Abstract
Tumor necrosis factor-alpha (TNF-α) exists in two forms: secretory TNF-α (sTNF-α) and transmembrane TNF-α (tmTNF-α). Although both forms of TNF-α induce tumor cell apoptosis, tmTNF-α is able to kill tumor cells that are resistant to sTNF-α-mediated cytotoxicity, indicating their differences in signal transduction. Here, we demonstrate that internalization of TNFR1 is crucial for sTNF-α- but not for tmTNF-α-induced apoptosis. sTNF-α induces binding of tumor necrosis factor receptor type 1-associated death domain protein (TRADD) to the death domain (DD) of TNFR1 and subsequent activation of nuclear factor kappa B (NF-κB), and the formation of death-inducing signaling complexes (DISCs) in the cytoplasm after internalization. In contrast, tmTNF-α induces DISC formation on the membrane in a DD-independent manner. It leads to the binding of signal transducer and activator of transcription 1 (STAT1) to a region spanning amino acids 319–337 of TNFR1 and induces phosphorylation of serine at 727 of STAT1. The phosphorylation of STAT1 promotes its binding to TRADD, and thus recruits Fas-associated protein with DD (FADD) and caspase 8 to form DISC complexes. This STAT1-dependent signaling results in apoptosis but not NF-κB activation. STAT1-deficiency in U3A cells counteracts tmTNF-α-induced DISC formation and apoptosis. Conversely, reconstitution of STAT1 expression restores tmTNF-α-induced apoptotic signaling in the cell line. Consistently, tmTNF-α suppresses the growth of STAT1-containing HT1080 tumors, but not of STAT1-deficient U3A tumors in vivo. Our data reveal an unappreciated molecular mechanism of tmTNF-α-induced apoptosis and may provide a new clue for cancer therapy.
Similar content being viewed by others
Main
Tumor necrosis factor-alpha (TNF-α) exists in two forms: a 26-kDa transmembrane form and a 17-kDa soluble form.1 Transmembrane TNF-α (tmTNF-α) is the precursor of soluble TNF-α (sTNF-α) and expressed on the surface of activated lymphocytes, macrophages and other cell types.2 Both forms of TNF-α mediate a variety of biological activities by binding to TNF receptor (TNFR) 1 and TNFR2, including cell apoptosis and proliferation.3, 4, 5 However, the bioactivities of two forms of TNF-α are not quite the same and the antitumor effects between both TNF-α forms are distinct. tmTNF-α is able to kill tumor cells that are resistant to sTNF-α-mediated apoptosis.6 In vivo, sTNF-α suppresses tumor growth by recruiting lymphocytes while tmTNF-α inhibits tumor growth by promoting factor associated suicide (Fas) expression and apoptosis.7 However, the molecular mechanisms underlying the different activities of sTNF-α and tmTNF-α are still not fully understood.
TNFR1 is a member of death receptors. The intracellular segment of TNFR1 includes three important and structured domains: the internalization domain (TRID),8 the neutral sphingomyelinase domain (NSD)9 and the death domain (DD)10, 11 which are required for apoptosis induction by TNFR1 ligands. The DD of TNFR1 is well characterized and mediates homotypic interactions with other DD-containing proteins. TNFR1 and tumor necrosis factor receptor type 1-associated death domain protein (TRADD) interact through their DDs in response to sTNF-α. This interaction is the basis for the formation of two alternative protein complexes (complex I and complex II). Complex I is formed at the cell membrane by recruiting receptor-interacting protein (RIP)1, TNF receptor associated factor (TRAF)2 and cellular inhibitor of apoptosis protein (cIAP)1 and cIAP2. This complex activates nuclear factor kappa B (NF-κB) and promotes cell survival and antiapoptotic activities. TNFR1 can be also internalized into the cytoplasm, which is dependent on the TRID domain. Subsequently, a cytosolic complex II can be formed by the recruitment of Fas-associated protein with DD (FADD) and procaspase 8 to the internalized TNFR1–TRADD