Abstract
A tunneling nanotube (TNT) is a newly discovered structure involved in cell–cell communication and is found in various types of cells. Here we identify S100A4 as an extracellular molecule and describe its role in attracting the growth direction of TNTs. Together with its putative receptor, receptor for advanced glycation end product, we demonstrate their involvement in TNT direction guidance. Our results further suggest a mechanism for direction guidance of TNTs. In TNT-initiating cells, p53 activates caspase-3, which leads to S100A4 cleavage and its subsequent decrease in cellular concentration. The decrease in cellular S100A4 induces the formation of a gradient of S100A4, from a low concentration in initiating cells toward a high concentration in target cells. This concentration gradient of S100A4 induces direction guidance for TNTs.
Similar content being viewed by others
Main
Tunneling nanotubes (TNTs) are described as thin membranous and hovering freely channels located between cells.1, 2 As a new principle in cell–cell communication, TNTs are found in various types of cells, such as rat pheochromocytoma PC12 cells,1 HEK293 cells,1, 3 EBV-transformed human B cell line,4 murine macrophage J774 cells,4 DU 145 human prostate cancer cells,5 THP-1 monocytes,6 hepatic HepG2 cells,7 TRVb-1 cells,7 bovine mammary gland epithelial cells,7 human monocyte-derived macrophages,4, 8 primary cultures of rat astrocytes,9 myeloid-lineage dendritic cells,6 hematopoietic stem and progenitor cells10 and bacteria.11 Similar cell–cell communication projections have been described as cytonemes in developing Drosophila imaginal discs12, 13, 14, 15 and MHC class II+ cells in mouse cornea16 in vivo. TNTs can transfer cellular organelles, surface receptors, GPI-anchored proteins and calcium fluxes.17 The cytoskeletal components of TNTs are reported to be F-actin and myosin Va.17, 18 In cultured rat hippocampal neurons and astrocytes, p53, epidermal growth factor receptor, Akt, phosphoinositide 3-kinase and mTOR are critical for TNT initiation.18 TNTs always develop from insulted cells toward non-insulted cells, and are used to transfer cellular contents to non-insulted cells. However, how TNTs find their destination to these healthy cells is still unknown. Herein, we identified the small calcium-binding protein S100A4 and described its role as a direction guidance molecule of TNT. Its putative receptor, receptor for advanced glycation end products (RAGE), was also determined to be involved in TNT guidance.
Results
TNT transferred cellular contents between cells
Rat hippocampal astrocytes were treated with H2O2 for 2 h to induce TNT development,18 and then transfected with EGFP or RFP for better visibility. A series of representative images of TNT were taken by moving the scanning plates (z axis) (Figures 1a1–a3). As the scanning plates changed, a TNT (arrow) could be clearly observed (Figures 1a1–a3). A super-resolution image of TNT with plasma membrane marker showed that TNT was a tube-like structure (Figure 1b). Different from other cellular extensions, the TNTs appeared hovering freely in the culture (Supplementary Movie S1). Scanning of the z axis showed that the TNT was a detached, ‘bridging’ structure positioned between two cells (*) (Figures 1c and c′). This was also demonstrated by the time-lapse movie (Figure 1d, Supplementary Movie S2).
TNT transferred cellular contents between cells. (a) A TNT between two astrocytes (transfected with EGFP) was scanned at different z-axis plates (a1–a3). Arrow: TNT. Scale bar: 5 μm. (b) A super-resolution image for TNT showed that TNT is a tubular structure. The cells were labeled with a membrane marker. Scale bar: 5 μm. (c) An image of the z-axis view of a TNT between two astrocytes (*) labeled with RFP, indicating that TNT was a detached, ‘bridging’ structure between cells. Scale bar: 20 μm. (c′) High-magnification image for c. Scale bar: 10 μm. (d) A TNT (arrow) between two astrocytes is detached from the bottom surface. Scale bar: 10 μm. (e) TNT (arrow) transferred cellular materials from one astrocyte (transfected with RFP) to another astrocyte (transfected with EGFP). IC, Initiating cell; TC, Target cell. Scale bar: 50 μm. (e′) High-magnification image for E. Scale bar: 10 μm. (f1–f6). Cellular content (arrowhead) was transferred from one astrocyte to another through a TNT (arrow) with time. Scale bar: 10 μm.
To distinguish the initial cell populations, the astrocytes were first cultured separately and then transfected with EGFP or RFP. The group of astrocytes that was treated with H2O2 for 2 h to induce TNT development18 was named ‘initiating cells’. These initiating cells were then trypsinized and put onto the other group of cells (‘target cells’) to coculture. TNTs (arrow) would usually develop within 24 h in the direction from the initiating cells toward the target cells as previously described18 (Figure 1e). In the high-power image, red fluorescence was observed in the green cell, indicating that cellular contents of the red cell could be transferred through the TNT to the green cell (Figure 1e′). In another example, the initiating and the target cells were undistinguishable, but the cellular content trafficking from one cell to another was also observed (Figures 1f1–f6, Supplementary Movie S3). Our results suggest that TNTs are tubular, ‘bridging’ structures that transfer cellular contents between cells.
S100A4 attracted the direction of TNTs in astrocytes and neurons
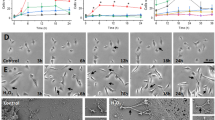
Two populations of astrocytes labeled with EGFP and RFP were cocultured, and the TNTs were induced by H2O2 treatment in RFP-transfected astrocytes. One target astrocyte was destroyed with microinjecting staurosporine. Interestingly, the direction of the established and developing TNTs changed to another astrocyte nearby (Supplementary Figure S1), suggesting that there might be a direction guidance molecule secreted by the target cells. The components of culture medium from the target cells were separated using a combination of ion exchange, hydrophobic and size-exclusion chromatography. The protein in each of the fraction was immobilized onto beads and introduced to astrocyte cultures (Supplementary Figure S2). Their respective ability of TNT guidance by each fraction was examined. The fractions that represented attraction roles toward TNTs were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE), and the two major bands (* and arrow) were isolated and sent for sequencing. The MALDI-TOF-MS sequencing results identified the protein band with a molecular weight of 10–16 kDa (arrow) to be S100A4 (Figure 2a).
S100A4 attracted TNT direction in astrocytes. (a) Two protein bands (* and arrow) after gel filtration were sent for sequencing. The protein band (arrow) in SDS–PAGE was identified as S100A4. (b) To determine the direction of TNTs, from the center of the cell body to the center of the coated bead, a line was drawn as axis x. Axis y was perpendicular to x. Therefore, x and y marked four quadrants: I, II, III and IV. Only the TNTs positioned (no matter where the initiation point is) on the same side with the bead were measured (e.g., in the example diagram, quadrant I and II, but not III and IV). Then a line was drawn from the initiating point of the TNT to the bead (line a). A line was drawn from the end point of the TNT to the bead (line b). The angle (α) was measured between a and b. α⩽5° was determined as ‘toward the bead’ (in the example diagram, the left panel, not the right panel). (c) Astrocytes were transfected with EGFP. The beads coated with recombinant S100A4 (50 μg/ml) attracted TNTs (arrow). (c′) High-magnification image for c. (d) Phase-contrast image of c. (d′) High-magnification image for d. Arrow: TNT. Scale bar: 5 μm. (e) Astrocytes were transfected with EGFP. The beads that were coated with PBS did not attract TNTs (arrow). (e′) High-magnification image for e. (f) Phase-contrast image of e. Scale bar (c–f): 50 μm. (f′) High-magnification image for f. Arrow: TNT. Scale bar (c′–f′): 5 μm. (g) The bead (white circle) coated with BSA (50 μg/ml) did not attract TNT (arrow). Scale bar: 50 μm. (g′) High-magnification image for g. Arrow: TNT. Scale bar: 5 μm. (g″) High-magnification image for g. Dashed arrow: potential TNT in extension. Scale bar: 5 μm. (h) The bead (white circle) coated with boiling denatured S100A4 (50 μg/ml) did not attract TNT (arrow). Scale bar: 50 μm. (h′) High-magnification image for h. Arrow: TNT. Scale bar: 5 μm. (i) Summary of direction guidance of TNT by various conditions. Data represent mean±S.E. (100 cells per each experiment for three independent preparations), **P<0.01 compared with control groups.
