Abstract
Mouse embryonic fibroblasts (MEFs) are commonly grown in cell culture and are known to enter senescence after a low number of passages as a result of oxidative stress. Oxidative stress has also been suggested to promote centrosome disruption; however, the contribution of this organelle to senescence is poorly understood. Therefore, this study aimed to assess the role of the centrosome in oxidative stress induced-senescence using MEFs as a model. We demonstrate here that coincident with the entry of late-passage MEFs into senescence, there was an increase in supernumerary centrosomes, most likely due to centrosome fragmentation. In addition, disrupting the centrosome in early-passage MEFs by depletion of neural precursor cell expressed developmentally downregulated gene 1 (NEDD1) also resulted in centrosomal fragmentation and subsequent premature entry into senescence. These data show that a loss of centrosomal integrity may contribute to the entry of MEFs into senescence in culture, and that centrosomal disruption can cause senescence.
Similar content being viewed by others
Main
Cells normally possess stringent checkpoint mechanisms to ensure that they do not replicate with damaged DNA or other cellular abnormalities. These checkpoints often manifest as cell-cycle arrest, wherein defective cells are either repaired or eliminated.1 However, under certain culture conditions and in response to particular forms of stress, cells enter senescence, a state of irreversible growth arrest in the G1 to S phase transition of the cell cycle,2 and can remain metabolically active for long periods of time.3 In addition to proliferation arrest, some senescent cells become resistant to apoptotic death and most show morphological and metabolic changes, including cellular enlargement, increased lysosome biogenesis and atypical senescence-associated β-galactosidase activity (SA-β-gal).4
Senescence can be triggered by many events including DNA damage, telomere shortening, cell culture or oxidative stress, cytotoxic drugs, overexpression of tumor suppressor genes such as p53, or as a protective cellular response to strong mitogenic signaling by oncogenes.5, 6, 7, 8, 9, 10 As the centrosome is the primary microtubule-organizing center of the cell and is therefore critical for correct cell-cycle progression,11 it is likely that defects in this organelle can also have a role in senescence. Indeed, although disruption of the centrosome has been shown to cause cell-cycle arrest, it can also predispose cells to undergo senescence.12, 13 Whether the centrosome is also involved in senescence caused by other factors, such as oxidative stress, remains unknown.
Mouse embryonic fibroblasts (MEFs) are commonly used as a model to study cellular senescence. These cells senesce as a result of oxidative stress in culture, which causes extensive DNA damage and resembles hydrogen peroxide-induced senescence in human cells, relying predominantly on the activation of p53 and p19ARF pathways.14, 15 To determine whether the centrosome contributes to senescence in MEFs, this study analyzed protein encoded by neural precursor cell expressed developmentally downregulated gene 1 (NEDD1), as a marker for the centrosome during senescence. This protein is important in the targeting of γ-tubulin to the centrosome and is therefore essential for correct centrosomal function.16, 17, 18, 19 This study showed that the entry of MEFs into senescence was coincident with an accumulation of centrosome abnormalities. Importantly, disrupting the centrosome in early-passage MEFs induced a loss of centrosome integrity and led to premature senescence, suggesting that centrosomes are involved in MEF senescence.
Results
MEFs enter senescence in culture after passaging
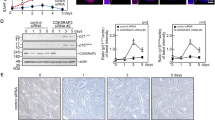
MEFs were used as a model system for cells that grow for a finite number of divisions in cell culture before entering senescence. In multiple experiments using different batches of cells, MEFs proliferated until passage 5–6, after which time their population doubling time was dramatically increased (example shown in Figure 1a). The population doubling time of these cells at passage 6 was ∼320 h as opposed to cells from passage 1 doubling every 28 h. After passage 6, the surviving cells recovered and the doubling time decreased again (data not shown). This is likely due to a population of cells escaping senescence and proliferating again as a result of spontaneous immortalization, a characteristic of MEFs in culture.20 However, this study only analyzed cells until the majority of the population first entered senescence (passage 6). To confirm that these cells were indeed entering senescence, a β-galactosidase assay was performed to stain cells that express SA-β-gal activity.4 Correlating with the doubling time, it was evident that the majority of cells were senescent at passage 6 (Figure 1b). In addition, the protein levels of p16, p19 and p21, which are known to be associated with senescence, were also analyzed. Expression of all of these proteins was increased from about passage 3, and stayed high as the cells entered senescence (passage 6) (Figure 1c). The level of p21 mRNA transcript was analyzed using quantitative real-time PCR (qPCR). This correlated well with the protein expression, as p21 mRNA was increased at passage 4 and again at passage 6 (Figure 1d).
