzed by western blotting with antibodies against to Yme1L or HSP60, and mitochondrial protein HSP60 was used as a loading control (b). (C) The role of Yme1L protease enzyme activity in the regulation of mitochondrial morphology. Human Yme1L and its mutant Yme1L-E600Q were expressed in WT or Yme1L knockdown MEF cells containing mito-DsRed, and mitochondrial morphology was scored according to the criteria detailed in Materials and Methods, standard deviations (S.D.) from three independent experiments in which 100 cell hybrids were scored. Statistical analysis was used by Student’s t-test (*P<0.05 versus control). A value of P<0.05 was considered significant. Statistical significant differences are shown with asterisks
To study the role of Yme1L in mitochondrial dynamics, we used short hairpin-mediated RNA interference (shRNAi) to reduce Yme1L efficiently and examined mitochondrial morphology in MEF cells (Figure 1B and b). Consistent with previous reports,21, 22 Yme1L knockdown (shYme1L (short hairpin-mediated RNA interference of Yme1L)) leads to remarkable mitochondrial fragmentation in about 70% of MEF cells, whereas wild-type (WT) MEF cells show almost all tubular mitochondria (Figures 1B and a and C).
To test whether the protease activity of Yme1L regulates mitochondrial morphology, we overexpressed hYme1L and its mutants hYme1L-E600Q (a point mutation in HEXXH motif, which is required for proteolysis) in MEF cells, hYme1L-E600Q, but not hYme1L, induces the partial fragmentation of mitochondria (Figure 1C), indicating that the proteolytic activity of Yme1L is required for regulation of mitochondrial dynamics. We also expressed shYme1L (against mouse Yme1L)-resistant proteins hYme1L or hYme1L-E600Q in shYme1L MEF cells; exogenous hYme1L expressed shYme1L cells show similar pattern of OPA1 processing to WT cells and thus restores partial proteolytic activity (Supplementary Figure 7). As shown in Figure 1C, the expression of hYme1L in shYme1L MEF cells reduces the fragmentation of mitochondria from 71% of cells (shYme1L cells) to 45% of cells; in contrast, 64% of cells expressing hYme1L-E600Q still exhibits fragmented mitochondria, further suggesting that the enzymatic activity of Yme1L is required for the maintenance of mitochondrial morphology.
Mitochondrial fusion ability is maintained in Yme1L knockdown cells but is biased toward the ‘kiss-and-run’ type and less fusion/fission rates
To examine whether the mitochondrial fragmentation induced by shYme1L is due to the deficiency of mitochondrial fusion, we checked the change of protein level of the mitochondrial fusion key proteins by western blot analysis. As shown in Figure 2A, the loss of Yme1L in MEF cells significantly decreases the cleavage of OPA1 at S2 site instead of increasing the processing at S1 cleavage site, similar results were found in HeLa cells (Supplementary Figure 1). In addition, no changes in protein levels of Mfn1 and Mfn2 were observed in shYme1L MEF or HeLa cells (Figure 2A and Supplementary Figure 1), indicating that the mitochondria still retain the capacity to fuse in shYme1L cells.
Yme1L knockdown leads to kiss-and-run type mitochondrial fusion. (A) Yme1L knockdown (shYme1L) and control MEF cell lysates were assessed by western blotting for changes in the protein levels of key factors (OPA1, Mfn1 and Mfn2) of mitochondrial fusion, and GAPDH and HSP60 were used as loading control. (B) The mitochondrial fusion activity in WT or shYme1L MEF cells were assessed by PEG fusion assay. The mitochondria in cells were imaged (a) by confocal microscope and were quantified (b) by estimating the percentage of mitochondria showing colocalization of fluorophores, error bars indicate S.D. from three experiments. (C) WT MEF cell expressing both mito-DsRed and mito-PA-GFP was photoactivated a small subset of mito-PA-GFP-labeled mitochondria by excitation and then the mitochondria were tracked by time-lapse microscopy for 20 min. The disperse of mito-PA-GFP were checked within 20 min (a), the major mitochondrial fusion type in WT cells is ‘Two combined into one’ (b). (D) The same assay in ‘C’ was analyzed in shYme1L MEF cells. The fluorescence of mito-PA-GFP is dispersed (a), and shYme1L cells proceed with mostly ‘kiss-and-run’ mitochondrial fusion type (b). (E) Comparison of mitochondrial fusion and fission between WT and shYme1L MEF cell. Ten photoactivated mitochondria labeled with mito-PA-GFP were tracked by time-lapse microscopy for 20 min in one cell. Three independent WT or shYme1L MEF cells were analyzed, the number of mitochondrial fission and two types of mitochondrial fusions within 20 min were counted. Bars represent means±S.D. of three independent experiments. Statistical significance analysis were used by Student’s t-test (*P<0.05 versus WT ‘kiss-and-run’ type). (F) Comparison of mitochondrial fusion type between WT and shYme1L MEF cells. Total 100 