complex. This complex is called death-inducing signaling complex (DISC) as it is crucial for apoptotic induction.8, 12, 13 In general, the default signaling pathway of sTNF-α is NF-κB pathway activated by complex I at the cell surface, which renders cells resistant to sTNF-α-induced cytotoxicity. However, in case of impaired NF-κB activation the cytosolic complex II is formed, thus switching the signaling from pro-survival to pro-death. In addition to DD and TRID of TNFR1, NSD is also involved in sTNF-α-mediated apoptosis through activation of neutral sphingomyelinase, when TNFR1 is defective in internalization.14
Our previous study demonstrated that tmTNF-α eliminates tumor cells that are resistant to sTNF-α-mediated cytotoxicity. This observation indicates that tmTNF-α and sTNF-α may have different signal transduction via the same receptor.5 In contrast to the well-understood TNFR1-mediated apoptotic signaling by sTNF-α, tmTNF-α-induced cell death signaling remains elusive.15, 16 Here we show that unlike sTNF-α-induced apoptosis, tmTNF-α-induced cell death is independent of the internalization of TNFR1, and does not require DD and the NSD of TNFR1. Instead, tmTNF-α induces the formation of DISC at the plasma membrane in a STAT1-dependent manner to mediate apoptosis but not NF-κB activation. In sum, we identified a distinct and novel TNFR1-dependent signaling transduction mechanism underlying tmTNF-α-mediated apoptosis.
Results
tmTNF-α-mediated cytotoxicity is independent of the TNFR1 internalization
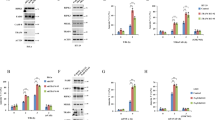
The internalization of TNFR1 is crucial for sTNF-α-induced cytotoxicity.8, 12, 17 To explore whether internalization of TNFR1 is also required for cytotoxicity initiated by tmTNF-α on fixed NIH3T3 cells, monodansylcadaverine (MDC) was used to inhibit the internalization of TNFR118 in U937 cells, expressing mainly TNFR1. As shown in Figure 1, both forms of TNF-α were cytotoxic to the target cells (Figure 1a), induced apoptosis (Figure 1b) and activated caspase 3 (Figure 1c). Caspase inhibitor z-VAD-FMK could block both forms of TNF-α-induced apoptosis (Supplementary Figure S1a). Interestingly, treatment with MDC significantly suppressed sTNF-α-induced cytotoxicity and apoptosis but did not affect tmTNF-α-mediated cell death. Neutralization of tmTNF-α by a specific antibody blocked the activities of tmTNF-α. These observations suggest that tmTNF-α-mediated apoptosis is independent of the internalization of TNFR1. This hypothesis was further confirmed by the evidence (Figure 1d) that sTNF-α induced a marked decrease of cell surface TNFR1 expression whereas tmTNF-α had no effect. MDC treatment of sTNF-α-stimulated U937 cells restored TNFR1 expression to normal. However, total protein expression of TNFR1 was not changed by either form of TNF-α regardless of the absence or presence of MDC (Supplementary Figure S1b), indicating that neither TNF-α nor MDC affected the production of TNFR1. In order to further verify that the TNFR1 internalization was not affected by tmTNF-α, double immunofluorescent staining was performed to check for colocalization of TNFR1 (red) and the endosomal marker EEA (green) by confocal microscopy. As shown in Figure 1e, sTNF-α induced colocalization of TNFR1 with EEA in the cytoplasm (yellow) close to the plasma membrane, whereas tmTNF-α did not cause any redistribution of TNFR1 in U937 cells. As expected, treatment with MDC prevented sTNF-α-induced TNFR1 internalization and colocalization with the endosomal marker. The same phenomena could be observed in HEK 293T cells ectopically expressing TNFR1 (Supplementary Figure S1c). Furthermore, expression of a TNFR1 mutant, Y236A-TNFR1 that fails to internalize8 significantly reduced the cytotoxicity of sTNF-α but not of tmTNF-α (Supplementary Figure S1d). These data indicate that the internalization of TNFR1 is not involved in the cytotoxicity of tmTNF-α.
