Abstract
Mitogen-activated protein kinase kinase kinase kinase-3 (MAP4K3) is a Ste20 kinase family member that modulates multiple signal transduction pathways. We recently identified MAP4K3 as proapoptotic kinase using an RNA interference screening approach. In mammalian cells, MAP4K3 enhances the mitochondrial apoptosis pathway through the post-transcriptional modulation of selected proapoptotic Bcl-2 homology domain 3-only proteins. Recent data suggest that MAP4K3 mutations contribute to pancreatic cancer, which highlights the importance of studying the in vivo function of this kinase. To determine whether the cell death function is conserved in vivo and which downstream signalling pathways are involved, we generated transgenic flies expressing happyhour (hppy), the Drosophila MAP4K3 orthologue. Here, we show that the overexpression of hppy promotes caspase-dependent apoptosis and that the hypothetical kinase domain is essential for inducing cell death. In addition, we show that hppy expression triggers the activation of both the c-Jun N-terminal kinase (JNK) and target of rapamycin (TOR) signalling pathways; however, only JNK signalling is required for apoptosis. Together, our results show that hppy has a JNK-dependent proapoptotic function in Drosophila, which reinforces the hypothesis that MAP4K3 might act as tumour suppressor by regulating apoptosis in higher eukaryotes.
Similar content being viewed by others
Main
Mitogen-activated protein kinases (MAPKs) are highly conserved in all eukaryotes and control various cellular processes, such as cell growth, differentiation and cell death. The three MAPK pathways, that is, those mediated by extracellular signal-regulated kinase (ERK), p38 and c-Jun N-terminal kinase (JNK), have been found to be deregulated in cancers.1 Therefore, a better understanding of the molecular mechanisms controlling MAPK signalling pathways is crucial for designing effective cancer therapies. However, the analysis of these pathways in mammalian models is complicated by the multiple forms of crosstalk between the MAPK signalling pathways and the functional redundancy of various MAPK isoforms.
Mitogen-activated protein kinase kinase kinase kinase-3 (MAP4K3), a Ste20 family member, was identified as a new component of the target of rapamycin (TOR) signalling pathway with an RNA interference (RNAi) screen in Drosophila S2 cells.2 In response to amino acids in cell culture, MAP4K3 modulates the phosphorylation of S6 protein kinase and the translation inhibitor, eukaryotic translation initiation factor 4E-binding protein (4E-BP), which are direct targets of TOR activity. Recently, the involvement of MAP4K3 in the regulation of TOR activity was confirmed in vivo with the characterisation of Drosophila MAP4K3 mutant flies.3 In addition, MAP4K3 activates the JNK pathway in mammalian cell lines upon stimulation with ultraviolet radiation and tumour necrosis factor-α.4 More recently, a genetic screen designed to identify novel modulators of ethanol sedation in flies identified loss-of-function mutations in Drosophila MAP4K3 that result in increased resistance to the sedative effects of ethanol. This result inspired the naming of the MAP4K3-coding gene as happyhour (hppy).5 Importantly, it was determined that Hppy regulates ethanol sensitivity by inhibiting epidermal growth factor receptor (EGFR)/ERK signalling in vivo. Taken together, these observations indicate that this kinase modulates multiple signal transduction pathways that affect cell survival.
We recently identified MAP4K3 as a proapoptotic kinase in an RNAi screen.6 Our data indicate that MAP4K3 regulates apoptosis by modulating selected proapoptotic Bcl-2 homology domain 3 (BH3)-only proteins.6, 7 This modulation seems to occur at the post-transcriptional level through both the mammalian TOR complex 1 (mTORC1) and JNK signalling pathways. Interestingly, our previous work showed a significant decrease in MAP4K3 protein levels in human pancreatic tumour samples,6 which is consistent with the previous finding that a somatic mutation in the MAP4K3 gene is associated with pancreatic cancer.8 This result strongly suggests that MAP4K3 has a tumour suppressor role. Together, these observations highlight the need for more information on the role of MAP4K3 in cell death in vivo.
