Abstract
Neural progenitor cells (NPCs) derived from human embryonic stem cells (hESCs) have great potential in cell therapy, drug screening and toxicity testing of neural degenerative diseases. However, the molecular regulation of their proliferation and apoptosis, which needs to be revealed before clinical application, is largely unknown. MicroRNA miR-195 is known to be expressed in the brain and is involved in a variety of proapoptosis or antiapoptosis processes in cancer cells. Here, we defined the proapoptotic role of miR-195 in NPCs derived from two independent hESC lines (human embryonic stem cell-derived neural progenitor cells, hESC-NPCs). Overexpression of miR-195 in hESC-NPCs induced extensive apoptotic cell death. Consistently, global transcriptional microarray analyses indicated that miR-195 primarily regulated genes associated with apoptosis in hESC-NPCs. Mechanistically, a small GTP-binding protein ADP-ribosylation factor-like protein 2 (ARL2) was identified as a direct target of miR-195. Silencing ARL2 in hESC-NPCs provoked an apoptotic phenotype resembling that of miR-195 overexpression, revealing for the first time an essential role of ARL2 for the survival of human NPCs. Moreover, forced expression of ALR2 could abolish the cell number reduction caused by miR-195 overexpression. Interestingly, we found that paraquat, a neurotoxin, not only induced apoptosis but also increased miR-195 and reduced ARL2 expression in hESC-NPCs, indicating the possible involvement of miR-195 and ARL2 in neurotoxin-induced NPC apoptosis. Notably, inhibition of miR-195 family members could block neurotoxin-induced NPC apoptosis. Collectively, miR-195 regulates cell apoptosis in a context-dependent manner through directly targeting ARL2. The finding of the critical role of ARL2 for the survival of human NPCs and association of miR-195 and ARL2 with neurotoxin-induced apoptosis have important implications for understanding molecular mechanisms that control NPC survival and would facilitate our manipulation of the neurological pathogenesis.
Similar content being viewed by others
Main
Successful differentiation of human embryonic stem cells (hESCs)1 or induced pluripotent stem cells2 into neural progenitor cells (NPCs) opens up fascinating prospects for human brain research and potential application in neural degenerative diseases. Clinical cell replacement-based therapies depend critically on the comprehensive understanding of donor cells. However, little is known about the cellular behaviors of NPCs derived from hESCs or the molecular mechanisms coordinating their proliferation and survival. Although numerous studies in rodent models have provided major molecular insights into these cellular processes,3 investigations performed directly on human cells are of great significance for developing effective therapeutic strategies.
During the past decade, a new class of regulatory genes known as microRNAs (miRNAs) has attracted a great deal of attention as an additional layer of mRNA regulation in eukaryotes. MicroRNAs are endogenous non-coding RNAs, approximately 22 nucleotides in length,4 and assembled into RNA-induced silencing complex (RISC), which facilitates their interaction with the 3′-untranslated region (3′-UTR) of target mRNAs through imperfect complementary base paring. The 2–8 bases of the 5′ portion of the miRNA sequence termed ‘seed region’ are critical in target recognition. Usually, miRNAs negatively regulate target gene expression through mRNA translation repression or degradation.5 The human genome is predicted to contain over 1000 miRNAs, about 3% of the total number of human genes.5 They are known to have important roles in diverse biological processes, including cancer pathogenesis,6 neural development,7 cell cycle regulation8 and so on. A feature of miRNAs is that they can modulate the expression of many genes simultaneously, making them powerful post-transcriptional regulators.9 Moreover, a single miRNA could execute different biological functions in different cell types. For this reason, the precise roles of miRNAs in various cell types should be investigated in specific situations if we want to understand miRNAs comprehensively.
miR-195 belongs to the miR-15/16 family, which consists of a group of miRNAs (miR-195, miR-15a, miR-15b, miR-16-1 and miR-16-2) sharing the same seed sequence.10 The mature sequences of miR-195 are evolutionarily conserved among mammalian species.11 Previous studies have shown that miR-195 is aberrantly expressed in multiple cell types of diseases including human hepatocellular carcinoma,12 glioblastoma,13, 14 colorectal cancer15 and cardiac hypertrophy.16 It was reported that, like other members of miR-15/16 family, miR-195 has various roles in the regulation of cell cycle progression17, 18 and cell apoptosis15, 19 in cancer progression and cardiovascular development. Notably, in different cell types, miR-195 may function oppositely. For instance, in DICER or DROSHA knockdown hESCs, miR-195 suppressed WEE1 to promote cell proliferation.17 However, in human glioblastoma cells, miR-195 targeted E2F3 directly leading to cell cycle arrest.14 In addition, miR-195 displays a moderate to low expression level in the mammalian embryonic brain, with the highest level at the preadult brain developmental stage.11 Despite all these progresses in miR-195 research, the functions of miR-195 in human NPCs have not been examined.
In this study, we demonstrated the role of miR-195 in coordinating NPC survival and apoptosis at the early stage of neural differentiation and identified a GTP-binding protein, ADP-ribosylation factor-like protein 2 (ARL2), as a genuine functional target of miR-195 in these biological processes. In addition, we found that the expression of miR-195 increased with the treatment of neurotoxin, paraquat and rotenone, implicating its potential involvement in regulating NPC response to neurotoxins.
Results
NPCs are generated from hESCs
To generate NPCs from hESCs, we used an adherent differentiation protocol modified from a previously reported approach.20 Undifferentiated hESCs of SHhES1 line,21 previously derived in our laboratory and maintained under a feeder-free condition (Figure 1a, upper schema), were treated with bone morphogenetic protein antagonist Noggin (100 ng/ml) for about 3 weeks. When the polarized neural epithelial structure became clearly visible, cells were picked up mechanically en bloc for neurosphere formation in the presence of basic fibroblast growth factor (bFGF) (Figure 1a, lower panel). After suspension culture, neurospheres were replated onto Matrigel-coated culture dishes. These cells, designated as passage 1 (p1) of SHhES1-NPCs, formed typical neural progenitor rosette structures. Later, NPCs were dissociated into single cells and replated at a high density for further expansion (Figure 1b). To characterize the properties of dissociated NPCs, expression levels of multiple neural lineage markers were examined by immunofluorescence staining. Nearly 90% of NPCs expressed NPC markers, SOX2 and Nestin (Figure 1c), whereas astrocyte marker GFAP and oligodendrocyte marker OLIG2 were rarely detected in p2 NPCs (Figure 1d). The high percentage of Ki-67-positive cells indicated that the majority of NPCs were still in cell cycle progression (Figure 1e). Moreover, dissociated NPCs were capable of generating neurospheres (Figure 1f). The differentiation potential of ShHES1-NPCs was tested by a spontaneous differentiation assay. Differentiated neural cells stretched from neurospheres (Figure 1g). Most of the cells were immunopositive to the antibody of neuron marker Tuj1 (Figure 1h), whereas a few of them were GFAP-positive glial cells (Figure 1i). SHhES1-NPCs generated with our protocol were expandable at a relatively high dividing rate and maintained the multipotent potential for at least 10 passages when bFGF was supplemented (data not shown). These NPCs hence became an ideal cellular tool for the study of molecular mechanisms governing NPC properties during early neural differentiation.