mitochondrial fusion events were assessed in both WT and shYme1L MEF cells, mitochondrial fission events of the same mitochondria were also calculated within the same time. (G) Behavior of matrix membrane markers mito-PA-GFP in WT and shYme1L MEF cells. mito-PA-GFP-based photoactivated mitochondrial assay was used as described in C. The photoactivated mito-PA-GFP-labeled mitochondria were traced every 20 s for 15 min by confocal microscope and the individual mitochondria analyzed by ImageJ software, the data from individual normalized mito-PA-GFP fluorescence traces of nine WT or shYme1L mitochondria from three independent experiments were averaged to yield the traces. The discontinuities in a few traces are due to brief defocusing during recording
To check the mitochondrial fusion activity, we performed PEG-induced cell hybrid assay. Both WT and shYme1L cells show larger number of merged yellow fluorescences (colocolization of mt-DsRed and mt-GFP) under confocal microscope (Figure 2B and a), and further quantified data demonstrate that 92% WT cells show mitochondrial full fusion and 84% shYme1L cells show full fusion (Figure 2B and b), indicating that shYme1L mitochondria have a little bit less fusion activity than WT mitochondria, but shYme1L cells still maintain normal mitochondrial fusion ability. To further test the mitochondrial fusion activity in shYme1L cells, we adopted an assay as described previously4 in living MEF cells that uses photoactivatible GFP (PA-GFP). Within a single cell, we photoactivated a small subset of mito-PA-GFP-labeled mitochondria by excitation and then tracked the mitochondria by time-lapse microscopy for 20 min. In WT cells, mito-PA-GFP in mitochondria were found to be transferred to non-photoactivated mitochondria because of the mitochondrial fusion (Figure 2C and a and Supplementary Movie 1). Similarly, in shYme1L cells, photoactivated mitochondria could transfer mito-PA-GFP to nonactivated mitochondria (Figure 2D and a and Supplementary Movie 2). Interestingly, the fusion type of most shYme1L mitochondria is different from the type in WT mitochondria, instead of the ‘two combining into one’ type in WT cells (Figure 2C and b and Supplementary Movie 1), photoactivated mitochondria immediately dissociated from the nonactivated mitochondria after transferring its mito-PA-GFP in shYme1L cells (Figure 2D and b, Supplementary Movie S2), this type of mitochondrial fusion is called ‘kiss-and-run’ type or ‘transient fusion’.24 We also tracked 10 photoactivated WT or shYme1L mitochondria labeled with mito-PA-GFP in one cell, and recorded the number of mitochondrial fission and two types of mitochondrial fusion. Figure 2E shows 17.3 ‘two combining into one’ type and 2.7 ‘kiss-and-run’ type of mitochondrial fusion events in WT cell, and only 4.3 ‘two combining into one’ type and 9.3 ‘kiss-and-run’ type of mitochondrial fusion events in shYme1L cell, suggesting that Yme1L knockdown leads to significant increased mitochondrial ‘kiss-and-run’ fusion type. In addition, Figure 2E shows 20 mitochondrial fusion and 21 fission events in WT cells and 13.6 fusion and 16.3 fission events in shYme1L cells, indicating that shYme1L results in less mitochondrial fusion/fission rates per mitochondrion, perhaps because shYme1L cells are unhealthy22 and may have less mitochondrial movement activity; or because shYme1L mitochondrion is fragmented and small and has less opportunity to fusion or fission. We also summed 100 mitochondrial fusion events for analysis in each cell line. As shown in Figure 2F, in WT cells, there were 100 mitochondrial fusion events and 91 fission events, fission events and fusion events are balanced, and only 15 out of 100 fusions belong to ‘kiss-and-run’ type; additionally, in shYme1L cells, 100 mitochondrial fusion events and 122 fission events happened, reveal that shYme1L cells exhibit more mitochondrial fission than fusion. Importantly, 63 out of 100 fusion events in shYme1L cells belong to ‘kiss-and-run’ type (Figure 2F), suggesting that shYme1L results in significant increased mitochondrial ‘kiss-and-run’ fusion type. From the time-lapse images of WT or shYme1L cells, we also tracked and quantified the GFP fluorescence intensity of individual photoactivated mitochondria as a function of time. As shown in Figure 2G, the averaged GFP fluorescence of WT photoactivated mitochondria declined by about 65% after 15 min of continual recording, and the averaged fluorescence of shYme1L photoactivated mitochondria declined by about 55%, indicating that shYme1L mitochondria still retain normal capacity to fusion although have a slightly declined fusion activity. Together, these results suggest that shYme1L significantly increases mitochondrial ‘kiss-and-run’ fusion type in MEF cells and that shYme1L-induced mitochondrial fragmentation is due to a little bit less fusion than fission events in cells.