MEFs undergo senescence after 5 passages. (a) At each passage (p), cells were counted and the doubling time was calculated. The p6 cells show greatly increased doubling time as cells enter senescence. (b) At each passage, the cells were analyzed for SA-β-gal staining. By passage 6, the majority of cells are blue, indicating that senescence has been activated. Scale bars=40 μm. (c) Equal amounts (50 μg) of MEF protein lysates loaded onto replicate gels were analyzed by immunoblotting. Typical markers of senescence p16, p19 and p21 are increased during passaging. (d) p21 mRNA expression in MEFs was quantitated by qPCR at passages 1, 4 and 6. Error bars show S.E.M. (n=3)
Cells with abnormal centrosome numbers increase in senescent MEFs
We next assessed whether there were also changes in centrosome structure or integrity in these cells. To test this, cells from an early passage (passage 1) and a late senescent passage (passage 6) were stained for centrosomal proteins. At passage 1, almost all cells presented the expected centrosomal staining (Figure 2a and b). NEDD1 and γ-tubulin always colocalized in these cells. However, at this passage, many cells displayed abnormal centrosomes (Figure 2c–e). In most cases, there was an increased number of centrosomal dots, often appearing smaller in size than the centrosomes at passage 1, sometimes distributed throughout the cell (Figure 2c and d) and sometimes clustered (Figure 2e). As in early-passage cells, NEDD1 and γ-tubulin always colocalized at the centrosomes.
Abnormal centrosome numbers in senescent MEFs. MEFs were stained for NEDD1 (green), γ-tubulin (red) and Hoechst 33342 (blue), at each passage until senescence. The centrosomes are enlarged in the insets. Where not obvious, the area enlarged is shown by a smaller box. (a and b) At passage 1 (p1), most cells display normal numbers of centrosomes with either one (panel a) or 2 (panel b) centrosomes costaining with NEDD1 and γ-tubulin. (c–e) By passage 6 (p6), many cells display more than two centrosomal structures either distributed throughout the cell (panels c and d) or in close proximity to each other (panel e). Scale bars=20 μm
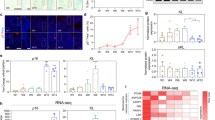
Quantification of the centrosomal dots in cells from passage 1 to passage 6 indicated that there was a gradual increase in the number of cells with more than two centrosomes from passage 1 (1%) to passage 5 (6%), and then a greater increase at passage 6 (16%) (Figure 3a). In the cells with more than two centrosomes, the actual number of centrosomes usually ranged from three to five; however, there was also a proportion of cells that displayed six or more centrosomes (Figure 3b). Although it was not feasible to costain cells for centrosomal markers and the senescent marker SA-β-gal, it appeared that abnormal centrosomes were occurring in morphologically senescent cells.
Quantification of abnormal centrosomes in senescent MEFs. The numbers of centrosomes in MEFs from passage 1 and passage 6 (examples shown in Figure 2) were counted. At least 300 cells were counted for each passage. (a) The percentage of MEFs with more than two centrosomes increases sharply between passages 5 and 6. (b) Of the cells displaying more than two centrosomes, the majority have three centrosomes, and some have four or more
The centrosomes in senescent MEFs appear to become fragmented
It was then assessed whether the increase in supernumerary centrosomes was due to fragmentation or overduplication of centrosomes. To address this question, cells at passage 1 and passage 6 were costained with NEDD1 and the centriolar marker, GT335, which detects glutamylated tubulin.21 At this resolution, it was often not possible to detect two centrioles within each centrosome; however, at passage 1, all NEDD1 and GT335 dots appeared normal with one to two centrosomal structures per cell (Figure 4a and b). At passage 6, there were many cells with more than two centrosomal structures, and they often appeared smaller in size (Figure 4c–e). In addition, although in most cells there was costaining of NEDD1 and GT335 in all dots (Figure 4c and d), there were some cells that contained dots only positive for NEDD1 and not GT335 (arrows, Figure 4e). This indicates that these dots likely represent fragments of centrosomal material, and did not contain an intact centriole. Although centrosome overduplication also results in multiple centrosomes, the smaller size and lack of complete localization of the centriole marker within all NEDD1-immunostained dots suggests that centrosome fragmentation is likely to be the primary mechanism for the supernumerary centrosomes in these cells.