SHhES1-NPCs are generated from human ESCs of SHhES1 line. (a) Schematic flow diagram (upper panel) and the representative morphology illustrating the sequential steps of neural differentiation (bottom panel). (b) Isolated rosettes were dissociated into single cells (p2) in the presence of bFGF (10 ng/ml). (c) Coimmunofluorescence staining for Nestin and SOX2 in p2 NPCs. (d) Coimmunostaining for GFAP and OLIG2. (e) Immunofluorescence staining for Ki-67. (f) Neurospheres formed from dissociated NPCs (p2) in the suspension culture with bFGF (10 ng/ml). (g) The morphology of adherent neurospheres after suspension for 2 weeks in the absence of bFGF. (h) and (i) Immunofluorescence staining of Tuj1 (h) and GFAP (i) in the cells indicated in (g). Scale bars in (a–i), 100 μm. In (c)–(e), (h) and (i) DAPI (blue) was used to highlight the nucleus
In addition, we generated NPCs from another independent hESC line (H9 line1) using a 3-inhibitor neural induction method, which was a modification of previously reported protocols.22 The three inhibitors included Noggin, SB431542 (pharmacological inhibitor of type I activin/nodal receptor signaling23) and compound C (a chemical inhibitor of transforming growth factor β (TGFβ) superfamily receptors24) (Supplementary Figure S1a). With this protocol, NPCs were efficiently and quickly generated from H9 hESCs (H9-NPCs). Immunofluorescence staining results showed that H9-NPCs expressed NPC markers such as CD133, DACH1, ZIC1, Nestin and SOX2 in a neural tube-like rosette structure (Supplementary Figure S1b). Expression of SOX2, Nestin and Ki-67 was also detected in dissociated H9-NPCs, whereas cells immunoreactive to antibodies of MAP2, GFAP, OLIG2 and LEX1 were rarely observed (Supplementary Figure S1c). Furthermore, flow cytometry analysis (FCA) showed that over 96% of H9-NPCs were SOX2+/OCT4−. Thus, H9-NPCs displayed the cell morphology and marker expression similar to SHhES1-NPCs and both of them were used in this study.
Overexpression of miR-195 in NPCs induces apoptosis
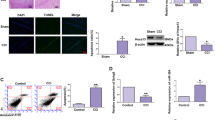
To explore whether miR-195 has a role in human embryonic stem cell-derived neural progenitor cell (hESC-NPC) proliferation and apoptosis, we overexpressed miR-195 in SHhES1-NPCs. Transfection of double-stranded miR-195 precursor (pre-miR-195) into SHhES1-NPCs reduced cell number by 30% as compared with the transfection of control scramble precursor (pre-miR-NC) (Figure 2a). As it has been reported that miR-195 affected cell cycle or cell apoptosis in other cell types,12, 15 we examined whether the NPC cell number reduction after overexpression of miR-195 was due to cell cycle alteration or increased cell apoptosis. FCA did not detect any discernible differences in the cell cycle profiles and 5-bromo-2′-deoxyuridine (BrdU) incorporation rate between miR-195-overexpressed NPCs and control cells (Supplementary Figures S2a and b), indicating that miR-195 might not control cell cycle distribution or proliferation of hESC-NPCs. Instead, Annexin V-PI staining assays detected more Annexin V-positive apoptotic cells in miR-195-overexpressed SHhES1-NPCs than in pre-miR-NC-transfected cells (Figure 2b), consistent with results of the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay (Figure 2c). In addition, the protein level of cleaved caspase-3 was upregulated after pre-miR-195 transfection in SHhES1-NPCs (Figure 2d). These results demonstrate that miR-195 can induce apoptosis in hESC-NPCs potently.
miR-195 induces cell apoptosis in hESC-NPCs. (a) Left panel: Cell images of SHhES1-NPCs 72 h after transfection with 50 nM of pre-miR-NC or pre-miR-195. Mock is the transfection reagent control. Scale bars, 100 μm. Right panel: The statistical analysis of cell numbers of NPCs shown in the left panel; n=3, *P<0.05. The value of pre-miR-NC was set as 1. (b) Left panel: Apoptosis was shown as the percentage of Annexin V-positive cells in cells indicated in (a) by FCA; Right panel: The statistical analysis of apoptotic cells; n=3, **P<0.01. (c) TUNEL assays were performed 48 h after transfection of 50 nM of pre-miR-195 or pre-miR-NC to detect apoptotic cell death. (d) A representative immunoblotting of cleaved caspase-3 in NPCs with 50 nM of pre-miR-195 or pre-miR-NC transfection for 48 h. For (c) and (d), the experiments were carried out for three times with similar results
To determine whether miR-195-induced apoptosis was a specific phenomenon to SHhES1-NPCs or was general to other hESC-NPCs, we examined the role of miR-195 in H9-NPCs. Similarly, TUNEL assays detected more apoptotic cells in pre-miR-195-transfected H9-NPCs (Supplementary Figures S2c and d), accompanied by a moderate but consistent reduction in the cell number (Supplementary Figure S2e). Moreover, caspase-3 was activated after transfection of pre-miR-195 in H9-NPCs (Supplementary Figure S2f). Taken together, miR-195 acted as a proapoptotic factor in hESC-NPCs.