The cytotoxicity of tmTNF-α was independent of the TNFR1 internalization. U937 cells were pre-treated with 100 μM MDC for 1 h and then stimulated with either 100 ng/ml sTNF-α or tmTNF-α on fixed NIH3T3 cells at an effector/target ratio of 10:1 for 24 h. For neutralization of tmTNF-α, effector cells were treated with anti-TNF-α (Ab) for 30 min prior to the addition to the target cells. (a) Cytotoxicity of TNF-α detected by MTT assays. (b) Apoptosis induced by TNF-α determined by Annexin V-PI staining. (c) Western blotting of caspase 3 activation. (d) TNFR1 expression analyzed by flow cytometry. (e) Confocal images of the colocalization of TNFR1 (red) and EEA (green) after a 30 min-stimulation of U937 cells with both forms of TNF-α (magnification, × 600). All the quantitative data represent means±S.D. of at least three independent experiments. *P<0.05, ***P<0.001 versus corresponding treatment in the control group
tmTNF-α-mediated cytotoxicity via TNFR1 is independent of DD and NSD
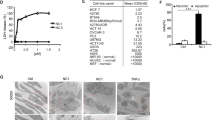
The DD and NSD of TNFR1 are involved in sTNF-α-induced apoptotic signaling.9, 10, 11 To investigate whether tmTNF-α-mediated cytotoxicity is also dependent on the DD or NSD of TNFR1, HEK 293T cells were as target cells transfected to express wild-type TNFR1, ΔDD-TNFR1 or ΔNSD-TNFR1, respectively (Figure 2a). In contrast to the blockage of sTNF-α-induced cytotoxicity in cells expressing ΔDD-TNFR1, tmTNF-α-mediated cytotoxicity was identical in target cells expressing wild type, ΔDD- or ΔNSD-TNFR1 (Figure 2b), indicating that tmTNF-α-mediated cytotoxicity is independent of the DD and the NSD within the cytoplasmic part of TNFR1.
tmTNF-α-mediated cytotoxicity via TNFR1 was independent of DD and NSD. HEK 293T cells were transfected with empty vector, wild-type TNFR1, ΔDD-TNFR1 or ΔNSD-TNFR1 containing plasmids. (a and c) The cell surface expression of TNFR1 and its mutants (a), and TNFR2 (c) detected by flow cytometry. (b) The transfected HEK 293T cells were stimulated for 24 h with 100 ng/ml sTNF-α or tmTNF-α on fixed NIH3T3 cells at an effector/target ratio of 10:1. For neutralization of tmTNF-α, effector cells were treated with anti-TNF-α (Ab) for 30 min prior to the addition to the target cells. The cytotoxicity was detected by MTT assays. (d) HEK 293T cells were co-transfected with control or TNFR2 siRNA and expression vectors for TNFR1 or its mutants as indicated. After 48 h, the cells were stimulated with tmTNF-α on fixed NIH3T3 for 24 h. The cytotoxicity of tmTNF-α was analyzed by MTT assays. (e) The 24 h-cytotoxicity of R32W-tmTNF-α expressed on COS-7 cells towards HEK 293T cells expressing TNFR1 or mutants thereof. (f) The 24 h-cytotoxicity of sTNF-α to HEK 293T cells co-treated with indicated concentrations of S1P. (g) The 24 h-cytotoxicity of tmTNF-α to HEK 293T cells expressing TNFR1 or its mutants in the presence of 10 μM S1P. All the quantitative data represent means±S.D. of at least three independent experiments. *P<0.05, **P<0.01, ***P<0.001 versus corresponding treatment in the control group
As tmTNF-α is the primary ligand of TNFR2, it is possible that tmTNF-α induces cytotoxicity via TNFR2 irrespective of the presence or absence of the DD or NSD of TNFR1. To test this possibility, we first analyzed whether exogenous expression of TNFR1 or its mutants affects TNFR2 expression. Indeed, TNFR2 expression was significantly elevated (Figure 2c). Therefore, we used siRNA to knockdown TNFR2 expression (Supplementary Figure S2a) and found that tmTNFα was still capable to induce cytotoxicity in cells expressing wild type, ΔDD-, or ΔNSD-TNFR1 (Figure 2d).