We characterised the role of the Drosophila orthologue of human MAP4K3, hppy, using a gain-of-function approach. Here, we show that hppy overexpression results in partial lethality during development and that the surviving flies have smaller wings due to the loss of epithelial cells. The ectopic expression of hppy in the larval wing disc induces cell death in a caspase-dependent manner. Furthermore, mutation of the putative adenosine triphosphate (ATP)-binding domain by the substitution of lysine 45 blocks apoptosis in hppy-overexpressing cells, indicating that the kinase activity of Hppy is required for cell death. Also, we demonstrate that Hppy induces the activation of TOR and JNK signalling pathways but only JNK signalling is required for its cell death function. Thus, we confirm that hppy, the Drosophila MAP4K3 orthologue, has a JNK signalling-dependent proapoptotic function in vivo.
Results
hppy expression induces defective wing phenotype in Drosophila
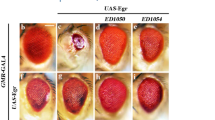
We previously identified human MAP4K3 as a potent cell death inducer.6 To determine the in vivo consequences of MAP4K3 expression, we cloned the Drosophila orthologue of human MAP4K3, hppy. The Drosophila genome encodes two hppy variant transcripts, hppyRA and hppyRB. We employed the UAS-Gal4 system to generate two independent fly lines expressing wild-type hppyRB and a kinase-dead version of this kinase (happyhour kinase dead (hppy KD)) and used real-time quantitative reverse transcription PCR (qRT-PCR) to assay transcript levels (Figure 1a). First, we noted that ubiquitous expression of hppy results in significant pupal lethality (Figure 1b) and that this lethality correlates with hppy expression levels (Figures 1a and b). Importantly, the surviving adult flies are smaller than the controls (data not shown), and have a reduced wing area (Figures 1c and d). As we previously showed that expression of human MAP4K3 in cultured cells results in apoptotic cell death,6 we tested whether the decrease in wing size upon hppy expression is related to cell loss during development by analysing the total number of bristle cells per wing in control flies and in flies expressing hppy and hppy KD (Figure 1e). We found a clear decrease in the total number of bristle cells in hppy-expressing flies, but not in flies expressing hppy KD. This result strongly suggests that the decrease in wing size results from significant cell loss caused by the kinase activity of Hppy.
Developmental consequences of hppy overexpression. (a) Analysis of hppy mRNA levels in transgenic flies. RNA was isolated from a single larva and analysed by qRT-PCR. The notations 4M, 7F, 2M and 8F refer to the independent transgenic lines employed in this study. The fold change in transcript levels is shown relative to controls (da>+). Mean values ±S.E.M. are shown. (b) Analysis of pupal lethality in hppy-expressing flies. Percentages correspond to the number of dead pupae relative to the total number of pupae. Values represent the average of three independent experiments. Mean values ±S.E.M. are shown. (c) Decreased wing area in adult hppy-expressing flies. Representative wings are shown. (d) Quantification of total wing area in hppy-expressing flies. (e) Analysis of epithelial cell number in hppy-expressing flies. Statistically significant values relative to controls (da>+) are indicated with asterisks (one-way ANOVA with Dunnett's post-test)
hppy expression induces apoptosis in Drosophila
To directly test whether the decrease in wing size observed in hppy-expressing flies is a consequence of cell loss, we analysed the levels of apoptosis in hppy-expressing tissues. The overexpression of hppy driven by en-Gal4 in the posterior compartment of the developing wing discs results in the appearance of cells that are terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL)-positive and display condensed chromatin (Figure 2a), confirming that this kinase induces apoptotic cell death. Importantly, we failed to detect significant levels of cells displaying nuclear markers of apoptosis in two independent lines expressing the kinase-inactive Hppy (hppy KD), indicating that the kinase activity is required for the induction of apoptosis (Figure 2a). Moreover, analysis of the adult wings of flies expressing hppy under the control of en-Gal4 showed a defect in vein formation and a clear reduction of the posterior area of the wing (Figure 2b), further suggesting that the decrease in wing size caused by hppy expression is a consequence of cell death in the developing wing.