To test whether other miR-15/16 family members could also promote apoptosis in hESC-NPCs, we overexpressed miR-15a, which differs from miR-195 most in this family (Supplementary Figure S2g), in SHhES1-NPCs. In a manner similar to miR-195, transfection of miR-15a precursor (pre-miR-15a) resulted in a cell number reduction (Supplementary Figure S2h). Moreover, the protein level of cleaved caspase-3 was elevated by miR-15a, but not by an unrelated miR-128 (Supplementary Figure S2i). Therefore, miR-15a appears to be a proapoptotic factor in human NPCs, as is miR-195.
To further characterize miR-195-induced apoptosis, we used locked nucleic acid (LNA)-195 to silence miR-195 in pre-miR-195-transfected SHhES1-NPCs. As shown by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) results, LNA-195 efficiently reduced the level of miR-195 (Supplementary Figure S3a) when pre-miR-195 and LNA-195 were introduced into NPCs simultaneously. In the presence of LNA-195, miR-195-mediated reduction in the cell number and apoptosis were completely abolished (Supplementary Figures S3b–f). These data provide strong evidence for the role of miR-195 in the induction of apoptotic cell death in hESC-NPCs.
Microarray analyses identify gene expression signatures regulated by miR-195 in hESC-NPCs
We were interested in the molecular basis underlying the specific role of miR-195. Previous studies identified several potential targets of miR-195, including BCL2, AKT3 and FGF2, and they were implicated in the functions of miR-195 in various cell types.15, 25 We examined whether the expression of these genes was regulated by miR-195 in human NPCs. However, neither mRNA levels of BCL2, AKT3 and FGF2 nor protein levels of BCL2 and AKT3 changed in pre-miR-195-transduced NPCs (Supplementary Figures S4a and b). Thus, it seems unlikely that miR-195 induced apoptosis in hESC-NPCs through modulating the expression of these previously reported target genes.
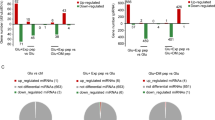
To identify genuine targets of miR-195 in NPCs, we carried out gene expression microarray analyses. Global gene expression profiles of NPCs transfected with equal doses of pre-miR-195+ LNA-NC (miR-195 group), pre-miR-195+LNA-195 (LNA-195 group) and pre-miR-NC+LNA-NC (control group) were measured. Analyses were made for differentially expressed genes with a P-value <0.05 and a fold-change >2.0, shown as the ratio of values in the miR-195 group versus those in the control group. In that case, 192 differentially expressed genes were identified. Gene ontology (GO) terms associated with these genes were enriched out of thousands of biological terms (Figure 3a). As ordered by P-value ranking, miR-195-regulated genes were related to cell proliferation and cell death biological processes. This finding was in agreement with functions of miR-195 reported in this and other studies.
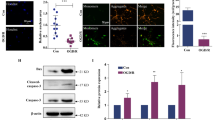
Microarray-based approach identifies ARL2 as a genuine target of miR-195. (a) Gene ontology (GO) analysis of differentially expressed genes (P<0.05, fold-change >2) in miR-195 or control groups of SHhES1-NPCs. The chart lists top 15 GO terms sorted by the P-value and involving at least 12 gene counts. (b) Upper panel: Schematic illustration of the 3′-UTR of ARL2 and its potential miR-195 binding sites predicted by TargetScan. Immediately below are sequence alignments of putative miR-195 binding sites in different species. The seed complementary sites are highlighted in red, and mutants are in green. (c) Dual luciferase reporter assays in 293T cells co-transfected with dual-luciferase constructs containing 0.05 μg wild-type (wt-ARL2) or the mutants (mu1-4) in the 3′-UTR of ARL2 along with 20 pmol of pre-miR-NC or pre-miR-195; n=3, *P<0.05, **P<0.01. The normalized luciferase activity of pre-miR-NC-transfected cells was set as 1. Mu – All mutant includes the mutation of all four identified miR-195 binding sites. (d) Upper panel: mRNA levels of ARL2 were determined by qRT-PCR in miR-195-overexpressed NPCs with LNA-195 or control LNA (LNA-NC); n=3, *P<0.05. The value of control was set as 1. Immediately below are the representative western blotting of ARL2 protein level. Three independent experiments were conducted with similar results
To narrow down the candidate genes of miR-195 targets, we overlapped differentially expressed genes in comparisons of miR-195/control and miR-195/LNA-195 and identified seven genes, which were downregulated by miR-195 overexpression but recovered by LNA-195 cotransfection. Of these genes, ARL2 was predicted as a candidate target of miR-195 by all three widely used programs, TargetScan, PicTar and miRanda (Table 1). Furthermore, four putative miR-195 binding sites were identified in the 3′-UTR of ARL2 as highly conserved among different species (Figure 3b). The functional interaction of miR-195 with putative target sites of ARL2 was examined by luciferase reporter assays. Ectopic expression of miR-195 significantly suppressed the activity of the reporter carrying the wild-type predicted target sites of ARL2. Morever, this inhibitory effect could be abolished only when all four sites were mutated. Mutation of any single site was unable to disrupt the interaction (Figure 3c). Importantly, overexpression of miR-195 in hESC-NPCs suppressed endogenous expression of ARL2 at both mRNA and protein levels (Figure 3d). Thus, our data provide the experimental evidence that ARL2 is a genuine target of miR-195 in hESC-NPCs.
Downregulation of ARL2 is primarily responsible for miR-195-mediated apoptosis in hESC-NPCs
ARL2 is known to encode a small GTP-binding protein of the RAS superfamily.26 However, its function in NPCs was unknown. We asked whether ARL2, as a direct downstream target of miR-195, had a function related to the regulation of apoptosis in hESC-NPCs. To this end, three different ARL2 small interfering RNAs (siRNAs) were used to silence the expression of ARL2 in SHhES1-NPCs specifically. Successful repression of ARL2 expression was verified by qRT-PCR and western blotting (Figures 4a and b). Morphologically, cells became round and bright after transfection of ARL2 siRNAs. Moreover, knockdown of ARL2 reduced the NPC number evidently in a manner resembling miR-195 overexpression (Figure 4c). Western blotting results revealed elevated protein levels of cleaved caspase-3 in all ARL2 siRNA-transfected NPCs, although to different extents (Figure 4d). Annexin V-PI analyses revealed an increase in the percentage of apoptotic cells when ARL2 was downregulated (Figures 4e and f). These results demonstrated that the silencing of ARL2 produced a phenotype similar to that of overexpression of miR-195 and that ARL2 was required for the survival of hESC-NPCs. Nevertheless, it remained unclear whether miR-195 induced apoptosis through suppressing ARL2 expression. We addressed this question by overexpressing ARL2 before introducing pre-miR-195 into NPCs and found that overexpression of ARL2 prevented miR-195-induced cell number reduction (Figures 4g–i). The finding supports the notion that downregulation of ARL2 may account for miR-195-mediated apoptosis in human NPCs.