To confirm the role of S100A4 in TNT guidance, the beads coated with recombinant S100A4 were added to the astrocytic culture. The heparin beads coated with 50 μg/ml S100A4 were placed at one corner of the culture dish. After a 24-h incubation, the direction of TNT growth of the EGFP-transfected cells toward the beads was observed. Briefly, a line was drawn from the center of the cell body to the center of the coated bead and labeled as axis x. Axis y is perpendicular to axis x, and therefore x and y marked four quadrants, which were labeled I, II, III and IV. Only the TNTs that transpired (not contingent on the initiation point) at the same side as the bead were measured (Figure 2b, e.g., in the provided diagram, quadrant I and II, but not III and IV). A line was then drawn from the initiating point of the TNT to the bead (line a). Another line was drawn from the end point of the TNT to the bead (line b). The angle (α) was measured between a and b. α⩽5° was determined as ‘toward the bead’ (in the example diagram, the left panel, not the right panel; Figure 2b).
S100A4 remarkably attracted the TNTs toward the direction of the beads when compared with phosphate-buffered saline (PBS)- or bovine serum albumin (BSA)-coated groups (Figures 2c–g and c′–g′). When S100A4 was denatured by boiling for 10 min, the denatured S100A4 did not induce guidance of TNTs (Figures 2h and h′). Our data indicate that S100A4 might indeed be the direction guidance cue molecule for TNTs (Figure 2i). In HEK293 cells, overexpression of S100A4 (red fluorescent cells) induced significant TNT targeting (Figures 3a, b). S100A4 was transfected into astrocytes and HEK293 cells, as they were both used as target cells in different experiments. Regardless of cell type, the initiating astrocytes always developed TNTs toward the S100A4-overexpressing cells (Figure 3d). This indicates that S100A4 was sufficient to attract TNT direction. In the TNTs established between HEK cells expressing S100A4 and astrocytes (Figures 3b and b′), cellular content transfer could be observed (Figures 3c1 and c2, arrow heads), implying that the TNTs were functional.
HEK-expressed S100A4 attracted TNT. (a) HEK293 cells overexpressing vector-only (red fluorescent cells) did not attract TNT (arrow) initiating from astrocytes (green fluorescent cells). Scale bar: 20 μm. (a′) High-magnification image for a. Scale bar: 5 μm. (b) HEK293 cells overexpressing S100A4 (red fluorescent cells) attracted TNT (arrow) initiating from astrocytes (green fluorescent cells). Scale bar: 20 μm. (b′) High-magnification image for b. Scale bar: 5 μm. (c1 and c2) Cellular content (arrowhead) transfer was observed with time in b′. Scale bar: 5 μm. (d) The initiating astrocytes always developed TNTs toward the S100A4-overexpressing cells, regardless the cell type. A, astrocytes; H, HEK293. Data represent mean±S.E. (100 cells per each experiment for three independent preparations), **P<0.01 compared with control groups.
RAGE was involved in TNT direction guidance
During TNT formation between two opposing astrocytes, microinjection of the S100A4-overexpressing construct, induced TNT growth from target cells toward the injected cells (Figures 4b and b′). In contrast, knocking down endogenous S100A4 by microinjection of S100A4 siRNA into the target cells inhibited TNT attraction (Figures 4e and e′). The culture media from the target cells was absorbed by either S100A4 antibody or control antibody rabbit serum IgG, and then placed back into to the target cell culture. The resulting S100A4 antibody-absorbed media exhibited significantly reduced TNT attraction by the target cells (Figures 4c, c′). Taken together, these data confirm that S100A4 in the culture medium attracts the directions of TNTs (Figure 4j).
RAGE was involved in TNT guidance. (a–i) In the TNTs between astrocytes (IC, initiating cell, green fluorescent) and astrocytes (TC, target cell, red fluorescent), S100A4 in the target cells and its putative receptor RAGE in the initiating cells were critical for TNT guidance demonstrated by applications of S100A4 (microinjected to the target cells, 50 μg/ml), S100A4 antibody (microinjected to the target cells, 25 μg/ml), control IgG (rabbit serum, microinjected to the target cells, 25 μg/ml), S100A4 or negative control (NC) siRNA (transfected to the target cells, 100 ng/μl), RAGE or NC siRNA (transfected to the initiating cells, 100 ng/μl), RAGE antibody (microinjected to the initiating cells, 25 μg/ml) and heparin (2 μg/ml in the culture medium). Scale bar: 50 μm. (a′–i′) High-magnification images for a-i. Arrow: TNT. Scale bar: 10 μm. (j and k) Summary of data showed that RAGE is involved in TNT direction guidance. Data represent mean±S.E. (100 cells per each experiment for three independent preparations), **P<0.01 compared with control groups.
RAGE is a putative receptor for S100A4.19, 20, 21 In the TNTs formed between two opposing astrocytes, knocking down RAGE greatly reduced the number of TNTs toward the target cells (Figures 4d, d′). RAGE effects were knocked down by either RAGE siRNA or its neutralizing antibody22 in initiating cells. Inclusion of the RAGE antagonist heparin in initiating cells also inhibited TNT guidance (Figures 4i and i′). Our data suggest that the function of RAGE in initiating cells is to act as the receptor for S100A4, which is specifically secreted by target cells to mediate TNT direction guidance (Figure 4k). Alterations of S100A4 and RAGE did not change the number of TNTs induced (Supplementary Figure S3). This excluded the possibility that S100A4 and RAGE affected the total number of TNT induction.
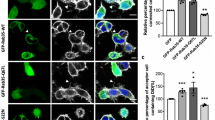
The presence and expression pattern of RAGE was determined by immunostaining in both initiating and target cells. Both initiating and target cells (separately cultured and cocultured) expressed RAGE (Figures 5a–d). In most of the initiating cells, RAGE expressed homogenously (Figure 5a′), whereas in some of the target cells, RAGE appeared as a punctured pattern (Figure 5b′). Western blot showed that RAGE was expressed in both initiating and target cells at a relatively similar quantity (Figure 5e). Quantitative RT–PCR also demonstrated that mRNA of RAGE in both initiating and target cells was also at similar quantity (Figure 5f).