The mitochondrial fragmentation (‘kiss-and-run’ type) induced by Yme1L knockdown is independent on OPA1 S2 or S1 processing
Yme1L is associated with proteolysis of OPA1 at the S2 site,11 we then examined whether the shYme1L-induced mitochondria fragmentation (‘kiss-and-run’ type) is due to impaired OPA1 S2 processing. OPA1−/− MEF cells show completely fragmented mitochondria due to loss of mitochondrial fusion (Figure 3A and a), expression of either isoform_1 (only S1 site exists) or isoform_7-ΔS1 (S1 site is deleted and only S2 site exists) causes partial tubulation of mitochondria (Figure 3A and a) and leads to the existence of long and short OPA1 forms (Figure 3A and b). In OPA1−/− cells expressing isoform_7-ΔS1, shYme1L results in a significant decrease of S2 processing (Figure 3A and b). Deficiency of Yme1L in OPA1−/− cells containing isoform_1 causes a slight increase of S1 processing (Figure 3A and b). Importantly, shYme1L does not result in complete loss of OPA1 long form or short form in OPA1−/− cells expressing isoform_1 or isoform_7-ΔS1 (Figure 3A and b). Surprisingly, shYme1L leads to almost all fragmented mitochondria in OPA1−/− cells expressing isoform_1 (Figure 3A and a), suggesting that fragmentation occurs independent of S2 processing as isoform_1 does not contain S2 site. Furthermore, shYme1L also causes significantly mitochondrial fragmentation in isoform_7-ΔS1-expressed OPA1−/− cells, indicating that shYme1L-induced fragmentation is also independent of OPA1 S1 processing. It should be noted that although shYme1L-induced mitochondrial fragmentation is independent of OPA1 S2 or S1 processing, it is dependent on processing as OPA1 processing exists in either isoform_1- or isoform_7-ΔS1-expressed OPA1−/− cells (Figure 3A and b) and OPA1 processing is essential for mitochondrial fusion.11
OPA1 S1 or S2 processing is not required for mitochondrial fragmentation induced by Yme1L knockdown. (A) Mitochondria morphology was scored in OPA1 null MEF cells expressing OPA1 isoform_1 or 7-ΔS1 with or without Yme1L knockdown (a), bars represent means±S.D. of three independent experiments, and statistical significance analysis were used by Student’s t-test (*P<0.05 versus control), a value of P<0.05 was considered significant. OPA1 processing in OPA1 null MEF cells expressing OPA1 isoform_1 or 7-ΔS1 with or without shYme1L was analyzed by western blotting with anti-OPA1 antibody (b). Isfm7-dS1 indicates ‘OPA1 isoform_7-ΔS1’. (B) The morphology of OMA1 null cells with or without Yme1L knockdown was analyzed by confocal microscope (a), and mitochondrial morphology were scored, bars represent means±S.D. of three independent experiments, and statistical significance were determined by Student’s t-test (*P<0.05 versus control) (b). (C) The mitochondrial fusion activity in OMA1 null or shYme1L OMA1 null MEF cells were analyzed by PEG fusion assay. The mitochondria in cells were imaged (a) by confocal microscope and were quantified (b) by estimating the percentage of mitochondria showing colocalization of red and green fluoresences
To further examine the effect of OPA1 S1 processing on shYme1L-induced mitochondrial fragmentation, we checked the mitochondrial morphology in shYme1L OMA1 null MEF cells. As shown in Figure 3B, OMA1 null cells show normal tubular mitochondria (97%), but shYme1L resulted in significant fragmented mitochondria (59%), suggesting that OPA1 S1 processing is not required for shYme1L-induced mitochondrial fragmentation as the OPA1 S1 processing is lost in OMA1 null cells (Figure 5C). As shYme1L results in less OPA1 processing in OMA1 null cells (Figure 5C), to exclude the possibility that the mitochondrial fragmentation is resulted from less mitochondrial fusion activity, we adopt a PEG assay to check the mitochondrial fusion activity; both OMA1 null and OMA1 null plus shYme1L cells show almost all full mitochondrial fusion activity (Figure 3C), indicating that shYme1L-induced mitochondrial fragmentation in OMA1 null cells is not due to the deficiency of mitochondrial fusion but is the result from more fission than fusion events and increased ‘kiss-and-run’ fusion type as mentioned previously. Taken together, shYme1L-induced mitochondrial fragmentation (‘kiss-and-run’ type) is independent of OPA1 S1 or S2 processing although OPA1 processing is required.
Exogenous OPA1 expression facilitates shYme1L-induced mitochondrial fragmentation
Greatly high expression of OPA1 in cells results in mitochondrial fragmentation and increases ‘kiss-and-run’ type of mitochondrial fusion.6, 25, 26 To check the role of OPA1 in shYme1L-induced mitochondrial fragmentation, we slightly increased the protein level of OPA1 by expressing OPA1 isoform_1 in MEFs, and the expression level of OPA1 isoform_1 is about 1.5-fold of endogenous isoform_1 (Figure 4b). The expression of exogenous isoform_1 results in elongated mitochondria and about 84% mitochondria show tubular, whereas 72% mitochondria were tubular in control cells (Figure 4a), these changes in mitochondrial morphology are consistent with observations by other groups.6 To test the effect of OPA1 on shYme1L-induced mitochondrial fragmentation, we then performed Yme1L knockdown with less efficiency to distinguish the differences of mitochondrial morphology between OPA1 expressed and control cells. As shown in Figure 4b, the efficiency of shYme1L was 60%, and this shYme1L causes only 35% fragmented mitochondria in control cells but results in 58% fragmented mitochondria in OPA1 isoform_1-expressing cells (Figure 4a), indicating that OPA1 isoform_1 expression facilitates shYme1L-induced mitochondrial fragmentation. We also examined the effect of L-OPA1 on shYme1L-induced mitochondrial fragmentation. As shown in Figure 4b, about 1.5-fold of exogenous L-OPA1 (OPA1 isoform_1-ΔS1) in cells leads to 83% tubular mitochondria (Figure 4a), indicating L-OPA1 expression increases mitochondrial fusion, which is consistent with previous report.27 Furthermore, 60% Yme1L knockdown results in 56% fragmented mitochondria in L-OPA1-expressing MEF cells (Figure 4a), suggesting that L-OPA1 expression increases shYme1L-induced mitochondrial fragmentation.