MEFs at passage 6 contain fragmented centrosomes. Cells were stained for NEDD1 (green), GT335 (red) and Hoechst 33342 (blue), at passages 1 and 6. The centrosomes are enlarged in the insets. Where not obvious, the area enlarged is shown by a smaller box. (a and b) At passage 1 (p1), most cells display normal numbers of centrosomal structures with either 1 (panel a) or 2 (panel b) pairs of centrioles costaining with NEDD1 and GT335. Each pair of centrioles normally appears as a single dot at this resolution. (c–e) By passage 6 (p6), some cells displaying more than two centrosomal structures have complete colocalization of NEDD1 and GT335 in all centriole-like dots (panels c and d), and other cells have some NEDD1-positive but GT335-negative dots (arrows, panel e). The dots are often smaller in centrosomal structures at p6. Scale bars=20 μm
A reduction of NEDD1 leads to premature entry into senescence
From these experiments, it was unclear whether centrosome disruption was a reflection of cells entering senescence, or whether these changes contributed to the fate of the cells entering senescence. To investigate this, centrosomes were disrupted by depletion of NEDD1 using siRNA in MEFs at an early passage (passage 1). No change was observed in NEDD1 or γ-tubulin levels in control siRNA-treated cells over the course of the experiment (Figure 5a–c). After a double transfection of NEDD1 siRNA over 5 days in culture, NEDD1 levels were drastically reduced (Figure 5d and i). After removal of the siRNA at day 5, the cells gradually regained NEDD1 expression over 8 and 11 days after the initial siRNA transfection (Figure 5e and f).
NEDD1 depletion causes premature entry of MEFs into senescence. Passage 1 MEFs were transfected with a control (scrambled) siRNA or NEDD1 siRNA and then stained for NEDD1 (green) and γ-tubulin (red) on day 5 when the siRNA was removed and again on day 8 and day 11. (a–c) The MEFs transfected with the control siRNA show no reduction of NEDD1 or γ-tubulin protein. (d) After 5 days of NEDD1 siRNA treatment, the levels of NEDD1 and γ-tubulin are dramatically reduced at the centrosome. (e and f) The levels of these proteins gradually increases 8 days after the initial transfection (3 days after siRNA removal) and again 11 days after the initial transfection (6 days after siRNA removal). (g) Cells treated with the control siRNA maintain low numbers of SA-β-gal-positive cells on day 5, 8 and 11. Cells treated with NEDD1 siRNA have relatively low levels of SA-β-gal staining after 5 days of NEDD1 depletion, but SA-β-gal-positive cells become more abundant 8 days after initial depletion, and again after 11 days. (h) The number of SA-β-gal-expressing cells was counted. At days 8 and 11, there is a significant increase in the number of senescent cells in the NEDD1 siRNA-treated cells compared with control siRNA-transfected cells. There is also a significant increase in the number of senescent cells between days 5 and 11 of the NEDD1 siRNA-treated group. Approximately 300 cells were counted for each group. Error bars show S.E.M. from three independent experiments. Scale bars=40 μm. (i) Immunoblotting shows protein levels of NEDD1 in MEFs from passage 1 transfected with control (scrambled) or NEDD1 siRNA for 72 h, and again for another 48 h before harvesting. β-Actin serves as a loading control
To investigate whether this reduction in NEDD1 could cause premature entry of early-passage MEFs into senescence, cells depleted of NEDD1 were assessed for SA-β-gal activity on various days after NEDD1 knockdown. Cells treated with control siRNA showed no increase in senescent cells (<5% SA-β-gal-stained cells at each day) (Figure 5g and h). However, cells depleted of NEDD1 showed an increase in senescent cells 8 and 11 days after the initial NEDD1 siRNA transfection (12 and 22% SA-β-gal-stained cells, respectively). This was significant at day 8 (P=0.01), and approaching significance at day 11 (P=0.05) (Figure 5h). In addition, there was a significant increase in the number of senescent cells between days 5 and 11 in the NEDD1 siRNA-treated group (P=0.02), indicating increased senescence over time, even when NEDD1 was becoming reexpressed in the cells.