RAGE expression in both initiating and target astrocytes. (a) RAGE expression was detected by immunostaining in the initiating astrocytes. Scale bar: 20 μm. (a′) High-magnification images for a. Scale bar: 5 μm. (b) RAGE expression was detected by immunostaining in the target astrocytes. Scale bar: 20 μm. (b′) High-magnification images for b. Scale bar: 5 μm. (c) TNTs (arrow) were established between the initiating astrocytes (green fluorescent) and target astrocytes (red fluorescent). (d) RAGE expression was detected by immunostaining in both cell populations. Scale bar (c and d): 20 μm. (e) Western blot shows that the protein levels of RAGE were similar in both initiating and target astrocytes. (f) Quantitative RT–PCR shows that the mRNA levels of RAGE are similar in both initiating and target astrocytes. Data represent mean±S.E. (in three independent preparations), **P<0.01 compared with control groups.
Chinese hamster ovary (CHO) cells, which lack endogenous RAGE,23 were used to confirm the role of RAGE in TNT guidance. Uninduced CHO cells were not able to develop TNTs (Figures 6a and b). When induced by treatment of H2O2 for 2 h, nontransfected wild-type (WT) CHO cells developed TNTs, although not toward S100A4-coated beads (Figures 6c and c′). The induced CHO cells transfected with RAGE construct developed TNTs toward the S100A4-coated beads (Figures 6d and d′). Our data suggest that RAGE may act as the receptor in the initiating cells and respond to S100A4 signaling (Figure 6e).
Transfected RAGE induces TNT guidance in CHO cells. (a) In nontransfected and uninduced CHO cells, there were few TNTs observed. The bead was coated with S100A4. (b) In the RAGE-transfected construct and uninduced CHO cells, there were few TNTs observed. The bead was coated with S100A4. (c) In nontransfected and induced CHO cells, TNTs were detected but not directed toward the S100A4-coated bead. Scale bar (a–c): 50 μm. (c′) High-magnification image for c. Arrow: TNT. Scale bar: 20 μm. (d) In RAGE-transfected construct and induced CHO cells, TNTs were detected toward the direction of the S100A4-coated bead. Scale bar: 50 μm. (d′) High-magnification image for d. Arrow: TNT. Scale bar: 20 μm. (e) Quantitative summary of the data. Data represent mean±S.E. (100 cells per each experiment for three independent preparations), **P<0.01 compared with control groups.
Neuronal activity was required for attracting TNTs between astrocytes and neurons
The TNTs located between astrocytes and neurons have also been investigated. Our data illustrated that, similar to TNTs between astrocytes, S100A4 and its putative receptor RAGE were essential for TNT guidance toward neurons (Figures 7a, left panel, and b) and between neurons and astrocytes (Figure 7a, right panel). Because neurons are excitable cells, we then addressed whether neuronal activity or spontaneous firing had a role in TNT guidance. Channelrhodopsin-2 (ChR2) was transduced to neurons by adeno-associated virus (AAV, Figure 7c). Because the major source of secreted S100A4 is astrocytes in the CNS, neurons produce and secrete very low levels of S100A4,24 and the conditioned medium from astrocytes was added to the neuronal culture (Figure 7c). Then H2O2-stimulated initiating astrocytes were cocultured with neurons. After being activated by ∼470 nm blue light (Figure 7c), the cation channels of ChR2 opened to allow Na+ influx to neurons and further activated the infected neurons (Figure 7d).25, 26 Neuronal activation induced a remarkable increase in TNT attraction, whereas coapplication of S100A4 siRNA inhibited such increase (Figure 7e). After the neurons were activated, the culture media of the target cells were then absorbed by the S100A4 antibody and placed back to the neuronal culture. The ability of attracting TNT direction was reversed to control the level by such treatment (Figure 7e), suggesting that neuronal activity and S100A4 both contributed to TNT guidance. Similar effects were also observed by applying different concentrations of KCl, which increased neuronal activity in general (Figure 7f). When neuronal activity was suppressed by NMDA and the non-NMDA receptor antagonists CNQX and MK-801, the attraction of TNT by neurons was greatly reduced (Figure 7g). These results indicate that both neuronal activity and S100A4 are critical for TNT guidance. The total numbers of TNT induced between astrocytes and neurons were not altered by S100A4, RAGE, ChR2 and CNQX+MK-801 (Supplementary Figure S3), suggesting that these treatments did not affect the total number of TNT induction.
S100A4 and neuronal activity attracted TNT direction in neurons. (a) In the TNTs located between astrocytes and neurons (left panel), as well as between neurons and astrocytes (right panel), S100A4 in the target cells was critical for TNT guidance. (b) S100A4 putative receptor RAGE in the initiating cells was critical for TNT guidance. (c) The target neurons were infected with adeno virus-packaged ChR2 and cultured with the conditional medium from astrocytes. After coculture with the induced initiating astrocytes, neurons were activated by ∼470 nm blue light at 10 Hz, 2 ms for 2 h. (d) Neuronal firing from an ChR2-infected neuron (circled) was recorded by patch clamp. Scale bar: 10 μm. Top panel: Neuronal firing in response to brief current injections (50 pA, 5 ms, 10 Hz). Bottom panel: Neuronal firing was induced by ( ) 470 nm light at 10 Hz, 2 ms. (e) Neuronal activity and S100A4 were critical for TNT guidance. (f) Different concentrations of KCl (1, 5, 10, 25, 50 and 100 mM) were applied to increase neuronal activity. (g) CNQX (10 μM) and MK-801 (10 μM) were applied to decrease neuronal activity. Data represent mean±S.E. (100 cells per each experiment for three independent preparations), **P<0.01 compared with control groups.
Caspase-3 cleaved S100A4, which induced a gradient for TNT guidance
Like many other proteins trafficked through the nonclassical export pathway,27, 28 S100A4 is a cytosolic protein and can be secreted to the medium by unknown mechanisms.29 Theoretically, if both initiating and target cells produce and secrete S100A4, one should assume that a similar chemical gradient would form around both groups of cells. If indeed this is true, then why do TNTs only grow toward the target cells, and not back toward the initiating cell itself or to another initiating cell (Figure 8a)? We measured the concentrations of S100A4 in the cytosol and in the culture media around both initiating and target cells by sucking media with a microinjection needle next to different groups of cells with a negative air press. These results demonstrate that S100A4 levels are significantly lower inside and around the initiating cells compared with the target cells (Figure 8b). The conditioned media was collected by placing the needle next to the target cells and evenly distributing it onto the coculture. The percentages of TNTs toward the target cells, and the initiating cells themselves or another initiating cell, were roughly same (Figure 8b, bottom panel). When these conditioned media were absorbed by S100A4 antibody and put back to the coculture, there was no TNT guidance toward the target cells (Figure 8b, bottom panel). Quantitative RT–PCR showed that S100A4 mRNA levels were about the same in both initiating and target cells (Figure 8c). Our data suggest that S100A4 was remarkably lower in and around the initiating cells than the target cells. Therefore, a relatively high S100A4 chemical gradient around the target cells attracted the direction of TNTs.