OPA1 facilitates shYme1L-induced mitochondrial fragmentation. (a) The mitochondrial morphology of WT, WT MEF cells expressing exogenous OPA1 isoform_1 or L-OPA1 (OPA1 isoform_1-ΔS1), shYme1L and shYme1L cells expressing exogenous OPA1 isoform_1 or isoform_1-ΔS1 were analyzed and quantified. Data represent means±S.D. of three independent experiments, and statistical significance were determined with Student’s t-test (*P<0.05 versus shYme1L). OPA1_Isfm1-dS1 indicates ‘OPA1 isoform_1-ΔS1’. (b) The protein level of OPA1 in cells expressing exogenous OPA1 isoform_1 or L-OPA1 was assessed by western blotting using anti-OPA1 antibody. Band ‘c’ indicates S1 products from OPA1 isoform 4,7,6,8; band ‘d’ indicates S2 products from OPA1 isoform 4,7,6,8; band ‘e’ indicates S1 products from OPA1 isoform 1,2,3,5
Exogenous OPA1 expression plus shYme1L reduces mitochondrial membrane potential and accelerates actinomycin D-indcuced apoptosis
To check the effect of the expression OPA1 isoform_1 or isoform_1-ΔS1 plus shYme1L on mitochondrial functions, we performed TMRM assay to measure mitochondrial membrane potential (Δψm) in OPA1−/− or OPA1−/− MEF cells expressing OPA1 isoform_1. As shown in Supplementary Figure 4A, the expression of OPA1 isoform_1 in OPA1−/− cells results in the increase of mitochondrial membrane potential, suggesting that OPA1 is required for maintaining of mitochondrial membrane potential. The data are consistent with previous observation.28 Interestingly, exogenous expression of isoform_1 or isoform_1-ΔS1 in WT cells does not lead to increase of Δψm, furthermore, exogenous expression of isoform_1 or isoform_1-ΔS1 in shYme1L MEF cells results in Δψm reduction (Supplementary Figure 4B), suggesting that the fragmented mitochondria induced by shYme1L plus expression of isoform_1 or isoform_1-ΔS1 shows lower Δψm.
We also checked cell apoptosis by annexinV-PE staining assay. ShYme1L increases actinomycin D-induced apoptosis in MEFs, and exogenous expression of OPA1 isoform_1 or isoform_1-ΔS1 inhibits actinomycin D-induced apoptosis (Supplementary Figure 5). These results are consistent with the previous reports.21, 22, 29 However, Supplementary Figure 5 shows that exogenous expression of OPA1 isoform_1 or isoform_1-ΔS1 facilitates actinomycin D-induced apoptosis in shYme1L MEF cells, indicating that in shYme1L cells, the exogenous OPA1 isoform_1 or isoform_1-ΔS1 may be not well processed or folded, thus the accumulation of L-OPA1 (non-processed OPA1) disrupts the balance of OPA1 long and short form and impairs the inhibition of OPA1 on apoptosis. We also analyzed oligomerization of OPA1 after crosslinking with EDC in WT and shYme1L mitochondria. Supplementary Figure 8 shows that the oligomerization of OPA1 is increased in shYme1L mitochondria. PARL have an important role in OPA1 processing, and the oligomerization of OPA1 is reduced in PARL null mitochondria.29, 30 The difference in the level of OPA1 oligomerization between PARL null and shYme1L mitochondria may be due to the improved stability of OPA1 by shYme1L, and the increased oligomerization of OPA1 in shYme1L mitochondria may be associated with increased ‘kiss-and-run’ mitochondrial fusion type and increased mitochondrial fragmentation rather than apoptosis.
We also assessed the mitochondrial cristae structure by TEM. WT and WT expressing exogenous OPA1 isoform_1 or isoform_1-ΔS1 MEF cells have normal organized mitochondrial cristae structure (Supplementary Figure 6); but shYme1L and shYme1L plus exogenous expression of OPA1 isoform_1 or isoform_1-ΔS1 MEF cells show disorganized abnormal mitochondrial cristae structure (Supplementary Figure 6). Exogenous expression of OPA1 isoform_1-ΔS1 partially restored normal cristae morphology in Prohibitin 2 (PHB2) null cells in which L-OPA1 is disappeared,31 but exogenous expression of OPA1 isoform_1-ΔS1 or isoform_1 does not restore normal cristae structure in shYme1L cells where OPA1 processing is impaired, indicating that balanced and well processed OPA1 is required for normal mitochondrial cristae morphology. These results suggest that shYme1L-induced disorganized mitochondrial cristae structure may be associated with unbalanced OPA1.
Loss of Yme1L results in the stabilization of L-OPA1
We found that o-phenanthroline, an effective inhibitor of mitochondrial metalloproteases, significantly prevents the L-OPA1’s degradation induced by carbonyl cyanide m-chlorophenylhydrozone (CCCP) treatment (Supplementary Figure 2), suggesting that the stability of L-OPA1 are controlled by the mitochondrial metalloproteases. To check whether Yme1L regulates the stability of L-OPA1, we then detected the protein level of L-OPA1 with or without CCCP treatment in shYme1L OPA1−/− MEF cells expressing OPA1 isoform_1-ΔS1. As shown in Figure 5A, shYme1L significantly inhibits the CCCP-induced L-OPA1’s degradation, suggesting that Yme1L regulates the stability of L-OPA1. It has been reported that the deficiency of PHB2 leads to the loss of L-OPA1.31 We found that PHB2 knockdown (shPHB2) results in the degradation of L-OPA1 (Figure 5B), and shYme1L could prevent the loss of L-OPA1 caused by shPHB2, further confirming that Yme1L could regulate the proteolytic degradation of L-OPA1.