To further confirm the induction of senescence after centrosome disruption, cells were also depleted of Cep192, a centrosomal protein which is a major regulator of pericentriolar material recruitment, centrosome maturation and centriole duplication that has been shown to be important for the centrosomal localization of NEDD1 and γ-tubulin.22, 23 As expected, cells treated with Cep192 siRNA with the same conditions as for NEDD1 siRNA displayed a reduction in NEDD1 and γ-tubulin centrosomal staining, and a similar increase in the number of senescent cells (data not shown).
A reduction of NEDD1 causes centrosome abnormalities
Given that senescence in late-passage MEFs was also accompanied by an increase in supernumerary centrosomes (Figure 3), we assessed whether the depletion of NEDD1 from early-passage MEFs could also induce centrosome abnormalities. Depletion of NEDD1 reduces the levels of γ-tubulin at the centrosome (Figure 5), but not the centriole marker GT335; hence, GT335 was used for staining in these cells. Indeed, although almost all cells in control siRNA-treated cells had one to two centriole pairs (Figure 6a–c, each pair only visible as one centrosomal dot), after 5 days of NEDD1 depletion, there were many cells that showed more than two centrosomal structures (Figure 6d). This was increased at both 8 and 11 days after initial NEDD1 depletion (Figure 6e–g). GT335-positive dots were sometimes smaller in NEDD1-depleted cells, suggesting that they are likely to be centrosomal fragments. When these were quantitated, the number of cells with more than two centrosomal structures in NEDD1-depleted cells was increased to 28% at day 5, 38% at day 8 and to 45% at day 11 (Figure 6g). This was significant at all time points when compared with control-treated cells which had ∼10% of cells with more than two centrosomal structures at all days (P=<0.05). Again, there was also a significant increase in the accumulation of centrosomal structures between days 5 and 11 in the NEDD1 siRNA-treated group (P=0.01), indicating that centrosome abnormalities increased over time, even when NEDD1 was becoming reexpressed in the cells. Similar results were observed for depletion of Cep192 (data not shown). These data suggest that a loss of centrosomal integrity, caused by a reduction in NEDD1, can lead to centrosomal abnormalities and premature senescence in early-passage MEFs.
NEDD1 depletion causes abnormal centrosomes. An aliquot of cells treated with control or NEDD1 siRNA as in Figure 5 were stained for NEDD1 (green) and GT335 (red) at day 5 when the siRNA was removed and again at days 8 and 11. (a–c) At all time points in the control (scrambled) siRNA-treated MEFs, NEDD1 and GT335 staining show normal centrioles (1–2 pairs per cell). (d) 5 days after the first NEDD1 siRNA transfection, the level of NEDD1 is dramatically reduced at the centrosome, and GT335 staining reveals some cells have more than two centrosomal structures. (e and f) At 8 and 11 days after NEDD1 siRNA transfection, there are more cells with more than two centrosomal structures even though NEDD1 is becoming reexpressed. (g) The number of centrosomal structures in control scrambled siRNA-treated and NEDD1 siRNA-treated cells was quantitated. At days 5, 8 and 11, there is a significant increase in the number of cells containing more than two centrosomal structures in the NEDD1 siRNA-treated cells compared with control cells. In addition, there is a significant increase in the number of cells with more than two centrosomes between day 5 and day 11 in the NEDD1 siRNA-treated cells. Approximately 300 cells were counted for each group. Error bars show S.E.M. from three independent experiments. Scale bars=40 μm
Discussion
The centrosome has been implicated in cell-cycle arrest,24 but its involvement in the irreversible state of senescence remains less clear. MEFs at a late passage are a model system of oxidative stress-induced senescence, as standard culture conditions include atmospheric (20%) oxygen which causes DNA damage and senescence.14 In this study, MEFs passaged in atmospheric oxygen grew for five passages before entry of the majority of cells into senescence. This correlated with an increase in cell-cycle proteins p16, p19 and p21, which are known markers of senescence in MEFs.14 Closer analysis of senescent MEFs showed the presence of centrosome abnormalities. At passage 6, when the majority of cells had entered senescence, many cells displayed abnormal supernumerary centrosomes. Although the percentage of cells with supernumerary centrosomes was still relatively low (16% at passage 6), this is likely to represent a much larger pool of the senescent population, for two reasons. First, not all cells at passage 6 were positive for SA-β-gal activity. Hence, a pool of the population had not yet entered senescence. Second, abnormal centrosomes can result in aberrant mitoses with nonviable daughter cells,25 and hence some of the cells with abnormal centrosomes may have been eliminated before analysis. This result correlates well with a study in human Chang liver cells, which identified that 20% of cells exposed to low doses of hydrogen peroxide, which causes an increase in reactive oxygen species, accumulated supernumerary centrosomes compared with 5–7% in controls.26 The observation that MEFs accumulate supernumerary centrosomes during passaging is important, as MEFs are commonly used for many types of studies. A detailed understanding of the events that occur during the culture of these cells is critical when making assumptions and interpreting results.