Caspase-3 cleaved S100A4 in initiating cells and induced the subsequent formation of a chemical gradient of S100A4, which resulted in its increased concentration around target cells. (a) If both the initiating and the target cells produce and secrete the same amount of S100A4, how exactly is it that TNTs have the ability to distinguish these two types of cells and grow from the initiating cells toward the target cells? (b) The target cells contain higher levels of S100A4 in the cytosol (left panel) and the surrounding medium (middle panel). Last panel: the conditioned media was collected by a microinjection needle with a negative air pressure placed next to the target cells and evenly distributed into the coculture. The percentages of TNTs directed toward the target cell (TC), the initiating cells themselves (Self) or another initiating cell (IC) were roughly same. When these conditioned media was absorbed by S100A4 antibody and placed back into the coculture, there was no apparent TNT guidance toward the target cells. (c) Quantitative RT–PCR data showed that mRNA levels in the initiating cells were about the same as in the target cells. (d) There was a putative caspase-3 cleavage site (highlighted, 68NEVD71) in S100A4 sequence. The 3D structure of S100A4 (dimer) demonstrated that the caspase-3 cleavage released the entire N-terminal α-helical segment of S100A4. (e) Caspase-3 activity was significantly higher in the initiating cells than in the target cells. (f) Caspase-3 could cleave S100A4 in vitro and in vivo. Top left panel: recombinant S100A4 was cleaved by recombinant caspase-3 in vitro. Caspase-3 cleaved S100A4 into two separate bands at about 7 and 4k, which could be inhibited specifically by caspase-3 inhibitor, but not by caspase-6 inhibitor. Mutant S100A4-D71A could not be cleaved by caspase-3. Top right panel: cleavage of S100A4 by caspase-3 in vitro with time. Bottom panel: S100A4 was cleaved by caspase-3 in vivo. Astrocytes were treated with conditions that were previously mentioned, and cell extracts were used to do western blots. siRNA-1 and -2: caspase-3 siRNAs. (g) With caspase-3 inhibitor (Z-DEVD-fmk, 5 μM) treatment for 24 h, the levels of S100A4 were about the same in (left panel) and around (middle panel) the initiating and target cells. The percentages of TNTs toward the target cell (TC), the initiating cells themselves (Self) or another initiating cell (IC) were roughly same (right panel). (h) When neurons were activated by light, S100A4 in the surrounding medium increased remarkably. Data represent mean±S.E. (100 cells per each experiment for three independent preparations). **P<0.01 compared with control groups.
We then addressed the question of why the concentration of S100A4 around the target cells is higher than around the initiating cells. By studying S100A4 sequence, we found a potential caspase-3 cleavage site (68NEVD71) (Figure 8d). The putative 3D structure showed that if S100A4 was cleaved by caspase-3 at D71, the entire N-terminal α-helical segment would be released (Figure 8d). Caspase-3 activity assays proved that there was a significant amount of caspase-3 activation in the initiating cells compared with the target cells (Figure 8e). Recombinant WT S100A4 could be cleaved by recombinant caspase-3, but not caspase-6, and two fragments of about 7 and 4k were produced, respectively (Figure 8f, the identity of both fragments was confirmed by peptide N-terminal sequencing, data not shown), which could be inhibited by the specific caspase-3 inhibitor Z-DEVD-fmk. In contrast, the mutant S100A4-D71A could not be cleaved by caspase-3 (Figure 8f).
When treated with caspase-3 inhibitor for 24 h, the levels of cellular and secreted S100A4 showed no difference between the initiating and target cells. There was no significant difference in the numbers of TNTs toward the target cells, the initiating cells themselves or another initiating cell (Figure 8g). According to the data from the experiment described above, neuronal activity greatly enhanced TNT guidance. We examined if neuronal activity also increased S100A4 concentration around the target neurons. Neurons were infected with ChR2 and activated by light. The level of S100A4 in the medium next to the activated neurons was remarkably higher than that of the nonactivated neurons (Figure 8h). This suggests that neuronal activity increases the concentration of S100A4 in the surrounding environment.
To further explore the physiological function of TNTs, the total numbers of TNTs were determined in the astrocyte culture from WT and APP/PS1 mice with or without the presence of H2O2. Our data showed that in both uninduced (−H2O2) and induced (+H2O2) conditions, APP/PS1 mice developed remarkably more TNTs than WT mice (Figure 9a), suggesting that TNT development might be a stress-responsive machinery. Astrocytes from WT and APP/PS1 mice were cultured, and various insults were applied to these cells. Under uninduced condition, spontaneously developed TNTs were observed between cultured astrocytes. Percentage of cell death was compared between cells receiving TNT inputs and cells with no TNT connections. In both WT and APP/PS1 mouse astrocytes, cells receiving TNT inputs were more resistant to the toxicity induced by STS, etoposide (Etop), kainic acid (KA), glutamate (Glu), H2O2 and serum deprivation than the ones without TNT inputs (Figure 9b). We hypothesize that cells may use TNTs to transfer undamaged cellular organelles, useful substance or energy to other cells when undergo apoptosis.18 Therefore, the cells receiving TNT inputs have greater resistance to insults. However, data show that some pathogens, including PrPSc, HIV-1, intracellular and extracellular amyloid β (Aβ) peptides, can ‘hijack’ TNTs for their cell to cell spreading.18, 30, 31, 32, 33 Our present data confirmed that, in contrast to other insults examined in our experiments, extracellular Aβ peptides facilitated cell death in the cells receiving TNT inputs significantly (Figure 9b). To further support this, cultured astrocytes were microinjected with S100A4, anti-S100A4 antibody, IgG control antibody, S100A4 siRNA or negative control siRNA. After spontaneous TNTs were developed, the cultured medium was treated with either STS or extracellular Aβ peptides. Our data showed that in STS treatment group, cell death decreased in the cells injected with S100A4, whereas cell death enhanced in the cells injected with anti-S100A4 antibody or S100A4 siRNA. From the results above (Figure 4j), cells injected with S100A4 received more TNT inputs, whereas cells injected with anti-S100A4 or S100A4 siRNA received few TNT. Our data suggested that cells targeted by TNT were more viable with STS insult (Figure 9c). However, with extracellular Aβ peptide treatment, the cells with more TNT inputs (S100A4-injected cells) showed more cell death than controls, suggesting that cells targeted by TNT were more vulnerable to Aβ peptide (Figure 9c).
Possible TNT implication in cell death. (a) The numbers of TNTs in the hippocampal astrocyte culture from WT and APP/PS1 mice with or without H2O2 treatment were determined. (b) In both WT and APP/PS1 mouse astrocyte culture, cells receiving TNT inputs were more resistant to the toxicity induced by STS, Etop, KA, Glu, H2O2 and serum deprivation (−S) than the ones without TNT inputs. Extracellular Aβ induced more cell death in both WT and APP/PS1 astrocytes. (c) Astrocytes cultured from WT mice were microinjected with S100A4, anti-S100A4 antibody, IgG control antibody, S100A4 siRNA or negative control siRNA. Cell death was induced by STS or extracellular Aβ. Data represent mean±S.E. (n=3). **P<0.01 compared with control groups. (d) In stress-initiating cells, p53 activation induces TNT development and also caspase-3 activity. Caspase-3 cleaves intracellular S100A4, which therefore induces a chemical gradient of a relatively high concentration of extracellular S100A4 around the target cells. TNTs can follow this gradient to find the target cells.