To confirm these observations with an independent experimental setup, wild-type tmTNF-α and its mutant, R32W-tmTNF-α that selectively binds to TNFR1,19, 20 were ectopically expressed in COS-7 cells and cell surface expression of the ligand was monitored by FACS-analysis (Supplementary Figure S2b). As expected, R32W-tmTNF-α was only cytotoxic to U937 cells that mainly express TNFR1 but not to HL-60 cells that chiefly express TNFR2 (upper panels of Supplementary Figures S2c and d); however, wild-type tmTNF-α was able to kill both the cell lines (lower panels). These observations indicate that tmTNF-α exerts its cytotoxicity via both TNFR1 and TNFR2, whereas R32W-tmTNF-α selectively displays its activity through TNFR1. Consequently, we tested the importance of the DD and NSD of TNFR1 for R32W-tmTNF-α-mediated, TNFR1-dependent cytotoxicity. Again, tmTNFα-induced cytotoxicity was equal in cells expressing wild type, ΔDD-, or ΔNSD-TNFR1 (Figure 2e).
Sphingosine-1-phosphate (S1P) is a downstream molecule of NSD-mediated signaling and known to counterbalance ceramide-induced apoptosis.21, 22, 23 We tested the effect of S1P on sTNFα- and tmTNF-α-mediated cytotoxicity. S1P inhibited efficiently sTNF-α-induced cytotoxicity (Figure 2f), whereas tmTNF-α-induced cytotoxicity through wild type or ΔNSD-TNFR1 was unaffected by S1P (Figure 2g). The results above strongly indicate that both DD and NSD of TNFR1 are required for sTNF-α-, but not for tmTNF-α-mediated cytotoxicity.
tmTNF-α induces DISC formation via TNFR1 on the plasma membrane
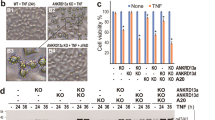
sTNF-α induces DISC formation in the cytoplasm via the DD of TNFR1.8 Here, we examined whether and where tmTNF-α induces the formation of DISC complexes. U937 cells were pre-treated with or without 100 μM MDC for 1 h, followed by stimulation with sTNF-α or R32W-tmTNF-α. As shown in Figure 3, both forms of TNF-α induced the formation of DISC complexes containing TRADD, FADD and caspase 8 as demonstrated by coimmunoprecipitation using an anti-TNFR1 antibody (Figure 3a). Knockdown of FADD expression significantly blocked cytotoxicity of both forms of TNF-α, indicating a requirement of DISC formation for their cytotoxicity (Figure 3b). Then, we checked DISC formation in cytoplasmic or membranous fractions and found that sTNF-α induced DISC formation in the cytoplasm (Figure 3c), whereas tmTNF-α induced DISC formation at the plasma membrane, as FADD and caspase 8 were present in the membrane fraction but not in the cytoplasmic fraction (Figures 3c and 3d). This was further confirmed by confocal microscopy, showing that tmTNF-α caused translocation of most of FADD and caspase 8 (green) from the cytoplasm to the cell surface, whereas FADD and caspase 8 were predominantly found in the cytoplasm of U937 cells treated with sTNF-α (Figure 3e). All these observations indicate tmTNF-α-induced DISC formation at the plasma membrane.