Given that these MEFs were acquiring centrosome abnormalities around the time they entered senescence, we wanted to assess whether centrosome disruption was a cause or a consequence of senescence. To test this, we disrupted the centrosome by depletion of one of its key components, NEDD1, in healthy MEFs and assayed for the induction of premature senescence. Indeed, after long-term NEDD1 depletion (8 days), an increase in the number of senescent cells was observed, which continued to increase even after NEDD1 levels had been restored. Hence, the depletion of NEDD1 for a sustained period of time induces senescence, most likely due to a disruption of centrosome function. Importantly, the depletion of Cep192, which also results in a reduction of NEDD1 and γ-tubulin at the centrosome22, 23 was also able to induce premature senescence. This provides further evidence that disruption of the centrosome resulting in a reduction in centrosomal NEDD1 and γ-tubulin can cause senescence.
To confirm whether the reduction of NEDD1 in senescent MEFs was also responsible for a lack of centrosome integrity and abnormalities, centrosome integrity was evaluated in these MEFs depleted of NEDD1. NEDD1 depletion resulted in an increase in the number of cells containing supernumerary centrosomes, which were often small in size indicating fragmented centrosomes. Given that the centrosome abnormalities in NEDD1-depleted cells increased 5 days after siRNA treatment, but senescent cells did not appear until day 8, it appears that centrosome fragmentation is an early event in this process, and this then leads to the induction of senescence. As NEDD1 is important in centriole assembly and duplication,16 it was expected that a reduction in NEDD1 levels may result in centrosome abnormalities due to a reduction in the number of centrosomes, rather than the supernumerary centrosomes observed in this study. In addition, centrosome amplification is commonly linked to tumorigenesis,27 and as such would not be expected in senescence cells. To explain the results observed, we hypothesized that the abnormal centrosomes in cell culture-induced senescent MEFs at passage 6 and NEDD1 depletion-induced senescent MEFs at an earlier passage may have been fragmented or incorrectly assembled, rather than overamplified. In populations displaying a high proportion of senescent cells, the centrosomal-like dots were often much smaller than in early-passage cells. In addition, not all NEDD1-positive dots costained for GT335, suggesting that they contained some centriolar material that did not include the centriole component glutamylated tubulin. Furthermore, Cep192 is important for centriole duplication,22, 23 and hence reduction of this protein should not allow centrosome overduplication, even though multiple centrosome-like dots were observed in these cells. Hence, it appears that senescent MEFs display fragmented rather than overduplicated centrosomes.
Loss of NEDD1 causing centrosome fragmentation has also been suggested in HeLa cells.17 This observation aligns well with a recent study on components of the human augmin complex (HAUS) which localizes to the centrosome and mitotic spindle.28 Depletion of components of the HAUS complex causes disorganized spindles and fragmented centrosomes.28 Importantly, FAM29A, one of these complex proteins, has been shown to interact with NEDD1 and recruits this protein and therefore also γ-tubulin to the centrosome and mitotic spindle.29 In the absence of HAUS components, the levels of NEDD1 and γ-tubulin at the centrosome are reduced, and there is a mis-localization of motor proteins away from the spindle.28 The centrosomes cannot sustain the resultant counterbalancing forces at the spindle pole and this leads to their fragmentation. The data in our study show that the depletion of NEDD1 in MEFs may contribute to counterbalancing forces at the centrosome as with HAUS depletion, which leads to centrosome fragmentation and senescence.