Discussion
This particular investigation has identified S100A4 as an extracellular attractive molecule, which has a role in TNT direction guidance. S100A4 belongs to a calcium-binding S100 protein family with at least 21 other members.29 S100A4 is localized in the cytoplasm, nucleus and the extracellular space.29 S100A4 has been extensively studied for its role in metastasis through its implication in angiogenesis, invasion and cell motility.29 As a cellular protein without a signal peptide at the N terminus, S100A4 is found secreted by a ‘nonclassical’ secretory pathway. The presence of extracellular S100A4 is evidenced in many systems.19, 29 Although the majority of extracellular proteins are secreted by the endoplasmic reticulum (ER)/Golgi-dependent secretory pathway, increasing data have showed that a group of nonclassical secreted proteins, such as S100A4, are exported through an ER/Golgi-independent pathway.27, 28 Several mechanisms have been suggested for such a nonclassical secretory pathway, including lysosome secretion,34 direct protein export,35 multivesicular body release36 and plasma membrane blebbing.37 However, how S100A4 is exported extracellularly is still unclear and needs to be further explored. S100A4 secretion is increased in astrocytes under stress, such as tumor migration, peripheral nerve or dorsal root injury.38, 39, 40
Our data suggest that for TNT between astrocytes and astrocytes, S100A4 is sufficient for TNT targeting. However, for TNT developed between astrocytes and neurons, the presence of S100A4 and neuronal activation are both required for TNT targeting. As excitable cells, neurons distinct themselves from other cells by their firing. Neuronal activity integrity is more sensitive than morphological integrity when neurons are stressed or during aging.41 Therefore, if TNTs transfer cellular contents from damaged cells to healthy cells as we hypothesized,18 TNTs need to successfully find the healthy target cells to build up connections. The requirement for both S100A4 and neuronal activity may be a way for TNTs to distinguish healthy neurons from others.
RAGE has been suggested as a putative cell surface receptor for S100A4.42 Our data confirm that S100A4 mediates TNT guidance through RAGE in initiating cells. Here we propose that the extracellular S100A4 indeed attracts TNT direction and, thus, has a role in TNT direction guidance. In the stressed initiating cells, p53 activation induces TNT development18 as well as caspase-3 activity. Caspase-3 cleaves intracellular S100A4, which therefore creates a chemical gradient around the target cells by inducing a relatively high concentration of S100A4. TNTs then follow this gradient to find the target cells (Figure 9d). It is widely accepted that caspase-3 activation is critical for apoptosis and cell death in many cells.43 However, recent evidence suggests that caspase-3 signaling is also important for development and differentiation of the neuronal system.44 In this particular study, whether caspase-3 activation induces cell death in initiating cells before they make TNTs with target cells has not been determined. It will be interesting to reveal the answer to this question by determining the role of TNT in cell death and neurodegeneration.
The in vivo evidence showing similar cell–cell communication projections has been described as cytonemes in developing Drosophila imaginal discs12, 13, 14, 15 and MHC class II+ cells in mouse cornea.16 TNT may indeed have an important role in neurodegenerative diseases. In Alzheimer’s disease (AD), both Aβ and tau are suggested to have a distinct impact on disease development and progression.45 Microinjection of Aβ into initiating cells is capable of inducing cell death in the target cells.18 One possible explanation for this is that Aβ can use TNTs as an ‘express way’ to spread to surrounding cells. This is supported by the fact that in TNTs, Aβ travels at a speed four-times faster than ER, Golgi, endosome and mitochondria.18 In addition, tau is recently found to be spread crossing synapses in brain tissues with early AD.46, 47 It is possible that tau is directly transfered by TNTs located between two cells and induces neurodegeneration. It would be of great significance to identify TNTs in different physiological and pathological tissues, which will help to understand the functional roles of TNTs and develop potential therapy based on inferring TNT development, targeting and cellular substance trafficking inside TNTs.
Materials and Methods
Chemicals, cDNA constructs, siRNAs and antibodies
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), CNQX (Sigma, St. Louis, MO, USA), MK-801 (Sigma), KCl (Sigma), STS (Sigma), Etop (Sigma), Glu (Sigma), KA (Sigma), heparin (Sigma), H2O2 (Beijing Chemicals Co., Beijing, China), recombinant caspase-3, caspase-6 (Pharmingen, San Diego, CA, USA), recombinant S100A4 (ProSpec, East Brunswick, NJ, USA), protein-A beads (Sigma), Z-DEVD-fmk (CalBiochem, Gibbstown, NJ) and Z-VEID-fmk (CalBiochem) were used in the experiments. Antibodies to S100A4 (CalBiochem), caspase-3 (Pharmingen) and IgG rabbit serum (Sigma) were used. RAGE antibody (25 μg/ml, N-16; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used as a neutralizing antibody.22 Organelle Light Intracellular Targeted Fluorescent Proteins for cell membrane were applied to label cellular membrane as described by the manufacturer (Invitrogen). Wild-type (WT) and mutated S100A4 were cloned in pET28a vector between NdeI and XhoI restriction sites. The primes used in cloning were 5′-GGAATTCCATATGGCGAGACCCTTGGAGG3′ (forward) and 5′-CCGCTCGAGTCACTTCTTCCGGGGCTCC-3′ (reverse). The S100A4-D71A mutant was generated using overlapping PCR mutagenic oligonucleotides, and the primers used for mutagenesis are 5′-CAATGAAGTTGCCTTCCAGGAG-3′ (forward) and 5′-CTCCTGGAAGGCAACTTCATTG-3′ (reverse). Heparin-coated acrylic beads (Sigma) were soaked with 50 μg/ml S100A4, BSA or PBS for overnight at 4 °C. All siRNAs and controls (Qiagen, Hilden, Germany) were diluted into 5 nM before injection or transfection by HiPerFect (Qiagen) described by the manufacturer. The silencing efficiency and off-target effects of all siRNAs were verified by Qiagen.
Cell cultures
Primary neurons and astrocytes were cultured from new-born Sprague Dawley rats or WT APP/PS1 mice following the regulations of Peking University Animal Care and Use Committee. In brief, fresh rat or mouse hippocampal tissues were washed with PBS (0.14 M NaCl, 0.003 M KCl, 0.01 M Na2HPO4 and 0.002 M KH2PO4, pH 7.2) dissociated with 0.25% trypsin (Invitrogen), which was then inactivated by 10% decomplemented fetal bovine serum (FBS; HyClone, Logan, UT, USA). The mixture was triturated through pipette to make a homogenous mixture. After filtering the mixture through 70-μm sterilized filters, the flow-through was centrifuged. The pellet was then washed once by PBS and once by Dulbecco’s modified Eagle’s medium (DMEM) solution containing 0.225% sodium bicarbonate, 1 mM sodium pyruvate, 2 mM L-glutamine, 0.1% dextrose, 1 × antibiotic Pen-Strep (all from Invitrogen) with 5% FBS. Cells were then plated on poly-L-lysine-coated (Sigma) plates at the density of 3 × 106 cells per ml. Neurons and astrocytes were incubated at 37 °C in DMEM with 5% FBS and with 5% circulating CO2. Cytarabine was added to culture media 24 h after plating at 10 μM to inhibit cell growth in neuronal cultures, but not in astrocytic cultures. Medium was changed every 48 h. To obtain astrocyte culture, when cells were confluent on the surface of culture flask, the cells were rocked in a covered culture flask at 250 g for 18 h. The cells were washed twice with DMEM and digested with 0.25% trypsin until the cells get rounded. The cells were transferred to another culture dish with DMEM and 5% FBS. Cells were treated for experiments at 7 days in culture. HEK293 and CHO cells were cultured with DMEM.