tmTNF-α induced formation of DISC via TNFR1 at the plasma membrane. U937 cells were pre-treated with 100 μM MDC for 1 h and then stimulated with 100 ng/ml sTNF-α or R32W-tmTNF-α on fixed COS-7 cells at an E/T ratio of 10:1 for 30 min. For neutralization of tmTNF-α, effector cells were treated with anti-TNF-α (Ab) for 30 min prior to the addition to the target cells. The total (a), cytoplasmic (c) and membrane (d and f) protein was immunoprecipitated with an anti-TNFR1 antibody and analyzed by immunoblotting with the indicated antibodies. (e) Confocal images of the cellular distribution of FADD or caspase 8 (green) in U937 cells in response to sTNF-α or R32W-tmTNF-α (magnification, × 400). (g) Degradation of Iκ-B was analyzed by western blotting. All immunoprecipitation (IP) or/and western data are representative of three independent experiments. (b and h) The 24 h-cytotoxicity of tmTNF-α (E/T: 10:1) or sTNF-α (100 ng/ml) towards U937 cells transfected with control or FADD siRNA (b) or pre-treated with PDTC (100 μM) for 1 h (h). The data represent means±S.D. of at least three independent experiments. ***P<0.001 versus corresponding treatment in the control group
In response to sTNF-α, TNFR1 also recruits TRAF2, RIP1 and cIAPs through TRADD to form membrane complexes to activate NF-κB.8, 12, 3 Thus, we examined whether this complex is also formed in tmTNF-α signaling. Interestingly, tmTNF-α did not induce recruitment of RIP1 and cIAP1, but a small amount of TRAF2, to TNFR1 (Figure 3f). In accordance with this observation TNFR1 activation by tmTNF-α did not cause IκB degradation (Figure 3g). However, sTNF-α still induced formation of complex I on the membrane and IκB degradation to some extent, although its dominant signaling was pro-apoptotic in U937 cells. NF-κB activation decreases cell sensitivity to apoptosis by promoting transcription of antiapoptotic genes.22 Consequently, sTNF-α-induced NF-κB activation is expected to limit its cytotoxicity. As demonstrated in Figure 3h, the NF-κB inhibitor PDTC significantly increased sTNF-α-mediated but not tmTNF-α-induced cytotoxicity, indicating that tmTNF-α induces apoptotic signaling without NF-κB activation as a limitation.
STAT1 is required for tmTNF-α-induced apoptosis
Our results indicate that the DD is not required for DISC recruitment to TNFR1 in tmTNF-α signaling. However, it remained mysterious how TNFR1 recruits DISC in this case. STAT1 directly interacts with TNFR1-bound TRADD, acting as a TNFR1 signaling component to suppress NF-κB activation in response to sTNF-α.24 As tmTNF-α could not activate NF-κB (Figures 3g and h), we assumed that STAT1 might be involved in tmTNF-α-induced DISC formation. Indeed, STAT1 was found in DISC complex through TNFR1 induced by both types of TNF-α. However, deletion of the DD of TNFR1 significantly inhibited sTNF-α-, but not tmTNF-α-, induced formation of DISC containing STAT1 (Figure 4a). In line with the results from Figure 3d, tmTNF-α-induced STAT1:DISC complex was specific for the membrane fraction and unaffected by ΔDD of TNFR1 (Figure 4b).
STAT1 was necessary for tmTNF-induced DISC formation. (a and b) HEK 293T cells were transfected to express wild-type TNFR1, or ΔDD-TNFR1 and then stimulated with 100 ng/ml sTNF-α or R32W-tmTNF-α for 30 min. For neutralization, R32W-tmTNF-α-expressing cells were treated with anti-TNF-α (Ab) for 30 min prior to the addition to the target cells. The total (a) and membrane (b) protein was immunoprecipitated with an anti-TNFR1 antibody and analyzed by immunoblotting. R1: TNFR1; ΔDD: ΔDD-TNFR1. U3A (c–e) or HT1080 (f) cells were transfected by STAT1-containing plasmid or siRNA against STAT1, respectively, followed by stimulation with R32W-tmTNF-α for 30 min. Immunoprecipitation (IP)/western blotting was performed in total (c and f), membrane (d) and cytoplasmic (e) protein with an antibody to TNFR1. Ab, anti-TNF-α antibody; C, Control; T, tmTNF-α. (g) U3A cells were transfected with His-tagged empty vector, His-TNFR1 or His-ΔDD-TNFR1 containing plasmids and stimulated with R32W-tmTNF-α for 30 min. DISC formation was analyzed by IP/western blotting using an anti-His antibody. All the IP/western data are representative of three independent experiments. (h) The 24 h-cytotoxicity of R32W-tmTNF-α to U3A cells pre-treated for 30 min with caspase 8 inhibitor Z-IETD-FMK (4 μM). The data represent means±S.D. of three independent experiments. ***P<0.001 versus corresponding treatment in the control group
To further elucidate the role of STAT1 in tmTNF-α-induced DISC formation and cytotoxicity, STAT1-deficient U3A-cell line and its parental cell line HT1080 were reconstituted by exogenous expression of STAT1 (Figures 4c–e) or transfected with siRNA to knock down STAT1 expression (Figure 4f), respectively. tmTNF-α-induced residual DISC formation in STAT1-deficient U3A cells could be boosted by exogenous expression of STAT1 (Figure 4c) and tmTNF-α-induced STAT1-dependent and -independent DISC were only detectable in the membrane fraction (Figure 4d), but not in the cytoplasmic fraction (Figure 4e). Conversely, evident DISC complexes were formed in parental HT1080 cells in response to tmTNF-α. However, knockdown of STAT1 expression markedly decreased the tmTNF-α-induced DISC (Figure 4f).