A functional centrosome is required for proper cell-cycle progression.13, 24 Our study provides evidence that a loss of centrosome integrity, which occurs in later-passage MEFs, can result in centrosomal fragmentation and senescence. In addition, depletion of core centrosomal proteins, such as NEDD1, can induce premature senescence. Given that MEFs are known to senesce as a result of oxidative stress and DNA damage,14 it is plausible that the oxidative stress is responsible for centrosome disruption, possibly by causing dysfunction of core centrosomal components such as NEDD1, and this then, in part, leads to senescence.
Materials and Methods
Cell lines and culture
MEFs were isolated from E14.5 C57Bl/6 embryos.30 MEFs, N18s and HeLa cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 50 Units/ml penicillin (Sigma, St. Louis, MO, USA), 0.05 mg/ml streptomycin (Sigma) at 37°C in 5% CO2. Other cells were retrieved from liquid nitrogen and not cultured. Population doubling time was calculated using the formula: ln2/(ln[A/A0])/t where A=cell number at t=0, A0=initial cell number, t=time (hours since last passage).
Cell staining
Cells were plated onto glass coverslips 24 h before staining, fixed in 100% methanol for 5 min at −20°C, washed in PBS and incubated in blocking solution (1% FBS/PBS) for 30 min. Primary antibodies in blocking solution were added for 2 h at RT: 1 : 50 rabbit α-NEDD1,31 1 : 500 mouse α-γ-tubulin (GTU-88, Sigma), 1 : 500 mouse α-GT335 (Gift from B Edde, CNRS, Montpellier, France) or 1 : 500 rabbit α-Cep192.23 Cells were washed in PBS and incubated with secondary antibodies: 1 : 1000 rabbit Alexa Fluor 488 and/or 1 : 1000 mouse Alexa Fluor 568 (Molecular Probes, Invitrogen) for 1 h at RT. After washes and staining with Hoechst 33342 (Molecular Probes, Invitrogen) coverslips were mounted in Prolong Gold Antifade reagent (Molecular Probes, Invitrogen). Images were obtained using a confocal microscope (Radiance 2100, Bio-Rad Laboratories, Hercules, CA, USA) as described previously32 and were processed using Adobe Photoshop CS2 Version 9.0 (Adobe, San Jose, CA, USA).
β-Galactosidase staining
Cells were plated onto glass coverslips 24 h before staining. Cells were fixed and stained using the Senescence β-galactosidase Staining Kit (Cell Signaling, Beverly, MA, USA). Cells were incubated for 24–48 h at 37°C before being imaged on a CKX41 microscope (Olympus, Center Valley, PA, USA), and photographed using a DP20 camera (Olympus).
Protein extraction and immunoblotting
Cells were lysed in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS, 50 mM Tris pH 7.4, 5 mM EDTA and protease inhibitor cocktail solution (Roche, Mannheim, Germany)) and lysates were cleared by centrifugation. In all, 50 μg of protein was separated by SDS-PAGE and transferred to PVDF membranes. Membranes were blocked in 5% skim milk powder in PBST (0.1% Tween20/PBS) for 1–2 h and incubated with primary antibodies overnight at 4°C: 1 : 500 rabbit α-p16 (M-156, Santa Cruz Biotechnology, Santa Cruz, CA, USA), 1 : 1000 rabbit α-p19 (Ab80, Abcam, Cambridge, MA, USA), 1 : 500 mouse α-p21 (F5, Santa Cruz Biotechnology), 1 : 200 rabbit α-NEDD131 or 1 : 5000 α-β-actin (AC-15, Sigma). After washing in PBST, the membranes were incubated with 1 : 2000 rabbit secondary ECL IgG horseradish peroxidase (HRP)-linked whole antibody (GE Healthcare, Piscataway, NJ, USA) to detect NEDD1 or 1 : 2000 IgG affinity isolated-alkaline phosphatase (AP) conjugated antibody (Chemicon, Temecula, CA, USA) to detect all other proteins, for 2 h at RT. Detection of bound HRP antibody was achieved using ECL Plus (GE Healthcare) and developed on X-ray film (Fuji, Dusseldorf, Germany). Detection of bound AP was achieved using ECF substrate (GE Healthcare) and developed using a Typhoon 9410 (Molecular Dynamics, Sunnyvale, CA, USA) and analyzed using ImageQuant software (GE Healthcare).