hppy promotes caspase-dependent apoptosis. (a) Ectopic expression of hppy causes cell death in Drosophila wing discs. The area of en-Gal4-driven gene expression is marked with GFP fluorescence (top). The region of cells with nuclear chromatin condensation, as revealed by Hoechst staining, is indicated with the dashed line (middle). TUNEL-positive cells colocalise with nuclear DNA condensation (bottom). (b) Ectopic expression of hppy in the posterior compartment of the wing causes a defect in the wing structure. The black line denotes the boundary between the anterior and posterior compartments of the adult wing. (c) hppy induces caspase activation. The area of en-Gal4-driven gene expression is marked with GFP fluorescence (top). Wing discs from larvae with the indicated genotypes were stained with an anti-active caspase-3 antibody (bottom). The scale bars represent 20 μm
We previously showed that apoptosis induced by human MAP4K3 can be blocked by expressing inhibitors of the mitochondrial (intrinsic) pathway such as the antiapoptotic B-cell lymphoma protein-2 (Bcl-2) family member B-cell lymphoma-extra large (Bcl-XL).6 To test the relevance of the intrinsic pathway in hppy-dependent apoptosis in flies, we expressed the Drosophila antiapoptotic Bcl-2 family protein Buffy 9 and examined the effect on caspase activation. We observed that the ectopic expression of hppy driven by en-Gal4 results in caspase activation in the posterior compartment of the developing wing discs (Figure 2c, left panel). Such caspase activation could be completely suppressed by the expression of the caspase inhibitor p35 10 (Figure 2c, middle panels), indicating that Hppy-induced cell death is caspase dependent. Buffy expression only modestly suppressed caspase activation (Figure 2c, right panels), however, suggesting that the mechanism of apoptosis induction in flies might be different from that in mammals.
JNK, but not TOR signalling, is required for Hppy-mediated apoptosis
Human Map4k3 promotes apoptosis through the post-transcriptional modulation of BH3-only proteins in the JNK and mTORC1 pathways.6 We therefore sought to determine the effect of hppy expression on the activation of the JNK and TOR pathways in Drosophila.
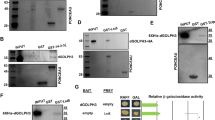
Monitoring of JNK activity in the wing disc by lacZ expression in a puckered (puc)-LacZ enhancer trap line11 revealed that hppy expression results in robust JNK activation and that this activation requires the kinase activity of Hppy (Figure 3a). To verify the involvement of the JNK signalling pathway in the apoptosis induced by hppy expression, we coexpressed bskDN, a dominant-negative form of Drosophila JNK,12 or puckered (puc), a JNK activity inhibitor.11 Inhibition of JNK signalling by the coexpression of bskDN or puc, marked by the suppression of puc-lacZ expression, suppresses the cell death induced by ectopic hppy expression (Figures 4a and b), indicating that JNK signalling is required for Hppy-mediated cell death.
hppy activates JNK and TOR signalling pathways. (a) Ectopic expression of hppy induces JNK activation in Drosophila wing discs. The area of en-Gal4-driven gene expression is marked with the GFP fluorescence (top). Wing discs from larvae with the indicated genotypes were stained for JNK activity with an anti-β-galactosidase antibody (bottom). The scale bars represent 20 μm. (b) Ubiquitous expression of hppy enhances phosphorylation of 4E-BP. Phopho-4E-BP levels were analysed with western blots of protein lysates of flies with the indicated genotypes and normalised against levels of tubulin
JNK is required for hppy-induced apoptosis. (a) Expression of a dominant-negative version of Drosophila JNK (bskDN) suppresses Hppy-mediated apoptosis. The area of en-Gal4-driven gene expression is identified by GFP fluorescence (top). Wing discs from larvae with the indicated genotypes were stained with an anti-β-galactosidase antibody (middle) and Hoechst (bottom). Condensed chromatin is indicated by the dashed line. (b) Expression of the JNK activity inhibitor, puckered (puc), suppresses Hppy-mediated apoptosis. The area of en-Gal4-driven gene expression is identified by GFP fluorescence (top). Wing discs from larvae with the indicated genotypes were stained with an anti-β-galactosidase antibody (middle) and Hoechst (bottom). Condensed chromatin is indicated by the dashed line. (c, d) The expression of either TSC1/2 (TSC) or 4E-BPTA fails to block Hppy-dependent apoptosis. The area of en-Gal4-driven gene expression is identified by GFP fluorescence (top). Wing discs from larvae with the indicated genotypes were stained with Hoechst (bottom). Condensed chromatin is indicated by the dashed line. Quantitative analysis of cell death was performed in wing discs from larvae with the indicated genotypes. The data presented are the mean values ±S.E.M.; n ⩾ 14 in each group. The scale bars represent 20 μm