Yme1L is responsible for stability of protein L-OPA1. (a) Control and Yme1L knockdown OPA1−/−MEF cells expressing L-OPA1 were incubated with CCCP, and then the stability of L-OPA1 was assessed by western blotting with anti-OPA1 antibody. (b) Control and shYme1L OPA1 null MEF cells expressing L-OPA1 were further induced by PHB2 knockdown (shPHB2), and the cell lysates were detected by western blotting with OPA1 antibody. (c) Control and Yme1L knockdown OMA1 null MEF cells were treated with or without CCCP for 4 h, and the stability of OPA1 were checked by western blotting using anti-OPA1 antibody
Mitochondrial protease OMA1 is associated with the stability of L-OPA1,9 to examine whether the regulation of L-OPA1 stability by Yme1L is dependent on OMA1; we checked the stability of L-OPA1 in OMA1 null cells. As shown in Figure 5C, CCCP treatment in OMA1 null cells also causes the degradation of L-OPA1, suggesting that OMA1 is not the only protease regulating the stability of L-OPA1. Interestingly, shYme1L significantly reduced the degradation of L-OPA1 by CCCP treatment in OMA1 null cells (Figure 5C), further confirming that Yme1L regulates the stability of L-OPA1.
To determine whether the effect of shYme1L on OPA1 processing and stability is associated with some other mitochondrial proteases, we assessed the change of protein levels by western blotting. Supplementary Figure 3 shows that shYme1L has no effect on protein level of some mitochondrial proteases, such as Omi, Afg3L2, Lon and OMA1. But OPA1 cleavages at S1 site by OMA1 are increased in shYme1L cells (Figure 2A), suggesting that Yme1L cooperates with OMA1 to regulate OPA1 processing and stability.
Drp1 and Mff are required for mitochondrial fragmentation induced by Yme1L deficiency
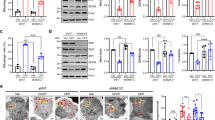
Mitochondrial fragmentation can be caused by either decreased mitochondrial fusion or increased mitochondrial fission. To clarify how mitochondrial fission machinery involves in shYme1L-induced mitochondrial fragmentation, we then checked the effect of shYme1L on the mitochondrial fission proteins Drp1, Mff, Mid49, Mid51 and mitochondrial fission protein 1 (Fis1) by western blotting. ShYme1L did not affect the protein levels of Drp1, Fis1 and Mid51 in MEF cells (Figure 6A); interestingly, shYme1L results in a slight increase of protein Mff and Mid49 (Figure 6A), indicating that mitochondrial fission appears to be promoted upon Yme1L deficiency. These results are consistent with our previous data that there is more fission than fusion events in shYme1L MEF cells (Figure 2E).
Drp1 and Mff are required for Yme1L knockdown-induced mitochondrial fragmentation. (A) Western blot analysis of control and shYme1L MEF cells using antibodies recognizing Drp1, Mff, Mid49, Mid51, Fis1, Yme1L and GAPDH. (B) The effect of shDrp1 (Drp1 knockdown) or shMff on shYme1L-induced mitochondrial fragmentation. Mitochondrial morphology was analyzed in control, shYme1L, shYme1L plus shDrp1 or shYme1L plus shMff MEF cells. Bars represent means±S.D. of three independent experiments, and statistical analysis were used by Student’s t-test (*P<0.05 versus WT_shYme1L). (C) The role of Mff in shYme1L induced mitochondrial fragmentation. Mff null cells were infected by control or shYme1L retrovirus for Yme1L knockdown, and cells were then fixed for imaging (a) by confocal microscope or were scored into three classifications (long tubular, tubular and >50% tubular) for analysis of mitochondrial morphology (b). Error bars represent means±S.D. of three independent experiments, and statistical significance analysis were used by Student’s t-test (*P<0.05 versus long tubular in Mff null cells)
To examine the importance of mitochondrial fission in shYme1L-induced mitochondrial fragmentation, we decreased mitochondrial fission in shYme1L MEF cells by Drp1 knockdown (shDrp1). Supplementary Figure 9 shows that shDrp1 efficiently reduces endogenous Drp1 in MEF cells. Importantly, compared with shYme1L cells, the deficiency of Drp1 in shYme1L cells dramatically reduced the fragmented mitochondrial from 64 to 19% (Figure 6B), suggesting that Drp1 is required for shYme1L-induced mitochondrial fragmentation.
Mff recruits Drp1 into mitochondria from cytosol,32 we performed Mff knockdown (shMff) assay in shYme1L cells, the efficiency of shMff is showed in Supplementary Figure 9. Figure 6B shows that the Mff knockdown in shYme1L cells reduced the fragmented mitochondrial from 64 to 17%. We also deplete Yme1L and assess mitochondrial morphology in Mff null MEF cells, as shown in Figure 6C, Mff null MEF cells showed about 83% long tubular mitochondria, and shYme1L Mff null MEF cells show 16% long tubular, 56% tubular and 28% ‘>50% tubular’ mitochondria, indicating that Yme1L knockdown promotes mitochondrial fragmentation in Mff null MEF cells. Compared with WT MEF cells (about 69% fragmented mitochondria in shYme1L MEF cells), shYme1L Mff null MEF cells show increased tubular mitochondria (Figure 1C and Figure 6C and b), suggesting Mff knockout could inhibit shYme1L-induced mitochondrial fragmentation.
Loss of Yme1L results in normal SIMH and the stabilization of SLP-2
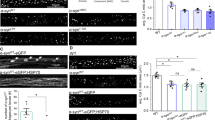
To test whether Yme1L is required for SIMH, we stimulated the SIMH with cycloheximide for 6 h in WT or shYme1L MEF cells. As shown in Figure 7A and a, after treatment of cycloheximide, both shYme1L cells (expressing nuclear targeted GFP) and WT cells showed abnormally elongated mitochondria because of SIMH. About 97% of WT cells contain abnormally long tubular mitochondria; about 70% shYme1L cells have abnormal long tubular mitochondria and about 22% shYme1L cells show tubular mitochondria (Figure 7A and b). These results suggest that shYme1L cells keep normal SIMH activity, although compared with WT cells, shYme1L cells show a slightly decreased long tubular mitochondria.