qPCR
In all, 2 μg of total RNA was used to synthesize cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Samples for qPCR were prepared from this cDNA using the RT2 SYBR Green qPCR Master Mix (SA Biosciences, Frederick, MD, USA) according to the manufacturers’ instructions. Primers for p21 and GAPDH mRNA expression were as follows: QPCRmp21f: 5′-AGTGTGCCGTTGTCTCTTCG-3′, QPCRmp21r: 5′-ACACCAGAGTGCAAGACAGC-3′, GAPDHf: 5′-AGACAGCCGCATCTTCTTGT-3′, GAPDHr: 5′-GAATTTGCCGTGAGTGGAGT-3′. Samples were run on a Rotor Gene 6000 machine (Corbett Research, NSW, Australia) with the following conditions: 2 min at 50°C, 15 min at 95°C, followed by 40 cycles of 95°C for 15 s, 60°C for 25 s and 72°C for 10 min, followed by a 3-min extension at 72°C. Melt curve was achieved by a ramp from 72 to 99°C with 5 s for each step. Reactions were performed in triplicate and the mRNA expression levels normalized against the internal control gene GAPDH using the ΔΔCT method. Data were analyzed using the Rotor-Gene 6000 Series Software.
siRNA transfection
NEDD1 siRNAs were designed by Invitrogen to target the following sequences: mNEDD1 siRNA no. 1: 5′-GAGACAUUGUGAAUCUGCAAGUGGA-3′, mNEDD1 siRNA no. 2: 5′-CCGGCACAUCAAGUACUCAUUGUUU-3′. Cep192 siRNA was designed to target the sequence 5′-GGAGGACUUUGUAAUCUCUtt-3′ (GenePharma, Shanghai, China), adapted from the study by Zhu et al.23 One day before transfection, cells were seeded in 6-well plates at 2 × 105 cells per well. siRNA transfections were conducted using 5 μl Lipofectamine 2000 reagent (Invitrogen), according to the manufacturers’ instructions, using 3 μl of each NEDD1 siRNA pooled, and 6 μl of Cep192 siRNA or Negative Universal Control (Medium GC). A second siRNA transfection was conducted 72 h after the first transfection, and left for a further 48 h.
Statistical analysis
To determine significance, a two-tailed t-test (two-sample assuming equal variances) was conducted. A P-value of <0.05 was considered significant.
Abbreviations
- NEDD1:
-
neural precursor cell expressed developmentally downregulated gene 1
- SA-β-gal:
-
senescence-associated β-galactosidase activity
- MEF:
-
mouse embryonic fibroblast
- qPCR:
-
quantitative real-time PCR
References
Pietenpol JA, Stewart ZA . Cell cycle checkpoint signaling: cell cycle arrest versus apoptosis. Toxicology 2002; 181–182: 475–481.
Sherwood SW, Rush D, Ellsworth JL, Schimke RT . Defining cellular senescence in IMR-90 cells: a flow cytometric analysis. Proc Natl Acad Sci USA 1988; 85: 9086–9090.
Goldstein S . Replicative senescence: the human fibroblast comes of age. Science 1990; 249: 1129–1133.
Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 1995; 92: 9363–9367.
Harley CB, Futcher AB, Greider CW . Telomeres shorten during ageing of human fibroblasts. Nature 1990; 345: 458–460.
Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW . Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev 1998; 12: 3008–3019.
Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW . Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997; 88: 593–602.
Zhu J, Woods D, McMahon M, Bishop JM . Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev 1998; 12: 2997–3007.
Ben-Porath I, Weinberg RA . The signals and pathways activating cellular senescence. Int J Biochem Cell Biol 2005; 37: 961–976.
Atadja P, Wong H, Garkavtsev I, Veillette C, Riabowol K . Increased activity of p53 in senescing fibroblasts. Proc Natl Acad Sci USA 1995; 92: 8348–8352.