Confocal imaging
Cells were imaged using a Zeiss LSM-510 inverted confocal microscope (Carl Zeiss, Oberkochen, Germany). All imaging data were taken in the line-scanning mode of the cells excited at 488 nm for EGFP or Alexa488 and 563 nm for RFP. X-Y and X-Z sections were taken 1 μm for every scanning step. To determine the direction of TNTs, from the center of the cell body to the center of the coated bead, a line was drawn and labeled as axis x. Axis y was perpendicular to axis x. Therefore, x and y marked four quadrants: I, II, III and IV. Only the TNTs end up (no matter where the initiation point is) at the same side with the bead measured (Figure 2b, e.g., in the example diagram, quadrant I and II, but not III and IV). A line was then drawn from the initiating point of the axon to the bead (line a). Another line was drawn from the end point of the TNT to the bead (line b). The angle (α) was measured between a and b. α⩽5° was determined as ‘toward the bead’ (in the example diagram, the left panel, not the right panel) (Figure 2b). Nikon Super Resolution N-SIM Microscope (Nikon, Tokyo, Japan) was used for super-resolution image capture. Each image was collected using a × 100 oil immersion objective.
Microinjection
Thin-walled Borosilicate glass capillaries (outer diameter=1.0 mm, inner diameter=0.5 mm) with microfilament (MTW100F-4; World Precision Instruments, Sarasota, FL, USA) were pulled with a Flaming/Brown Micropipette Puller (P-97; Sutter Instruments, Novato, CA, USA) to obtain injection needles with a tip diameter of about 0.5 μm. Microinjections were performed in the cytosol of each cell using the Eppendorf Microinjector FemtoJet and Eppendorf Micromanipulator (Eppendorf, Hamburg, Germany). Neurons were injected with 25 fl per shot at an injection pressure of 100 hPa, a compensation pressure of 50 hPa and an injection time of 0.1 s. The solutions were injected at the indicated concentrations with 100 μg/ml dextran Texas Red or Alexa488 (Molecular Probes, Eugene, OR, USA) as a fluorescent marker to recognize the injected cells. AirTram (Eppendorf) was used to provide a negative air pressure to collect the culture media near the initiating or target cells.
AAV infection
ChR2 cDNA was subcloned from pEGFP-N3 into pAdTrack with BglII and XhoI digestions. AAV was packaged in HEK293 cells, and the infectious particle was measured as 2 × 106/ml (MOI=1.33). The purified virus supernatant was added to cell culture medium at the dilution of 1 : 500 for 24 h.
Medium absorption
The protein-A sepharose beads were soaked with S100A4 antibody or control IgG overnight over a rotor. The culture medium was then absorbed by constantly rotating with the coated beads overnight. The absorbed medium was then placed back to the culture.
Protein preparation
The WT S100A4 and S100A4-D71A were overexpressed in Escherichia coli strain Rosetta (DE3) as N-terminal (His)6-tagged proteins. Bacterial cells harboring the S100A4 plasmids were cultured in LB media supplemented with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol at 37 °C to an OD600 of 0.6–0.8. Protein expression was induced with 0.5 mM at 18 °C for 20 h. Cells were harvested and resuspended in 20 mM Tris-HCl, pH 7.5, 500 mM NaCl and 50 mM imidazole (equilibration buffer). Cells were sonicated and centrifuged at 50 000 × g for 60 min at 4 °C, and soluble fractions were loaded onto a 5-ml HisTrap HP column (GE Healthcare/Amersham, Uppsala, Sweden) equilibrated with equilibration buffer. The columns were washed with equilibration buffer containing 100 mM imidazole, and the target proteins were eluted with equilibration buffer containing 500 mM imidazole. Purified proteins were further purified via gel filtration utilizing buffer containing 20 mM Tris-HCl pH 7.5 and 150 mM NaCl. Purified proteins were concentrated for crystallization trials using the Amicon Ultra Centrifugal Filter Devices of a 3000 MWCO (Millipore Corporation, Billerica, MA, USA).
Electrophysiology recording
Whole-cell patch-clamp recordings were acquired from rat cultured hippocampal neurons 24 h post infection using standard whole-cell patch-clamp techniques.48 Patch pipettes were pulled from thin-walled borosilicate glass (Sutter Instruments) with a micropipette puller (P-2000; Sutter Instruments) to a resistance of 3–5 MΩ. A MultiClamp 700B amplifier (Axon Instrument, Sunnyvale, CA, USA) was used, and currents were low-pass filtered at 1 kHz and then digitalized online at 10 kHz using a Digidata 1440A (Axon Instrument). Data were acquired and stored with the use of pClamp 10 software (Axon Instrument). Photocurrents were measured while holding neurons in voltage clamp at −65 mV. Whole-cell capacitance and series resistance was measured from the amplifier, and compensation was adjusted to 80%. For whole-cell patch-clamp studies, the standard extracellular solution contained (in mM) 125 NaCl, 2 KCl, 3 CaCl2, 1 MgCl2, 30 glucose and 25 HEPES (pH 7.3 with NaOH). The recording pipette solution consisted of (in mM) 97 potassium gluconate, 38 KCl, 0.35 EGTA, 20 HEPES, 4 magnesium ATP, 0.35 sodium GTP, 6 NaCl (pH 7.25 with KOH). For all experiments, raw data were analyzed and plotted using Origin 7.5 software (Microcal Software, Sunnyvale, CA, USA).
Optical stimulation
DG-4 high-speed optical switch with 300-W xenon lamp (Sutter Instruments) was used to deliver the light pulses for ChR2 activation. This system could steer a laser beam at a maximum rate of 10 ms per site, and formed a flexible pattern of stimulation with the help of the image acquired by the CCD camera. The laser beam was steered with a two-dimensional acousto-optic deflector, and thus the intensity and dwell time at each site could be finely tuned.
Coomassie blue staining
The protein solution was denatured at 100 °C for 5 min and was separated on a 12% SDS–PAGE at 100 V for about 1.5 h. The gel was then stained in the Coomassie Brilliant Blue solution (0.1% Coomassie Brilliant Blue R-250, 25% isopropyl alcohol and 10% acetic acid) and rocked at room temperature for 4 h. The gel was destained in the destain solution (40% ethanol, 10% acetic acid and 50% ddH2O) and shaken at room temperature for about 3 h, while the gel solution was changed five times during this period.
Chromatography
Ion exchange, hydrophobic and size-exclusion chromatography were used to separate the components of culture medium. After passing the SepharoseQ anion exchange column (Amersham Pharmacia Biotech, Arlington Heights, IL, USA) and HiLoad (GE Healthcare) column, each fraction was coated to the beads to examine the TNT guidance function. The functional fractions of the medium were then injected into a size-exclusion chromatography column Superdex 75 HR 10/30 (GE Healthcare) equilibrated with 10 mM Tris–HCl, pH 7.4. The medium was fractionated at a flow rate of 0.5 ml/min and eluted in 1.5-column volumes. The fractions after gel filtration were then coated to the beads to examine the TNT guidance function.
Measurement of cell death
Cells were fixed in fresh 4% paraformaldehyde and 4% sucrose in PBS for 20 min at room temperature and permeablized in 0.1% Triton X-100 and 0.1% sodium citrate in PBS for 2 min on ice. Terminal deoxynucleotidyl transferase-biotin dUTP nick-end labeling (TUNEL) staining was performed using the in situ cell death detection kit I as described by the manufacturer (Roche, Basel, Switzerland). The coverslips were washed once in distilled water for 5 min and then mounted on glass slides to be observed under a fluorescence microscope. The percentage of cell death was determined by the ratio of the number of TUNEL-positive cells over the total of 100 cells in one count. The average of five counts was calculated as the percentage of neuronal cell death in a certain treatment.