Yme1L is responsible for the stability of SLP-2. (A) WT and Yme1L knockdown MEF cells were mixed, and treated with cycloheximide (CHX, 50 μg/ml) for 6 h, the morphology of mitochondria was imaged (a) by confocal microscope and scored (b) into three classifications (long tubular, tubular and >50% tubular). Error bars indicate S.D. from three experiments. (B) Control WT and shYme1L MEF cell lysates were detected by western blotting with anti-SLP-2 antibody, GAPDH was used as loading control. (C) Myc-tagged Yme1L-E600Q or Myc-tagged Yme1L-E600Q plus Flag-tagged SLP-2 was transiently expressed in 293t cells and immunoprecipitated with anti-Flag M2 resin (Sigma-Aldrich), eluted protein samples were analyzed by western blotting with anti-Yme1L or anti-SLP-2 antibody
As L-OPA1 and SLP-2 are required for SIMH,5 to further check the effect of Yme1L on SIMH, we examined the stabilization of SLP-2 in shYme1L cells. As shown in Figure 7B, Yme1L deficiency in MEF cells leads to slightly increase of SLP-2 protein compared with control WT cells, suggesting that SLP-2 is stabilized by Yme1L knockdown. To test whether SLP-2 interacts with of Yme1L, we co-expressed SLP-2-Flag and Yme1L-E600Q-Myc in 293T cells and immunoprecipitation was performed with anti-Flag M2 beads. As shown in Figure 7C, SLP-2-Flag successfully precipitated exogenous Yme1L-E600Q-Myc and endogenous Yme1L, indicating that SLP-2 could efficiently interact with Yme1L. These results suggest that Yme1L is able to bind to SLP-2 and regulates SIMH in MEF cells.
Discussion
Mitochondrial AAA proteases (m-AAA and i-AAA protease) are key components of a quality control system in mitochondria.33 OPA1 is processed by mitochondrial protease OMA1 and i-AAA protease Yme1L at S1 and S2 sites.9, 10, 11, 21 OMA1 knockdown or knockout cells remain tubular mitochondria and resistant to CCCP-induced mitochondrial fragmentation;9, 10, 12 differently and interestingly, the depletion of Yme1L leads to obvious mitochondrial fragmentation and results in significantly increased ‘kiss-and-run’ type of mitochondrial fusion (Figures 2E and F). The reason may be as follows: first, compared with OMA1, Yme1L has a different cleave site (S2) on OPA1, and appears to have different ability to control the stability of L-OPA1; Second, mitochondrial ‘kiss-and-run’ type also involves in mitochondrial fission machinery, OMA1 locates at mitochondrial inner membrane, but the protease domain of Yme1L locates at mitochondrial inner membrane space, so Yme1L has more chance to be involved in mitochondrial fission in the help of other associated proteins, indeed, we found shYme1L results in slight increase of Mid49 and Mff in the protein level (Figure 6A).
Yme1L regulates the processing and stability of OPA1 (Figures 4b and 5), exogenous expression of OPA1 isoform1 or L-OPA1 accelerates the shYme1L-induced mitochondrial fragmentation (Figure 4a). In addition, exogenous expression of OPA1 isoform_1 or L-OPA1 plus shYme1L results in the reduction of mitochondrial membrane potential (Supplementary Figure 4) and increase of actinomycin D-induced apoptosis (Supplementary Figure 5) in MEFs. Our results suggest the importance of balanced OPA1 forms in mitochondrial morphology and function. WT cells have eight OPA1 isoforms, these isoforms form a precise balance among OPA1 long and short forms through processing and degradation regulated by mitochondrial proteases such as Yme1L and OMA1. In shYme1L cells, endogenous OPA1 processing and degradation were impaired, thus endogenous OPA1 isoforms especially S2 site containing isoforms such as isoform_7 are not well processed (Figure 4b), the exogenous expression of OPA1 isoform_1 or isoform_1-ΔS1 further leads to accumulation of L-OPA1 (non-processed OPA1) and disrupts the balance of OPA1 long and short forms (Figure 4b). In addition, exogenous isoform_1 or isoform_1-ΔS1 may compete to contact with other factors with endogenous OPA1 and may interfere with the processing, stability and function of endogenous OPA1 isoforms. In other words, exogenous isoform_1 or isoform_1-ΔS1 may have dominant negative effect on endogenous OPA1 isoforms. Indeed, Figure 4b shows that exogenous expression of isoform_1 or isoform_1-ΔS1 in shYme1L cells leads to increase of OPA1 band ‘c’ (S1 site processing products from isoform 4,7,6,8) and ‘e’ (S1 site processing products from isoform 1,2,3,5), suggesting that the processing and function of endogenous OPA1 isoforms are impaired under this condition. It should be noted that the expression of OPA1 isoform_1 alone in OPA1 null cells only recovers about half of mitochondria from fragmented to tubular morphology,11 it is consistent with our theory that the precise balances of endogenous OPA1 isoforms processing and stability are required for the optimal functions of OPA1.