Doxsey S, Zimmerman W, Mikule K . Centrosome control of the cell cycle. Trends Cell Biol 2005; 15: 303–311.
Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S . Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol 2007; 9: 160–170.
Srsen V, Gnadt N, Dammermann A, Merdes A . Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J Cell Biol 2006; 174: 625–630.
Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J . Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol 2003; 5: 741–747.
Busuttil RA, Rubio M, Dolle ME, Campisi J, Vijg J . Oxygen accelerates the accumulation of mutations during the senescence and immortalization of murine cells in culture. Aging Cell 2003; 2: 287–294.
Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A . NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J Cell Biol 2006; 172: 505–515.
Luders J, Patel UK, Stearns T . GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol 2006; 8: 137–147.
Manning JA, Lewis M, Koblar SA, Kumar S . An essential function for the centrosomal protein NEDD1 in zebrafish development. Cell Death Differ 2010; In press (doi: 10.1038/cdd.2010.12).
Manning JA, Shailini S, Risk JM, Day CL, Kumar S . A direct interaction with NEDD1 regulates γ-tubulin recruitment to the centrosome. PLoS ONE 2010; 5: e9618.
Todaro GJ, Green H . Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol 1963; 17: 299–313.
Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M . Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol 1998; 143: 1575–1589.
Gomez-Ferreria MA, Rath U, Buster DW, Chanda SK, Caldwell JS, Rines DR et al. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol 2007; 17: 1960–1966.
Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C et al. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol 2008; 18: 136–141.
Doxsey S, McCollum D, Theurkauf W . Centrosomes in cellular regulation. Annu Rev Cell Dev Biol 2005; 21: 411–434.
Fukasawa K . Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer 2007; 7: 911–924.
Chae S, Yun C, Um H, Lee JH, Cho H . Centrosome amplification and multinuclear phenotypes are induced by hydrogen peroxide. Exp Mol Med 2005; 37: 482–487.
Fukasawa K . Centrosome amplification, chromosome instability and cancer development. Cancer Lett 2005; 230: 6–19.
Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol 2009; 19: 816–826.
Zhu H, Coppinger JA, Jang CY, Yates III JR, Fang G . FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J Cell Biol 2008; 183: 835–848.
Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S . A tumor suppressor function for caspase-2. Proc Natl Acad Sci USA 2009; 106: 5336–5341.
Manning JA, Colussi PA, Koblar SA, Kumar S . Nedd1 expression as a marker of dynamic centrosomal localization during mouse embryonic development. Histochem Cell Biol 2008; 129: 751–764.
Foot NJ, Dalton HE, Shearwin-Whyatt LM, Dorstyn L, Tan SS, Yang B et al. Regulation of the divalent metal ion transporter DMT1 and iron homeostasis by a ubiquitin-dependent mechanism involving Ndfips and WWP2. Blood 2008; 112: 4268–4275.
Acknowledgements
This study was supported by funds from the Cancer Council of South Australia. JAM was supported by an Australian Postgraduate Award. SK is a Senior Principal Research Fellow of the National Health and Medical Research Council (Grant ID 399300). We thank D Cakouros for performing the qPCR analysis, B Edde for the gift of the GT335 antibody, L Pelletier for the gift of the Cep192 antibody and members of our laboratory for comments on the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Rights and permissions
This article is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 license. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Manning, J., Kumar, S. A potential role for NEDD1 and the centrosome in senescence of mouse embryonic fibroblasts. Cell Death Dis 1, e35 (2010). https://doi.org/10.1038/cddis.2010.12
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2010.12
Keywords
This article is cited by
-
Loss of fragile WWOX gene leads to senescence escape and genome instability
Cellular and Molecular Life Sciences (2023)
-
Centrosome dysfunction: a link between senescence and tumor immunity
Signal Transduction and Targeted Therapy (2020)
-
Global gene expression profiling and senescence biomarker analysis of hESC exposed to H2O2 induced non-cytotoxic oxidative stress
Stem Cell Research & Therapy (2017)
-
Oncogenic PIK3CA induces centrosome amplification and tolerance to genome doubling
Nature Communications (2017)
-
Analysis of apoptosis methods recently used in Cancer Research and Cell Death & Disease publications
Cell Death & Disease (2012)