To assess the ability of Hppy to activate the TOR pathway in flies, we assessed the phosphorylation status of the TOR substrate, 4E-BP, in adult flies expressing hppy, as well as in appropriate controls, under the ubiquitous driver da-Gal4 (Figure 3b). We found that hppy expression results in enhanced phosphorylation of 4E-BP. Surprisingly, the expression of hppy KD failed to suppress the enhanced phosphorylation of 4E-BP, suggesting that Hppy kinase activity is not critical for TOR activation. To determine the role of the TOR pathway in Hppy-induced apoptosis, we coexpressed hppy with either tuberous sclerosis complex (TSC)1/2 (TSC), an inhibitor of TOR activity, or the TOR substrate 4E-BP, driven by en-Gal4. With both approaches, we failed to observe a significant reduction of cell death in the posterior compartment of the wing discs (Figures 4c and d). Thus, the TOR pathway is unlikely to have a significant role in Hppy-dependent cell death.
Together, these results demonstrate that hppy requires the JNK, but not TOR signalling pathway, to induce apoptotic cell death in Drosophila.
Discussion
The work reported here identifies Hppy, the Drosophila orthologue of human MAP4K3, as an in vivo modulator of apoptosis. In mammalian cells, MAP4K3 promotes apoptosis by inducing the post-transcriptional activation of a subset of BH3-only Bcl-2 family proteins. Expression of MAP4K3 leads to cell death through the mitochondrial (intrinsic) pathway; this action is suppressed by JNK and TOR inhibition. In this study, we provide the first in vivo evidence that the fly orthologue of MAP4K3, Hppy, is a death-inducing kinase that promotes caspase-dependent apoptosis. Mitochondria have a crucial role in the intrinsic apoptosis pathway in vertebrates. In Drosophila, however, the role of mitochondria in apoptosis is unclear, as mutants of the Drosophila bcl-2 homologues debcl and buffy show no obvious defects in developmental apoptosis12 and this form of cell death is not blocked in cells with depleted Cyt-C.13 Our data suggest that Hppy-mediated cell death occurs independently of the mitochondrial pathway; buffy expression failed to block Hppy-dependent apoptosis, and negative modulation of the TOR pathway in flies failed to suppress Hppy-dependent cell death. Given that the human orthologue of Hppy employs the TOR pathway to stimulate the mitochondrial pathway of apoptosis, we propose that suppression of this pathway in flies fails to affect Hppy-dependent cell death because the mitochondrial apoptosis pathway may be of limited importance in flies. Our genetic analysis suggests that the JNK pathway positively regulates Hppy-dependent cell death. As mentioned above, it is unlikely that the JNK pathway induces cell death by activating the mitochondrial pathway in Drosophila. In flies, the genes reaper, grim and hid (RGH) promote caspase activation by antagonising the Drosophila inhibitor of apoptosis proteins. Although reaper and grim are only expressed in cells that are destined for death, hid expression is controlled at both the transcriptional and post-transcriptional levels (reviewed in Bilak and Su14). Hid has been shown to be transcriptionally activated in a JNK- and Foxo-dependent manner. Additionally, the proapoptotic activity of Foxo is opposed by the action of receptor tyrosine kinases (RTKs) such as EGFR and insulin-like growth factor receptor. Given the recent observation that Hppy is a potent negative modulator of EGFR signalling,5 it is tempting to propose that this kinase might promote apoptosis by modulating signalling pathways that affect the levels of RGH proteins. This idea is consistent with the observation that another Ste20 family member, Hippo, promotes hid expression.15
A single amino-acid substitution in MAP4K3 has been associated with pancreatic cancer, suggesting that this kinase might be an important modulator of tumourigenesis.8 We also previously found that the levels of MAP4K3 are significantly reduced in pancreatic tumour samples.6 Given that in Drosophila, Hppy acts to both stimulate JNK signalling (this study) and dampen the EGFR pathway,5 it is tempting to propose that mammalian MAP4K3 might act as a tumour suppressor, not only by promoting JNK-dependent apoptosis but also by blocking the prosurvival effects of EGFR signalling mechanisms.