Materials and Methods
Cell culture and transfection
All cell lines (MEFs, HeLa and 293T) were maintained in Dulbecco’s modified Eagle’s medium containing 10% FBS, penicillin/streptomycin and 1 mM L-glutamine. All cells were cultured in a 5% CO2 atmosphere at 37 °C. Lipofectamine 2000 and Opti-MEM I (Invitrogen, Carlsbad, CA, USA) were used for transient transfection with expression constructs according to the manufacturer’s protocol.
Plasmid and shRNAi construction
The isoforms of human OPA1cDNA and its mutants were subcloned into pMSCV-puro vector; Human Yme1L cDNA and its mutant Yme1L-E600Q were cloned into the retrovirus vector pCL βW containing a C-terminal 9xMyc tag, and human SLP-2 cDNA was insert into pMSCV-puro vector with a C-terminal 3xFlag tag.
shRNAi was performed by lentivirus vector pLKO.1, the vectors constructed with oligonucleotides 5′-CCGGAAGAGCTCTTCAAAGCATTTGCTCGAGCAAATGCTTTGAAGAGCTCTTTTTTTG-3′ and 5′-AATTCAAAAAAAGAGCTCTTCAAAGCATTTGCTCGAGCAAATGCTTTGAAGAGCTCTT-3′ for human Yme1L; shRNAi against mouse Yme1L and mouse Mff were performed as described previously,11 the oligonucleotides 5′-GATCCCCGTGGCAGAGGAATTCATATTTCAAGAGAATATGAGTTCCTCTGCCACTTTTTGGAAA-3′ and5′-AGCTTTTCCAAAAAGTGGCAGAGGAACTCATATTCTCTTGAAATATGAATTCCTCTGCCACGGG-3′ for mouse Yme1L; the oligonucleotides 5′-ATAGGATCCCCGTCGAATTCAGTATGAAATTTCAAGAGAATTTCATACTGAATTCGACTTTTTGGAAA-3′ and5′-GCGAAGCTTTTCCAAAAAGTCGAATTCAGTATGAAATTCTCTTGAAATTTCATACTGAATTCGACGGG-3′ for mouse Mff.
Antibodies and reagents
The following antibodies were used in this study: anti-OPA1 (BD Biosciences, San Jose, CA, USA), anti-Drp1 (BD Biosciences), anti-Flag (Sigma-Aldrich, St. Louis, MO, USA) and anti-GFP (Santa Cruz, Dallas, TX, USA); polyclonal anti-Yme1L, anti-SLP-2, anti-Mid49, anti-Mid51, anti-Mff and anti-Fis1 were purchased from Proteintech (Wuhan, China), polyclonal anti-GAPDH and anti-HSP60 were from Santa Cruz Biotechnology. The reagents used in this paper were: CCCP, o-phenanthroline and M2-Flag beads were purchased from Sigma-Aldrich, Lipofectmine 2000 is from Invitrogen.
PEG fusion assay
WT or Yme1L knockdown MEF cells stably expressing mitochondrially targeted EGFP were co-cultured with MEF cells stably expressing mitochondrially targeted DsRed. The next day, the PEG fusion assays were then performed as described previously.11
Co-immunoprecipitation
293T cells were transiently transfected with Lipofectamine 2000 (Invitrogen) by following the manufacturer’s protocols and transiently expressed GFP-tagged proteins plus Myc-tagged or Flag-tagged proteins. Approximately 36 h post-transfection, cells were lysed in a Triton X-100-based lysis buffer (1% Triton X-100, 10% glycerol, 150 mM NaCl, 20 mM Tris, pH 7.4, 2 mM EDTA, protease inhibitor mixture) for 1 h, and the nuclear and cellular debris were cleared by centrifugation. Then the cytosolic lysates were incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) for 4 h at 4 °C. The resin was washed five times with lysis buffer, and proteins were recovered by boiling the beads in SDS sample buffer and analyzed on SDS-PAGE followed by western blot analysis.
Immunostaining
Mitochondrial-targed GFP (mito-GFP)-expressed MEF cells were cultured on coverslips, then cells were fixed with 10% formalin for 10 min and washed with PBS buffer for three times, and cells were incubated with PBS plus 0.1% Triton X-100 buffer for 10 min. Later cells were blocked with PBS plus 5% FBS buffer at room temperature (RT) for 1 h. Later, cells were incubated with anti-Yme1L primary antibody diluted in PBS plus 5% FBS buffer for 2 h at RT. Cells were then washed with PBS buffer three times. Then, PBS buffer containing Cy3-conjugated anti-rabbit second antibody was used to incubate cells for 1 h at RT. Finally, cells were washed three times with PBS and slides were analyzed by using an Olympus confocal microscope (Olympus Corporation, Tokyo, Japan).
Confocal microscopy and image processing
Confocal microscopy (FV1000) was performed with an Olympus IX81 microscope with a × 100 UPL SAPO NA 1.4 oil objective. MEF or HeLa cells were cultured on coverslips and fixed with 10% formalin. Live MEF cells were maintained at 37 °C in a 5% CO2 atmosphere. In live MEF cells expressing mito-PA-GFP and mito-DsRed, mitochondria within a region of interest were photoactivated with a 405-nm laser light. EGFP or photoactivated PA-GFP was excited with a 25-mW 488-nm argon laser, and DsRed was excited with a 1-mW 543-nm helium/neon laser. Image processing and analysis were performed by FV10-ASW 3.0 software (Olympus Corporation). Mitochondria were identified by mito-GFP or mito-DsRed. To determine mitochondrial morphology, cells were randomly selected for quantitative analysis and visually scored into four classifications (tubular, >50% tubular, <50% tubular and fragmented) as described previously.11
Electron microscopy
Cells were fixed for 1 h at 4 °C in phosphate buffer (PB) with 2.5% glutaraldehyde. After five washes in PB buffer (pH 7.4) for 5 min each, cells were post-fixed for 40 min at RT in 1% osmium tetroxide (OSO4). After washes with PB buffer, cells were dehydrated in 70, 80, 95 and 100% ethanol (10 min for each procedure), and then the cells were further dehydrated in 100% acetone two times for 10 min each at RT. The samples were then infiltrated sequentially in 1 : 1 (vol/vol) actetone/epoxyresin (30 min), 1 : 3 acetone/epoxy resin (30 min), 100% epoxy resin (3 h) and finally 100% epoxy resin (48 h) at 60 °C for polymerization. The sections were supported on copper grids. The 80-nm sections were post-stained in Sato lead for 1 min, and the stained sections were imaged onto negatives using a Jeol electron microscope operated at 80 kV (Joel Ltd, Tokyo, Japan).