ARL2 is a functional downstream target of miR-195 during proapoptotic process. (a) QRT-PCR analysis of ARL2 expression in SHhES1-NPCs 72 h post-transfection of 25 nM of ARL2 siRNAs. The value of control (ctrl) siRNAs was set at 1; n=3, *P<0.05, **P<0.01. (b) Western blot analyses of ARL2 expression in cells transfected with ARL2 siRNA. (c) Phase-contrast images of SHhES1-NPCs transfected with ARL2 siRNA or control oligonucleotides. The scale bar is 100 μm. (d) A representative western blotting of cleaved caspase-3 in NPCs transfected with siRNAs indicated in (c). (e) Representative plots of Annexin V/PI double staining assay following ARL2 RNAi in NPCs. Flow cytometry profile represents Annexin-V-fluorescein isothiocyanate (FITC) staining on the x axis and PI on the y axis. (f) Statistical analysis of the percentage of Annexin V-positive apoptotic cells in NPCs indicated in (c); n=3, *P<0.05 and **P<0.01. (g) Both exogenous and endogenous ALR2 protein levels in cells co-transfected with 50 pmol of pre-miR-195 and 5 μg of Flag-tagged ARL2-pPy plasmids (F-ARL2) were measured by western blotting. Three independent assays were conducted and similar results were obtained. (h) Transfection of ARL2 restored the cell number of miR-195-overexpressed NPCs compared with control group. The value of control group was set at 1. n=3, **P<0.01. (i) The representative cell image of (h). The scale bars are 100 μm
Neurotoxin-induced apoptosis is related to miR-195 and ARL2
To learn physiological or pathological processes associated with miR-195, we examined the expression pattern of miR-195 during human ESC differentiation into neural progenitors at induction days 0, 5, 10 and 25. Results of qRT-PCR analysis showed a gradually increasing trend in the level of miR-195 (Figure 5a), implicating the potential association of miR-195 with this process.
miR-195 and ARL2 might be related to neurotoxin-induced apoptosis. (a) The expression levels of miR-195 were measured by qRT-PCR during H9 ESC neural differentiation at different days, including D0, 5, 10 and 25; n=3. The value of D0 sample was set at 1. (b) QRT-PCR analysis of miR-195 expression levels in NPCs treated with 500 μM of paraquat or 500 nM of rotenone. The value of dimethylsulfoxide (DMSO) treatment was set at 1; n=3, *P<0.05. (c) The cell number of NPCs decreased under the treatment of paraquat (500 μM) or rotenone (500 nM). The scale bars are 100 μm. (d) The statistical analysis of the cell number in cells indicated in (c); n=4, **P<0.01. (e) and (f) The statistical analysis and representative plots of PI staining of NPCs treated as indicated; n=3, *P<0.05, **P<0.01. (g) The representative western blotting of ARL2 and cleaved caspase-3 in cells indicated in (e). Two independent experiments were performed. (h) The representative western blot analyses of GFP and cleaved caspase-3 in NPCs infected with RLL or Sponge-195 lentivirus particles with or without paraquat (PQ). Three independent experiments were conducted. (i) The representative plots of PI staining analysis of NPCs as indicated in (h). Three independent experiments had similar results
Recently, Zhang and Pan27 reported that a commonly used toxicant stressor and potential carcinogen, hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX), induced upregulation of miR-195 in the mouse brain and liver, suggesting possible involvement of miR-195 in the neurotoxin-induced apoptosis process. Therefore, we explored this possibility by examining the apoptosis and expression patterns of both miR-195 and ARL2 after treating hESC-NPCs with neurotoxins, paraquat and rotenone. Paraquat, an herbicide and suspected etiologic factor in the development of Parkinson’s disease, was reported to induce apoptosis in human neuroblastoma.28 Mitochondrial complex I inhibitor rotenone could be used to activate apoptotic pathways in neural cells.29 Here, we found that both paraquat and rotenone increased the miR-195 expression level in SHhES1-NPCs (Figure 5b). Simultaneously, they caused an evident reduction in the NPC number (Figures 5c and d). In addition, FCA revealed that the percentage of apoptotic cells increased when cells were treated with either paraquat or rotenone (Figures 5e and f). Interestingly, paraquat, but not rotenone, activated caspase-3 and reduced the ARL2 protein level (Figure 5g). It is possible that paraquat and rotenone activated distinct apoptotic pathways. Rotenone might induce apoptosis through a caspase-3 independent pathway, which is in agreement with the study in human neural stem cells.23 Taken together, miR-195 and ARL2 may have roles in apoptosis induced by neurotoxins such as paraquat.
At last, we determined whether miR-195 was required for paraquat-induced apoptosis. To this end, LNA-195 was used to block the effect of miR-195 in the cells treated with paraquat. However, paraquat-induced cell death could not be prevented by LNA-195 (data not shown). Considering that other members of the miR-195 family, such as miR-15a, might also function as proapoptosis factors to mask the effect of LNA-195, we engineered a lentiviral vector (named RLL) to an miRNA sponge plasmid30 (named Sponge-195) with seven tandems of a sequence, complementary to the seed sequence of the miR-195 family, in the 3′-UTR of a green fluorescence protein (GFP) reporter gene to suppress the function of the whole miR-195 family (Supplementary Figure S5a). The efficacy of Sponge-195 was validated in the following two assays. First, Sponge-195 was transfected into 293T cells together with pre-miR-195 or pre-miR-15a. FCA showed that both miRNAs were able to reduce the expression of GFP, indicating that members of the miR-195 family could bind the seven tandems in the Sponge-195 to reduce the GFP expression (Supplementary Figure S5b). Second, we examined whether the presence of Sponge-195 could affect the luciferase activity controlled by the 3′-UTR sequence of the ARL2 gene. Inhibition of the luciferase activity by miR-195 and miR-15a was completely abrogated when pre-miR-195 or pre-miR-15a was co-transfected with Sponge-195, indicating that Sponge-195 could efficiently sequester the members of the miR-195 family and maintain the target gene expression level (Supplementary Figure S5c). Subsequently, Sponge-195 was utilized to silence the effect of miR-195 family members in SHhES1-NPCs. RLL, the parental vector without the seven tandems, was used as a control. Western blotting showed that the enhanced active caspase-3 level caused by the paraquat treatment in RLL-transfected NPCs was eliminated by Sponge-195 (Figure 5h). Moreover, PI staining analysis revealed that Sponge-195 rescued paraquat-induced NPC apoptosis (Figure 5i). These observations clearly show that the miR-195 family has an important role in neurotoxin paraquat-promoted human NPC apoptosis.