Real-time PCR
Cells were harvested and the total RNA was isolated with TRIGene reagent (GenStar BioSolutions Co., Ltd., Beijing, China). Total RNA (2 μg) was reversely transcribed using TransScript II First-Strand cDNA Synthesis SuperMix (Beijing TransGen Biotech Co., Ltd., Beijing, China). Real-time PCRs were done by using TransStart Green qPCR SuperMix UDG (Beijing TransGen Biotech Co., Ltd.). Sequences of primers for rat S100A4 used were as following: 5′-TGTAATAGTGTCCACCTTCC-3′ (forward) and 5′-CATTGTCCC TGTTGCTGT-3′ (reverse). Sequences of primers for rat RAGE used were as following: 5′-GCCTGTCGTGGACTTGGT-3′ (forward) and 5′-TTCCGATGTTCAGAATGATGTT-3′ (reverse). Real-time PCR quantifications were run in triplicate for each sample and the average were determined. In order to use the comparative Ct method for relative quantification, the amplification efficiency of target and housekeeping gene must be approximately equal. Quantification was done using the comparative Ct method, and expression levels for the target gene was normalized to the GAPDH of each sample  Amplification was done for 45 cycles at 95 °C for 30 s, 59 °C for 30 s, 72 °C for 30 s, 95 °C for 1 min, 59 °C for 30 s and 95 °C for 30 s.
Amplification was done for 45 cycles at 95 °C for 30 s, 59 °C for 30 s, 72 °C for 30 s, 95 °C for 1 min, 59 °C for 30 s and 95 °C for 30 s.
Enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assay was used to determine the concentrations of S100A4. Conditioned culture medium (∼5 μl) was collected by a microinjection needle next to initiating or target cells by a negative pressure produced by AirTram (Eppendorf), and a kit to determine S100A4 (Uscn Life Science Inc., Wuhan, China) was used to measure their concentrations described by the manufacturer. Then, the relative optical intensity of each well was read out by microplate reader BioRad 480 (Bio-Rad, Hercules, CA, USA).
Immunohistochemistry
For labeling of RAGE, sections were incubated with 10% normal donkey serum (Jackson Immunoresearch, West Grove, PA, USA) in PBS/Triton for 40 min. Then, the serum was extracted out and a primary antibody against RAGE (1 : 200) was added to incubate the sections 4 °C overnight. After extensive washing, the Cy5-conjugated donkey-anti-rabbit secondary antibodies (diluted by 1 : 200; Jackson Immunoresearch) were added together to incubate the sections for 1 h at room temperature in darkness. Sections were mounted in 1 : 1 PBS/glycerol solution before examining with a confocal laser scanning microscope.
Caspase activity assay
Cellular proteins were extracted in the cell lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP-40 and 0.1% SDS) after treatments. Caspase-3 activities were measured with the cell extracts added by CaspACE Caspase-3 Colorimetric Assay kit (Promega, Madison, WI, USA) described as the manufacturer.
Western blots
Neuronal proteins were extracted in the cell lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40 and 0.1% SDS) and protein concentrations were measured by bicinchoninic acid assay (Pierce, Rockford, IL, USA). Protein extracts were denatured at 100 °C for 5 min and separated on 18% SDS–PAGE at 70 V for about 1 h. Proteins were transferred to Immobilon-P polyvinylidene fluoride membrane (Millipore Corporation) at 100 mA for 2 h. The membrane was blocked with 5% nonfat milk in Tris-buffered saline (TBS) with 0.1% Tween-20 (TBS-T) at room temperature for 1 h. Anti-S100A4, anti-caspase-3, anti-RAGE, anti-GAPDH and anti-actin antibodies were diluted at 1 : 1000 for western blots as primary antibodies. After three washes of 10 min each with TBS-T, goat anti-rabbit IgG conjugated with horseradish peroxidase (HRP) was added in a dilution of 1 : 2500 as the secondary antibody. The secondary HRP was detected by enhanced chemiluminescence. The optical density was analyzed by BioRad ChemiDox (Bio-Rad).
Statistical evaluation
The statistical significance was assessed by one-way analysis of variance (ANOVA). The Sheffé’s test was applied as a post-hoc for the significant difference shown by ANOVA. A P-value <0.05 was used as an indication of statistical significance.
Abbreviations
- Aβ:
-
amyloid β
- AD:
-
Alzheimer’s disease
- ANOVA:
-
analysis of variance
- BSA:
-
bovine serum albumin
- CHO:
-
Chinese hamster ovary
- ChR2:
-
channelrhodopsin-2
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- ER:
-
endoplasmic reticulum
- Etop:
-
etoposide
- FBS:
-
fetal bovine serum
- Glu:
-
glutamate
- HRP:
-
horseradish peroxidase
- KA:
-
kainic acid
- PBS:
-
phosphate-buffered saline
- SDS–PAGE:
-
sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- RAGE:
-
receptor for advanced glycation end product
- TBS:
-
Tris-buffered saline
- TBS-T:
-
TBS with Tween-20
- TNT:
-
tunneling nanotube
- TUNEL:
-
terminal deoxynucleotidyl transferase-biotin dUTP nick-end labeling
- WT:
-
wild-type
References
Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH . Nanotubular highways for intercellular organelle transport. Science 2004; 303: 1007–1010.
Lim YS, Tang BL . Intercellular organelle trafficking by membranous nanotube connections: a possible new role in cellular rejuvenation? Cell Commun Adhes 2012; 19: 39–44.
Onfelt B, Davis DM . Can membrane nanotubes facilitate communication between immune cells? Biochem Soc Trans 2004; 32 (Pt 5): 676–678.
Onfelt B, Nedvetzki S, Yanagi K, Davis DM . Cutting edge: membrane nanotubes connect immune cells. J Immunol 2004; 173: 1511–1513.
Vidulescu C, Clejan S, O’Connor KC . Vesicle traffic through intercellular bridges in DU 145 human prostate cancer cells. J Cell Mol Med 2004; 8: 388–396.
Watkins SC, Salter RD . Functional connectivity between immune cells mediated by tunneling nanotubules. Immunity 2005; 23: 309–318.
Wustner D . Plasma membrane sterol distribution resembles the surface topography of living cells. Mol Biol Cell 2007; 18: 211–228.
Onfelt B, Nedvetzki S, Benninger RK, Purbhoo MA, Sowinski S, Hume AN et al. Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J Immunol 2006; 177: 8476–8483.
Zhu D, Tan KS, Zhang X, Sun AY, Sun GY, Lee JC . Hydrogen peroxide alters membrane and cytoskeleton properties and increases intercellular connections in astrocytes. J Cell Sci 2005; 118 (Part 16): 3695–3703.
Freund D, Bauer N, Boxberger S, Feldmann S, Streller U, Ehninger G et al. Polarization of human hematopoietic progenitors during contact with multipotent mesenchymal stromal cells: effects on proliferation and clonogenicity. Stem Cells Dev 2006; 15: 815–829.
Dubey GP, Ben-Yehuda S . Intercellular nanotubes mediate bacterial communication. Cell 2011; 144: 590–600.
Ramirez-Weber FA, Kornberg TB . Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell 1999; 97: 599–607.
Hsiung F, Ramirez-Weber FA, Iwaki DD, Kornberg TB . Dependence of Drosophila wing imaginal disc cytonemes on decapentaplegic. Nature 2005; 437: 560–563.
Kornberg T . Pictures in cell biology. cytonemes. Trends Cell Biol 1999; 9: 434.
Roy S, Hsiung F, Kornberg TB . Specificity of Drosophila cytonemes for distinct signaling pathways. Science 2011; 332: 354–358.