Materials and Methods
Antibodies
The primary antibodies employed in this study are listed in Supplementary Table S1.
Drosophila strains and phenotypic analysis
Fly stocks and crosses were maintained on standard cornmeal agar media at 25°C. Genotypes of flies employed in this study are listed in Supplementary Table S2. Drosophila MAP4K3 (hppy) was PCR-amplified from BDGP clone RH10407 and subcloned into the pUAST vector. A kinase-dead hppy mutant was made by mutating lysine 55 to glutamic acid in the ATP-binding domain of this kinase using the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA, USA). The corresponding transgenic flies were generated by BestGene Inc. (Chino Hills, CA, USA; http://www.thebestgene.com). The en-Gal4, UAS-gfp/CyO, da-Gal4, UAS-puc and UAS-TSC1/2 (kind gifts from N Tapon), UAS-Buffy (Quinn et al.9), UAS-p35 (Hay et al.10), UAS-4E-BPTA (Imai et al.17) and all others were obtained from the Bloomington Stock Center (Bloomington, IN, USA). Wing phenotypes were analysed at 25°C. Wings were dissected from 3- to 4-day-old females and mounted for microscopic examination in halocarbon oil. Images were acquired using a Leica (Wetzlar, Germany) stereomicroscope ( × 40) equipped with a Leica colour digital camera. Images are presented in the orientation of proximal to the left and anterior to the top.
RNA extraction and real-time PCR
Isolation of total RNA was performed using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Quantitative real-time RT-PCR was performed on an Mx4000 (Stratagene) real-time cycler using the QuantiTect SYBR Green RT-PCR system (Qiagen). Gene-specic primers for the actin 79B, buffy, puc and TSC genes were obtained from Qiagen (QuantiTect Primer Assays). Primers recognising hppyRA and hppyRB transcripts were described in a previous study.5 The relative transcript levels of the target genes were normalised against actin mRNA levels; quantification was performed using the comparative Ct method.16 The mRNA expression levels of hppy, buffy, puc or TSC in transgenic flies used in this study and driven by the ubiquitous da-Gal4 driver are shown in Supplementary Figure S1.
Immunohistochemistry and TUNEL
Inverted third-instar larvae were fixed in 3% paraformaldehyde in PBS for 20 min at room temperature and extensively washed in PBS containing 0.2% Triton X-100 (PBT). Tissues were then blocked for 1 h in PBT containing 1% bovine serum albumin (BSA). Primary antibodies diluted in 1% PBT/BSA were incubated overnight at 4oC. Dissected larvae were washed and then incubated for 4 h at room temperature with secondary antibodies (1/200) and Hoechst 33342 (1/500) (Invitrogen, Carlsbad, CA, USA) diluted in 1% PBT/BSA. After washing, wing discs were dissected and mounted in Vectashield (Vector Laboratories, Burlingame, CA, USA). Fluorescence images were acquired with a Zeiss LSM510 confocal microscope. Images are presented in the orientation of anterior to the left and ventral to the top. TUNEL staining was performed with the ApopTag red in situ apoptosis detection kit (Millipore, Billerica, MA, USA) according to the manufacturer's instructions.
Western blotting
In all, 100 μg of protein lysates was resolved by SDS-PAGE and transferred to Immobilon PVDF membranes (Millipore). The membranes were blocked in 5% milk and subsequently incubated with the indicated primary antibody (see Supplementary Table S1) before being incubated with the appropriate HRP-conjugated secondary antibody. Antibody complexes were visualised by enhanced chemiluminescence.
Statistical analysis
Mean values are presented with error bars corresponding to ±S.D. or ±S.E.M. as indicated. Statistical analysis was performed using Prism statistical analysis software (http://www.graphpad.com). Significance is indicated as ***P<0.001, **P<0.01 and *P<0.05.