Discussion
In this study, we report the functional role of miR-195 in regulating apoptosis of NPCs derived from two independent lines of hESCs. Overexpression of miR-195 or its family member miR-15a induced significant apoptotic cell death. Consistent with this function, genome-wide gene expression analyses uncovered that cell death was the major biological process regulated by miR-195. Mechanistically, ARL2 was an authentic downstream functional target of miR-195. We provide the first evidence that ARL2 is essential for the survival of hESC-NPCs. Pathologically, we found that miR-195 and ARL2 might be involved in the neurotoxin-induced apoptosis process. The direct link between miR-195 and ARL2 and the functional role of ARL2 established here provide new insights into molecular regulation of cell survival and apoptosis in human NPCs and may help find new strategies to improve the survival of NPCs when they are used in the disease treatment.
Several groups have independently reported that members of miR-15/16 family have important roles in regulating a wide variety of biological and pathological processes.31 In this study, we showed that overexpression of miR-195 evoked cell apoptosis in hESC-NPCs. Silencing of miR-195 alone in SHhES1-NPCs did not produce discernible phenotypes under normal culture conditions and could not prevent paraquat-induced NPC apoptosis (data not shown). However, silencing all members of the miR-195 family considerably reduced NPC apoptosis induced by the neurotoxin. The finding suggests the existence of redundant roles among members of the miR-195 family and reveals the critical role of this family for neurotoxin-mediated apoptosis or neural cell degeneration in human NPCs.
It has been reported that different hESC lines may display different potentials for differentiation into particular cell lineages.27 Therefore, NPCs derived from different hESC lines may respond differentially to proapoptotic stimuli. We noticed that the apoptotic response of SHhES1-NPCs to miR-195 overexpression was stronger than H9-NPCs, although apoptosis occurred in both lines of NPCs. As the two NPC lines were initially generated by two different induction protocols, we needed to distinguish whether the differential response was due to the differentiation methods used or their derivation from two different hESC lines. To address this issue, we generated H9-NPCs by the same Noggin induction method used to generate SHhES1-NPCs. Interestingly, the apoptotic response of H9-NPCs generated by the same protocol with SHhES1-NPCs exhibited weaker response to miR-195 overexpression, as did H9-NPCs generated by three inhibitors (data not shown). Therefore, miR-195 provoked apoptosis to a lesser extent in H9-NPCs than in SHhES1-NPCs, regardless of the differentiation strategy.
The molecular mechanisms through which miR-195 executes its biological functions are largely unknown, although several potential targets were reported. In this study, we first reported the genome-wide gene expression signature regulated by miR-195 in human NPCs. The information will help us to understand better how miR-195 functions. Notably, in hESC-NPCs, miR-195 did not affect the expression of several known targets which were previously identified in other cell types. Instead, miR-195 specifically targeted ARL2 and downregulation of ARL2 phenocopied the proapoptotic effect of overexpression of miR-195 in hESC-NPCs. Importantly, ARL2 overexpression abrogated the phenotype of miR-195 overexpression. In consistent with our finding, two groups recently reported that miR-195 directly regulated ARL2 expression.32, 33 Nishi et al. found that miR-195 modulated ATP levels and degenerated the mitochondria via ARL2 in cardiac myocytes.32 However, whether ARL2 was associated with apoptosis in cardiac myocytes was not mentioned in their study. Differently, they found that mutation of any of the four miR-195 binding sites could block the effect of miR-195, whereas in our study miR-195 lost its inhibitory activity to ARL2 only when all of the four putative binding sites were mutated. Therefore, the function of a miRNA and the manner by which it acts are cellular context-dependent.
There have been few studies of ARL2 and its biological functions so far. It is considered to be involved in tubulin folding and microtubule dynamics in mammalian cells.34, 35 ARL2 can form a complex with tubulin-binding cofactor D36 and the tumor suppressor protein phosphatase 2A37 to regulate the phosphorylation of p53, which binds to the microtubule to affect the chemotherapy sensitivity38 and tumor aggressivity39 in breast cancer. In these studies, suppression of ARL2 expression in breast cancer inhibited apoptosis and tumor regression.38, 39 Their result is opposite to our finding that silencing of ARL2 leads to apoptosis in NPCs. Moreover, silencing of ARL2 expression was reported to decrease cell spreading.34 We observed similar phenomena in NPCs transfected with ARL2 siRNAs. Here, we provide direct evidence that silencing ARL2 expression leads to apoptosis in human NPCs. Therefore, ARL2 is a new factor required for NPC survival. Further investigations are warranted to address how ARL2 takes part in the control of cell survival.
Materials and Methods
Cell culture
Human ESCs of SHhES1 line (karyotype XX, p33–40) were cultured on irradiated mouse embryonic fibroblasts (MEFs) with a daily change of the medium consisting of knockout Dulbecco’s modified Eagle’s medium (Gibco/Life Technologies, Grand Island, NY, USA) supplemented with 20% serum replacement (Gibco), 1 mM glutamine, 0.1 mM β-mercaptoethanol, 0.1 mM non-essential amino acid, 50 U/ml penicillin plus 50 μg/ml streptomycin and 4 ng/ml bFGF (Invitrogen/Life Technologies, Grand Island, NY, USA).21 Human ESCs of H9 line (WA-09, karyotype XX, p48–56) were maintained as described previously.40
Generation and culture of NPCs
SHhES1 cells were cultured on Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) in the MEF-conditioned medium (MEF-CM) supplemented with 8 ng/ml of bFGF before neural differentiation. After culturing for one day with MEF-CM, cells were induced into neural lineages by culturing with the N2B27 medium (1 : 1 mixture of DMEM/F12 supplemented with N2 supplement and Neurobasal medium supplemented with B27 supplement; Gibco) plus 100 ng/ml rmNoggin (R&D System, Minneapolis, MN, USA). Upon the appearance of polarized cells after induction for about 3 weeks, the cells were mechanically picked up and cultured in suspension to form neurospheres in the N2B27 medium supplemented with 10 ng/ml bFGF. When the neurospheres were attached, the cells were defined as p1. From p2 onwards, cells were dissociated into single cells by 0.05% trypsin (Gibco). All later passages of NPCs were maintained in the N2B27 medium with 10 ng/ml of bFGF.