Chinnery HR, Ruitenberg MJ, Plant GW, Pearlman E, Jung S, McMenamin PG . The chemokine receptor CX3CR1 mediates homing of MHC class II-positive cells to the normal mouse corneal epithelium. Invest Ophthalmol Vis Sci 2007; 48: 1568–1574.
Gurke S, Barroso JF, Gerdes HH . The art of cellular communication: tunneling nanotubes bridge the divide. Histochem Cell Biol 2008; 129: 539–550.
Wang Y, Cui J, Sun X, Zhang Y . Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ 2011; 18: 732–742.
Garrett SC, Varney KM, Weber DJ, Bresnick AR . S100A4, a mediator of metastasis. J Biol Chem 2006; 281: 677–680.
Donato R . RAGE: a single receptor for several ligands and different cellular responses: the case of certain S100 proteins. Curr Mol Med 2007; 7: 711–724.
Heizmann CW, Ackermann GE, Galichet A . Pathologies involving the S100 proteins and RAGE. Subcell Biochem 2007; 45: 93–138.
Takuma K, Fang F, Zhang W, Yan S, Fukuzaki E, Du H et al. RAGE-mediated signaling contributes to intraneuronal transport of amyloid-beta and neuronal dysfunction. Proc Natl Acad Sci USA 2009; 106: 20021–20026.
Lu C, He JC, Cai W, Liu H, Zhu L, Vlassara H . Advanced glycation endproduct (AGE) receptor 1 is a negative regulator of the inflammatory response to AGE in mesangial cells. Proc Natl Acad Sci USA 2004; 101: 11767–11772.
Mense SM, Sengupta A, Zhou M, Lan C, Bentsman G, Volsky DJ et al. Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol Genomics 2006; 25: 435–449.
Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K . Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 2005; 8: 1263–1268.
Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N et al. Multimodal fast optical interrogation of neural circuitry. Nature 2007; 446: 633–639.
Cleves AE . Protein transports: the nonclassical ins and outs. Curr Biol 1997; 7: R318–R320.
Nickel W . The mystery of nonclassical protein secretion. A current view on cargo proteins and potential export routes. Eur J Biochem 2003; 270: 2109–2119.
Boye K, Maelandsmo GM . S100A4 and metastasis: a small actor playing many roles. Am J Pathol 2009; 176: 528–535.
Gousset K, Schiff E, Langevin C, Marijanovic Z, Caputo A, Browman DT et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol 2009; 11: 328–336.
Gousset K, Zurzolo C . Tunnelling nanotubes: a highway for prion spreading? Prion 2009; 3: 94–98.
Davis DM, Sowinski S . Membrane nanotubes: dynamic long-distance connections between animal cells. Nat Rev Mol Cell Biol 2008; 9: 431–436.
Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Kohler K et al. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat Cell Biol 2008; 10: 211–219.
Andrei C, Margiocco P, Poggi A, Lotti LV, Torrisi MR, Rubartelli A . Phospholipases C and A2 control lysosome-mediated IL-1 beta secretion: Implications for inflammatory processes. Proc Natl Acad Sci USA 2004; 101: 9745–9750.
Nickel W . Unconventional secretory routes: direct protein export across the plasma membrane of mammalian cells. Traffic 2005; 6: 607–614.
Lancaster GI, Febbraio MA . Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins. J Biol Chem 2005; 280: 23349–23355.
Tournaviti S, Hannemann S, Terjung S, Kitzing TM, Stegmayer C, Ritzerfeld J et al. SH4-domain-induced plasma membrane dynamization promotes bleb-associated cell motility. J Cell Sci 2007; 120 (Part 21): 3820–3829.
Takenaga K, Kozlova EN . Role of intracellular S100A4 for migration of rat astrocytes. Glia 2006; 53: 313–321.
Takenaga K, Nygren J, Zelenina M, Ohira M, Iuchi T, Lukanidin E et al. Modified expression of Mts1/S100A4 protein in C6 glioma cells or surrounding astrocytes affects migration of tumor cells in vitro and in vivo. Neurobiol Dis 2007; 25: 455–463.
Kozlova EN, Lukanidin E . Metastasis-associated mts1 (S100A4) protein is selectively expressed in white matter astrocytes and is up-regulated after peripheral nerve or dorsal root injury. Glia 1999; 27: 249–258.
Hoyer S . Brain glucose and energy metabolism during normal aging. Aging (Milano) 1990; 2: 245–258.
Helfman DM, Kim EJ, Lukanidin E, Grigorian M . The metastasis associated protein S100A4: role in tumour progression and metastasis. Br J Cancer 2005; 92: 1955–1958.
Nicholson DW, Thornberry NA . Apoptosis. Life and death decisions. Science 2003; 299: 214–215.
D’Amelio M, Cavallucci V, Cecconi F . Neuronal caspase-3 signaling: not only cell death. Cell Death Differ 2010; 17: 1104–1114.
Price DL, Sisodia SS, Borchelt DR . Alzheimer disease – when and why? Nat Genet 1998; 19: 314–316.
de Calignon A, Polydoro M, Suarez-Calvet M, William C, Adamowicz DH, Kopeikina KJ et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 2012; 73: 685–697.
Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C et al. Trans-synaptic spread of tau pathology in vivo. PLoS One 2012; 7: e31302.
Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ . Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch 1981; 391: 85–100.
Acknowledgements
We thank Dr. Yi Rao (Peking University) for providing optogenetical equipment, Dr. Shiqiang Wang and Ms. Juanjuan Zhao (Peking University) for their assistance of confocal imaging and Dr. Lorenzo Finci (Peking University) for the editing of this manuscript. This work was supported by the National Program of Basic Research sponsored by the Ministry of Science and Technology of China (2009CB941301), National Science Foundation of China (NSFC) Major Research Grant (91132718) to YZ, and NSFC Major Research Grant (NSFC: Grant No. 91132306), General Funds (NSFC: Grant No. 30970942), National Basic Research Program of China (973 program: 2010CB529605), Instrument Developing Project of the Chinese Academy of Sciences (Grant No. 2010019) and ‘Hundred Talents Program’ of Chinese Academy of Science to LW.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Verkhratsky
Author Contributions
XS and YW performed all the experiments, except where otherwise noted, and analyzed the data. JZ and JT performed patch-clamping experiments. X-JW produced S100A4 mutant D71A. X-DS supervised the S100A4 mutant experiment. LW and YZ conceptualized the study, performed analyses and drafted the manuscript.
Supplementary Information accompanies the paper on Cell Death and Disease website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Sun, X., Wang, Y., Zhang, J. et al. Tunneling-nanotube direction determination in neurons and astrocytes. Cell Death Dis 3, e438 (2012). https://doi.org/10.1038/cddis.2012.177
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2012.177
Keywords
This article is cited by
-
In vitro and in vivo detection of tunneling nanotubes in normal and pathological osteoclastogenesis involving osteoclast fusion
Laboratory Investigation (2021)
-
Mitochondrial donation in translational medicine; from imagination to reality
Journal of Translational Medicine (2020)
-
Mitochondrial Dysfunction in Stroke: Implications of Stem Cell Therapy
Translational Stroke Research (2019)
-
Tunneling nanotubes mediate intercellular communication between endothelial progenitor cells and osteoclast precursors
Journal of Molecular Histology (2019)
-
Tunneling nanotube (TNT)-mediated neuron-to neuron transfer of pathological Tau protein assemblies
Acta Neuropathologica Communications (2016)