References
Bereiter-Hahn J, Voth M . Dynamics of mitochondria in living cells: shape changes, dislocations, fusion and fission of mitochondria. Microsc Res Tech 1994; 27: 198–219.
Youle RJ, van der Bliek AM . Mitochondrial fission, fusion, and stress. Science 2012; 337: 1062–1065.
Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV . A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med 2007; 356: 1736–1741.
Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC . Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell 2009; 20: 3525–3532.
Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J 2009; 28: 1589–1600.
Liu X, Weaver D, Shirihai O, Hajnóczky G . Mitochondrial 'kiss-and-run': interplay between mitochondrial motility and fusion-fission dynamics. EMBO J 2009; 28: 3074–3089.
Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet 2000; 26: 211–215.
Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet 2004; 36: 449–451.
Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol 2009; 187: 1023–1036.
Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM . Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol 2009; 187: 959–966.
Song Z, Chen H, Fiket M, Alexander C, Chan DC . OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol 2007; 178: 749–755.
Quirós PM, Ramsay AJ, Sala D, Fernández-Vizarra E, Rodríguez F, Peinado JR et al. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J 2012; 31: 2117–2133.
Coppola M, Pizzigoni A, Banfi S, Bassi MT, Casari G, Incerti B . Identification and characterization of YME1L1, a novel paraplegin-related gene. Genomics 2000; 66: 48–54.
Shah ZH, Hakkaart GA, Arku B, de Jong L, van der Spek H, Grivell LA et al. The human homologue of the yeast mitochondrial AAA metalloprotease Yme1p complements a yeast yme1 disruptant. FEBS Lett 2000; 478: 267–270.
Truscott KN, Lowth BR, Strack PR, Dougan DA . Diversefunctions of mitochondrial AAA+ proteins: protein activation, disaggregation, and degradation. Biochem Cell Biol 2010; 88: 97–108.
Leonhard K, Stiegler A, Neupert W, Langer T . Chaperonelike activity of the AAA domain of the yeast Yme1 AAA protease. Nature 1999; 398: 348–351.
Schreiner B, Westerburg H, Forné I, Imhof A, Neupert W, Mokranjac D . Role of the AAA protease Yme1 in folding of proteins in the intermembrane space of mitochondria. Mol Biol Cell 2012; 23: 4335–4346.
Campbell CL, Thorsness PE . Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J Cell Sci 1998; 111 (Pt 16): 2455–2464.
Baker BM, Haynes CM . Mitochondrial protein quality control during biogenesis and aging. Trends Biochem Sci 2011; 36: 254–261.
Koppen M, Langer T . Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol 2007; 42: 221–242.
Griparic L, Kanazawa T, van der Bliek AM . Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol 2007; 178: 757–764.
Stiburek L, Cesnekova J, Kostkova O, Fornuskova D, Vinsova K, Wenchich L et al. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol Biol Cell 2012; 23: 1010–1023.
Graef M, Seewald G, Langer T . Substrate recognition by AAA+ ATPases: distinct substrate binding modes in ATP-dependent protease Yme1 of the mitochondrial intermembrane space. Mol Cell Biol 2007; 27: 2476–2485.
Nijtmans LG, de Jong L, ArtalSanz M, Coates PJ, Berden JA, Back JW et al. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J 2000; 19: 2444–2451.
Chen H, Chomyn A, Chan DC . Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 2005; 280: 26185–26192.
Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 2008; 27: 433–446.
Ishihara N, Fujita Y, Oka T, Mihara K . Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J 2006; 25: 2966–2977.
Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem 2003; 278: 7743–7746.
Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 2006; 126: 177–189.
Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1 dependentcristae remodeling. Cell 2006; 126: 163–175.
Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Löwer B, Wunderlich FT et al. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev 2008; 22: 476–488.
Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol 2010; 191: 1141–1158.
Martinelli P, Rugarli EI . Emerging roles of mitochondrial proteases in neurodegeneration. Biochim Biophys Acta 2010; 1797: 1–10.
Acknowledgements
We thank Dr David Chan (California Institute of Technology) for communicating results before publication and for providing the OPA1−/− and Mff−/− MEF cell lines, we thank Dr Carlos López-Otín (Universidad de Oviedo) for gift of OMA1−/− MEF cells. This work was supported by grants from the National Natural Science Foundation of China (31171357), National Key Basic Research Program of China (2013CB531200).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Agostini
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Ruan, Y., Li, H., Zhang, K. et al. Loss of Yme1L perturbates mitochondrial dynamics. Cell Death Dis 4, e896 (2013). https://doi.org/10.1038/cddis.2013.414
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.414