H9 cells were firstly transferred to the mTesR1 medium (STEMCELL Technologies, Vancouver, BC, Canada) as described.41 Then, they were processed as reported,22 except that we added another TGFβ inhibitor, compound C (10 μM; Sigma, St. Louis, MO, USA), together with 500 ng/ml rmNoggin and 10 μM SB431542 (Tocris Bioscience, Bristol, UK) to induce neural differentiation. Similar to SHhES1 cell neural differentiation, we picked up cells in clumps and plated them onto Matrigel-coated dishes when polarized cells were visible. Later, NPCs were dissociated into single cells in the N2B27 medium plus 10 ng/ml bFGF and passaged by 0.05% trypsin.
Transient transfection of cells with pre-miR or LNA-miR
Cells were transient transfected with synthetic pre-miRNAs (Ambion/Applied Biosystems, Foster City, CA, USA) and/or LNA-miRNAs (Exiqon A/S, Vedbaek, Denmark) by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Their concentrations used in experiments were provided in the figure legends. Cells were harvested 72 h after transfection or as indicated.
Neurosphere-forming assay
NPCs were digested by 0.05% trypsin and cultured in suspension in poly-HEME (Sigma)-coated dishes. Spherical structures formed after 4- to 6-day culture in the presence of 10 ng/ml of bFGF. When bFGF was withdrawn, NPCs spontaneously differentiated into neuronal cells. Differentiated spheres were attached onto coverslips (Fisherbrand/Fisher Scientific, Pittsburgh, PA, USA) coated with Matrigel for immunofluorescence staining.
Immunofluorescence staining
NPCs and rosettes attached on coverslips were stained as described previously.42 The confocal images were captured using a confocal microscope (TCS SP5; Leica Microsystems, Wetzlar, Germany). Primary antibodies were used with following dilutions: Nestin (1 : 200; Chemico/MILLIPORE, Temecula CA, USA), SOX2 (1 : 500, rabbit polyclonal antibody raised and affinity-purified in our laboratory), GFAP (1 : 200; Chemicon), OLIG2 (1 : 250; Chemicon), Ki-67 (1 : 200; Chemicon), Tuj1 (1 : 500; Promega, Madison, WI, USA), MAP2 (1 : 500; Santa Cruz Biotechnology, Santa Cruz, CA, USA), LEX1 (1 : 100; Chemicon), CD133 (1 : 50; Miltenyi Biotec, Bergisch Gladbach, Germany), DACH1 (1 : 100; Proteintech, Chicago, IL, USA) and ZIC1 (1 : 100; Novus Biologicals, Littleton, CO, USA).
RNA extraction and real-time qRT-PCR
Total RNA was isolated from NPCs with TRIzol reagent (Invitrogen) and reverse-transcribed into cDNA using oligo (dT)15 and ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan). NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA) was used to quantify amounts of mRNA. QRT-PCR was performed using the ABI PRISM 7900 Fast Real-Time PCR system (Applied Biosystems) and the Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s instructions. The primer sequences used for qRT-PCR are provided in Supplementary Table S1. Mature miR-195 was detected by stem-loop qRT-PCR analysis using the Taqman Human MicroRNA Assay kits (Applied Biosystems). Small nucleolar RNU48 served as an endogenous reference RNA for normalizing the cellular content of other miRNAs.
Western blotting
Western blot analysis was performed under standard denaturing conditions using following antibodies at 1 : 1000 dilutions: cleaved caspase-3 (Cell Signalling Technology (CST), Denver, MA, USA), BCL2 (Cell Signalling Technology), α-tubulin (Sigma), GAPDH (Abmart, Arlington, MA, USA), AKT (Cell Signalling Technology), AKT3 (Cell Signalling Technology) and GFP (Roche, Indianapolis, IN, USA). ARL2 antibody (Abgent, San Diego, CA, USA) was used at 1 : 200 of dilution. All experiments were performed at least three times.
Flow cytometry analysis
NPCs were cultured on Matrigel-coated dishes for at least 48 h before experiments. NPCs were incubated for 2 h with BrdU (10 μM; BD Biosciences) and then fixed with 70% ethanol before FCA by a BD FACSAria Cell Sorter (Becton Dickinson, San Jose, CA, USA). Transfected NPCs were stained with Annexin V and propidium iodide (PI) according to the manufacturer’s manual. Cell cycles were analyzed by PI staining using a BD FACSAria Cell Sorter or BD Accuri C6 flow cytometer (BD Accuri Cytometers, Ann Arbor, MI, USA). After the treatment of paraquat (Sigma) or rotenone (Sigma), a simplified protocol for apoptosis assay by DNA content analysis was used as described.43
TUNEL assay
Transfected NPCs were fixed in 4% paraformaldehyde for 1 h at room temperature and then permeabilized in 0.2% Triton X-100/phosphate-buffered solution solution for 15 min. TUNEL staining was performed according to the manufacturer’s instructions (In Situ Cell Death Detection Kit, POD; Roche). Stained cells were analyzed using the BD FACSAria Cell Sorter.
Luciferase reporter assays
The 3′-UTR of human ARL2 (NM_001667) was cloned into a vector containing two Luciferase open reading frames, Renilla and firefly (psiCHECK2; Promega). The target regions were mutated by PCR with mutant primers as listed in Supplementary Table S1. Specifically, four miR-195 complementary sites, TGCTGCTA(G), were changed to TGCTGggA(G). The 293T cells were co-transfected with the reporter construct and pre-miR-195 using Lipofectamine 2000 (Invitrogen). Luciferase reporter assays were performed 48 h post-transfection using the Dual-Luciferase Assay System (Promega) according to the manufacturer’s instructions.