Abbreviations
- ATP:
-
adenosine triphosphate
- Bcl-2:
-
B-cell lymphoma protein-2
- Bcl-XL:
-
B-cell lymphoma-extra large
- BH3:
-
Bcl-2 homology domain 3
- da :
-
daughterless
- 4E-BP:
-
eukaryotic translation initiation factor 4E-binding protein
- EGFR:
-
epidermal growth factor receptor
- en :
-
engrailed
- ERK:
-
extracellular signal-regulated kinase
- GFP:
-
green fluorescent protein
- hppy KD:
-
happyhour kinase dead
- JNK:
-
c-Jun N-terminal kinase
- MAPK:
-
mitogen-activated protein kinase
- MAP4K3:
-
mitogen-activated protein kinase kinase kinase kinase-3
- mTORC1:
-
mammalian TOR complex 1
- puc :
-
puckered
- RGH, reaper, grim:
-
hid
- RNAi:
-
RNA interference
- TOR:
-
target of rapamycin
- TSC:
-
tuberous sclerosis complex
- TUNEL:
-
terminal deoxynucleotidyl transferase dUTP nick-end labeling
- UAS:
-
upstream activation sequence
References
Wagner EF, Nebreda AR . Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 2009; 9: 537–549.
Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF . A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J 2007; 403: 13–20.
Bryk B, Hahn K, Cohen SM, Teleman AA . MAP4K3 regulates body size and metabolism in Drosophila. Dev Biol 2010; 344: 150–157.
Diener K, Wang XS, Chen C, Meyer CF, Keesler G, Zukowski M et al. Activation of the c-Jun N-terminal kinase pathway by a novel protein kinase related to human germinal center kinase. Proc Natl Acad Sci USA 1997; 94: 9687–9692.
Corl AB, Berger KH, Ophir-Shohat G, Gesch J, Simms JA, Bartlett SE et al. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell 2009; 137: 949–960.
Lam D, Dickens D, Reid EB, Loh SH, Moisoi N, Martins LM . MAP4K3 modulates cell death via the post-transcriptional regulation of BH3-only proteins. Proc Natl Acad Sci U S A 2009; 106: 11978–11983.
Lam D, Martins LM . MAP4K3 enhances the expression of the BH3-only protein BID. Cell Cycle 2009; 8: 3248–3249.
Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008; 321: 1801–1806.
Quinn L, Coombe M, Mills K, Daish T, Colussi P, Kumar S et al. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J 2003; 22: 3568–3579.
Hay BA, Wolff T, Rubin GM . Expression of baculovirus P35 prevents cell death in Drosophila. Development 1994; 120: 2121–2129.
Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM et al. Puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev 1998; 12: 557–570.
Sevrioukov EA, Burr J, Huang EW, Assi HH, Monserrate JP, Purves DC et al. Drosophila Bcl-2 proteins participate in stress-induced apoptosis, but are not required for normal development. Genesis 2007; 45: 184–193.
Zimmermann KC, Ricci JE, Droin NM, Green DR . The role of ARK in stress-induced apoptosis in Drosophila cells. J Cell Biol 2002; 156: 1077–1087.
Bilak A, Su TT . Regulation of Drosophila melanogaster pro-apoptotic gene hid. Apoptosis 2009; 14: 943–949.
Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G . Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol 2003; 5: 914–920.
Schmittgen TD, Livak KJ . Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3: 1101–1108.
Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E et al. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J 2008; 27: 2432–2443.
Acknowledgements
We thank Leonie Quinn, Alex Whitworth, Pascal Meier, Nic Tapon and the Bloomington Stock Center for fly stocks.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies the paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Lam, D., Shah, S., de Castro, I. et al. Drosophila happyhour modulates JNK-dependent apoptosis. Cell Death Dis 1, e66 (2010). https://doi.org/10.1038/cddis.2010.44
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2010.44
Keywords
This article is cited by
-
Slik maintains tissue homeostasis by preventing JNK-mediated apoptosis
Cell Division (2023)
-
MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB
Nature Communications (2018)
-
Overlapping functions of the MAP4K family kinases Hppy and Msn in Hippo signaling
Cell Discovery (2015)
-
Analysis of apoptosis methods recently used in Cancer Research and Cell Death & Disease publications
Cell Death & Disease (2012)
-
Role of the promyelocytic leukaemia protein in cell death regulation
Cell Death & Disease (2012)