Gene expression microarray and data analysis
Affymetrix HG-U133 plus 2.0 chips were used for each group of NPCs with three biological replicates. RNA samples were prepared from SHhES1_NPCs 48 h post-transfection under the following conditions: (1) 50 pmol pre-miR-NC (25 nM) plus 50 pmol LNA-NC (25 nM); (2) 50 pmol pre-miR-195 (25 nM) plus 50 pmol LNA-NC (25 nM); and (3) 50 pmol pre-miR-195 (25 nM) plus 50 pmol LNA-195 (25 nM). RNAs were extracted by TRIzol, purified by a QIAGEN RNAeasy kit (QIAGEN, Valencia, CA, USA) and subjected to Affymetrix HG-U133 plus 2.0 chips (Shanghai Biochip Co. Ltd, Shanghai, China). Data were analyzed by DAVID Gene ID Conversion Tool (http://david.niaid.nih.gov) and the SBC Analysis System (http://sas.ebioservice.com).
RNA interference by oligonucleotides
ARL2 was knocked down using Stealth RNA interference (RNAi) Duplex Oligonucleotides (Invitrogen) in SHhES1_NPCs. The sequences of ARL2 stealth RNAi oligos were as follows: ARL2 i-1 (5′-TGCGCTGTCCACTACCCAGATGAGG-3′), ARL2 i-2 (5′-TCTTCAGGATGGTTGTCTTTCCAGC-3′) and ARL2 i-3 (5′-TGTTCAGCTTGAATCCTCGGTGCTC-3′). The negative control was purchased from Invitrogen (Stealth RNAi Negative Control Med GC and High GC).
Plasmid construction and nucleofection
The ARL2-PET plasmid is a kind gift from Dr. Nicholas J. Cowan. The DNA fragment containing the coding sequence of ALR2 (NM_001667), digested from ARL2-PET by NdeI and BamHI, were inserted into pPyCAGIP-Flag vector using the same enzyme sites. Then, 5 μg of ARL2-pPy or pPy-vector were nucleofected into 2 × 106 NPCs according to the manufacturer’s instructions (Amaxa Nucleofector, Amaxa Biosystems, Cologne, Germany). The program used is A33. After 48 h, 25 nM of pre-miR-NC or 25 nM of pre-miR-195 were transfected by Lipofectamine 2000. Cells were harvested after an additional 48 h for cell number counting (Countess; Invitrogen) and western blotting analysis.
The seven tandems of the miR-195 family targeting site sequence (Supplementary Figure S5a), synthesized by Sangon (Shanghai, China), were inserted into RLL vector (gift from Dr. Holm Zaehres) by SalI and KpnI enzyme sites. Then, 0.05 μg psiCHECK or ARL2 3′-UTR reporter plasmids, 10 pmol pre-miR-NC, pre-miR-195 or pre-miR-15a, and 0.8 μg RLL or Sponge-195 plasmids were transfected into 293T cells as indicated in Supplementary Figure S5c by Lipofectamine 2000 reagent according to the manufacturer’s instructions (Invitrogen).
Generation of lentiviral particles and virus infection
We obtained RLL and Sponge-195 lentiviral particles as described previously.44 The titer of lentiviral particles was detected in the 293T cells. The multiplicity of infection of NPCs was set at 2.
Statistical analysis
All values are shown as means±S.D. The Student’s t-test was used to determine the significance of differences in comparisons between two groups. The 0.05 level of confidence was considered statistically significant.
Abbreviations
- NPC:
-
neural progenitor cell
- hESC:
-
human embryonic stem cell
- hESC-NPC:
-
human embryonic stem cell-derived neural progenitor cell
- ARL2:
-
ADP-ribosylation factor-like protein 2
- miRNA:
-
microRNA
- 3′-UTR:
-
3′-untranslated region
- bFGF:
-
basic fibroblast growth factor
- FCA:
-
flow cytometry analysis
- TUNEL:
-
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
- qRT-PCR:
-
quantitative reverse transcription-polymerase chain reaction
- LNA:
-
locked nucleic acid
- GO:
-
gene ontology
- siRNA:
-
small interfering RNA
- PI:
-
propidium iodide
- RDX:
-
hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine
- GFP:
-
green fluorescence protein
- MEF:
-
mouse embryonic fibroblast
- TGF-β:
-
transforming growth factor-β
- BrdU:
-
5-bromo-2′-deoxyuridine
References
Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS et al. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282: 1145–1147.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–872.
Gaspard N, Vanderhaeghen P . Mechanisms of neural specification from embryonic stem cells. Curr Opin Neurobiol 2010; 20: 37–43.
Ambros V . MicroRNAs: tiny regulators with great potential. Cell 2001; 107: 823–826.
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297.
Lee YS, Dutta A . MicroRNAs in cancer. Annu Rev Pathol 2009; 4: 199–227.
Fineberg SK, Kosik KS, Davidson BL . MicroRNAs potentiate neural development. Neuron 2009; 64: 303–309.
Bueno MJ, Perez de Castro I, Malumbres M . Control of cell proliferation pathways by microRNAs. Cell Cycle 2008; 7: 3143–3148.
Bushati N, Cohen SM . microRNA functions. Annu Review Cell Dev Biol 2007; 23: 175–205.
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ . MiRBase: tools for microRNA genomics. Nucleic Acids Res 2008; 36: D154–D158.
Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT . The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol 2010; 402: 491–509.
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM . MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 2009; 50: 113–121.
Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 2007; 67: 2456–2468.
Zhang QQ, Xu H, Huang MB, Ma LM, Huang QJ, Yao Q et al. MicroRNA-195 plays a tumor-suppressor role in human glioblastoma cells by targeting signaling pathways involved in cellular proliferation and invasion. Neuro-Oncology 2012; 14: 278–287.
Liu L, Chen L, Xu Y, Li R, Du X . MicroRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun 2010; 400: 236–240.
van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci US A 2006; 103: 18255–18260.
Qi J, Yu JY, Shcherbata HR, Mathieu J, Wang AJ, Seal S et al. MicroRNAs regulate human embryonic stem cell division. Cell Cycle 2009; 8: 3729–3741.
Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circulation Res 2011; 109: 670–679.
Zhu H, Yang Y, Wang Y, Li J, Schiller PW, Peng T . MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovasc Res 2011; 92: 75–84.
Gerrard L, Rodgers L, Cui W . Differentiation of human embryonic stem cells to neural lineages in adherent culture by blocking bone morphogenetic protein signaling. Stem Cells 2005; 23: 1234–1241.
Sun BW, Yang AC, Feng Y, Sun YJ, Zhu Y, Zhang Y et al. Temporal and parental-specific expression of imprinted genes in a newly derived Chinese human embryonic stem cell line and embryoid bodies. Hum Mol Genet 2006; 15: 65–75.
Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L . Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 2009; 27: 275–280.
Li J, Spletter ML, Johnson DA, Wright LS, Svendsen CN, Johnson JA . Rotenone-induced caspase 9/3-independent and -dependent cell death in undifferentiated and differentiated human neural stem cells. J Neurochem 2005; 92: 462–476.
Zhou J, Su P, Li D, Tsang S, Duan E, Wang F . High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells 2010; 28: 1741–1750.
Chen YQ, Wang XX, Yao XM, Zhang DL, Yang XF, Tian SF et al. MicroRNA-195 promotes apoptosis in mouse podocytes via enhanced caspase activity driven by BCL2 insufficiency. Am J Nephrol 2011; 34: 549–559.
Clark J, Moore L, Krasinskas A, Way J, Battey J, Tamkun J et al. Selective amplification of additional members of the ADP-ribosylation factor (ARF) family: cloning of additional human and Drosophila ARF-like genes. Proc Natl Acad Sci USA 1993; 90: 8952–8956.
Zhang B, Pan X . RDX induces aberrant expression of microRNAs in mouse brain and liver. Environ Health Perspect 2009; 117: 231–240.
Yang W, Tiffany-Castiglioni E . Paraquat-induced apoptosis in human neuroblastoma SH-SY5Y cells: involvement of p53 and mitochondria. J Toxicol Environ Health Part A 2008; 71: 289–299.
Pei W, Liou AK, Chen J . Two caspase-mediated apoptotic pathways induced by rotenone toxicity in cortical neuronal cells. FASEB J 2003; 17: 520–522.
Ebert MS, Neilson JR, Sharp PA . MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007; 4: 721–726.
Esquela-Kerscher A, Slack FJ . Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–269.
Nishi H, Ono K, Iwanaga Y, Horie T, Nagao K, Takemura G et al. MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J Biol Chem 2010; 285: 4920–4930.
Wang K, Li P, Dong Y, Cai X, Hou D, Guo J et al. A microarray-based approach identifies ADP ribosylation factor-like protein 2 as a target of microRNA-16. J Biol Chem 2011; 286: 9468–9476.
Beghin A, Honore S, Messana C, Matera EL, Aim J, Burlinchon S et al. ADP ribosylation factor like 2 (Arl2) protein influences microtubule dynamics in breast cancer cells. Exp Cell Res 2007; 313: 473–485.
Zhou C, Cunningham L, Marcus AI, Li Y, RA Kahn . Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell 2006; 17: 2476–2487.
Bhamidipati A, Lewis SA, Cowan NJ . ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. Jf Cell Biol 2000; 149: 1087–1096.
Shern JF, Sharer JD, Pallas DC, Bartolini F, Cowan NJ, Reed MS et al. Cytosolic Arl2 is complexed with cofactor D and protein phosphatase 2A. J Biol Chem 2003; 278: 40829–40836.
Beghin A, Matera EL, Brunet-Manquat S, Dumontet C . Expression of Arl2 is associated with p53 localization and chemosensitivity in a breast cancer cell line. Cell Cycle 2008; 7: 3074–3082.
Beghin A, Belin S, Hage-Sleiman R, Brunet Manquat S, Goddard S, Tabone E et al. ADP ribosylation factor like 2 (Arl2) regulates breast tumor aggressivity in immunodeficient mice. PLoS One 2009; 4: e7478.
Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA . In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol 2001; 19: 1129–1133.
Ludwig TE, Levenstein ME, Jones JM, Berggren WT, Mitchen ER, Frane JL et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol 2006; 24: 185–187.
Xu HM, Liao B, Zhang QJ, Wang BB, Li H, Zhong XM et al. Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J Biol Chem 2004; 279: 23495–23503.
Wang R, Shi YF . A simplified protocol for apoptosis assay by DNA content analysis. BioTechniques 2002; 33: 88–91.
Li C, Yu H, Ma Y, Shi G, Jiang J, Gu J et al. Germline-competent mouse-induced pluripotent stem cell lines generated on human fibroblasts without exogenous leukemia inhibitory factor. PLoS One 2009; 4: e6724.
Acknowledgements
We thank Nicholas J. Cowan and Holm Zaehres for generously providing plasmids and Shengdi Chen for kindly providing paraquat and rotenone. We also thank ErBo Xu for critical reading of the manuscript. This study was supported by grants of the National Natural Science Foundation (91019023 and 30900858), National High Technology Research and Development Program of China (2010CB945200, 2011DFB300100, 2010CB965101 and 2009CB941103), Chinese Academy of Science (XDA01010102), Shanghai Leading Academic Discipline Project (S30201) and grants from Ministry of Education of China (20110073110082).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by P Salomoni
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Zhou, Y., Jiang, H., Gu, J. et al. MicroRNA-195 targets ADP-ribosylation factor-like protein 2 to induce apoptosis in human embryonic stem cell-derived neural progenitor cells. Cell Death Dis 4, e695 (2013). https://doi.org/10.1038/cddis.2013.195
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.195
Keywords
This article is cited by
-
Mesenchymal Stem Cell-Derived Exosomes are Effective for Radiation Enteritis and Essential for the Proliferation and Differentiation of Lgr5+ Intestinal Epithelial Stem Cells by Regulating Mir-195/Akt/β-Catenin Pathway
Tissue Engineering and Regenerative Medicine (2023)
-
RETRACTED ARTICLE: microRNA-195 attenuates neuronal apoptosis in rats with ischemic stroke through inhibiting KLF5-mediated activation of the JNK signaling pathway
Molecular Medicine (2020)
-
ARL2 overexpression inhibits glioma proliferation and tumorigenicity via down-regulating AXL
BMC Cancer (2018)
-
Identification of key regulators of pancreatic cancer progression through multidimensional systems-level analysis
Genome Medicine (2016)
-
Horizontal transfer of exosomal microRNAs transduce apoptotic signals between pancreatic beta-cells
Cell Communication and Signaling (2015)