These data indicate that STAT1 is indispensable to tmTNF-α-mediated DISC formation that is independent of the DD of TNFR1. As tmTNF-α-induced residual DISC formation was observed in STAT1-deficient U3A cells, we hypothesized that the DD of TNFR1, although not necessary (Figure 4b), may still enable to form STAT1-independent residual DISC. To test this idea, His-tagged TNFR1 or ΔDD-TNFR1 was expressed in STAT1-deficient U3A cells, which allows specific analysis of DISC formation at the exogenous receptor proteins. As expected, TNFR1-ΔDD completely failed to recruit tmTNF-α-induced DISC molecules (Figure 4g) and caspase 8 inhibitor Z-IETD-FMK abolished tmTNF-α-mediated cytotoxicity in STAT1-deficient U3A cells (Figure 4h), indicating a requirement of TNFR1 DD to recruit DISC for tmTNF-α-induced, STAT1-independent cytotoxicity.
To further explore the role of STAT1 in tmTNF-α-mediated cytotoxicity, the STAT1-deficient U3A cells and parental HT1080 cells were used as target cells. The cytotoxicity of tmTNF-α was markedly blocked in STAT1-deficient U3A cells or in STAT1 knockdown HT1080 cells compared with that in parental HT1080 cells, but restored by reconstitution of U3A cells with STAT1 (Figure 5a). To analyze the STAT1 function in tmTNF-α-mediated cytotoxicity in vivo, U3A and HT1080 cells were injected into nude mice to induce tumors. Five days after tumor inoculation, expression vector for tmTNF-α was injected into the tumor site. As demonstrated by Figure 5b, tmTNF-α was overexpressed in the tumors. However, only the growth of HT1080 tumors, but not of STAT1-deficient U3A tumors, was significantly suppressed by tmTNF-α (Figure 5c). Furthermore, overexpression of tmTNF-α induced apoptosis (Figure 5d) and caspase 3 activation (Figure 5e) in HT1080 tumors. In contrast, these activities of tmTNF-α were significantly suppressed in U3A tumors. These data imply that STAT1 is required for tmTNF-α-induced apoptosis via TNFR1 in a DD-independent manner.
STAT1 was required for tmTNF-induced apoptosis. (a) The 24 h-cytotoxicity of R32W-tmTNF-α to U3A, STAT1-expressing U3A, STAT1 knockdown HT1080 or parental HT1080 cells. The data represent means±S.D of at least three independent experiments. ***P<0.001 versus corresponding treatment in the control group. (b–e) 5 × 106 HT1080 or U3A cells were subcutaneously injected into the armpit of forelimb of nude mice. At day 5 after tumor cell inoculation, 100 μg of expression vector for R32W-tmTNF-α or empty vector was injected into the tumor site. (b) The expression of tmTNF-α was analyzed by immunohistochemistry (magnification, × 200). (c) Tumor growth curves (n=8 each group). *P<0.05, ***P<0.001 versus HT1080+vector. (d) Apoptosis in situ detected by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay (magnification, × 100). (e) Caspase 3 activation analyzed by western blotting that is representative of three independent experiments
STAT1 serves as an adaptor to recruit DISC to TNFR1 in tmTNF-α-mediated apoptotic signaling
STAT1 functions as an adaptor in activation of multiple signaling pathways or as a transcriptional activator to induce gene expression.25 Our result that tmTNF-α-induced DISC could be recruited to ΔDD-TNFR1 suggests that STAT1 may act as an adaptor to mediate the interaction between TNFR1 and TRADD/FADD/caspase 8 in this case. To test this hypothesis, we deleted different fragments in the region between TRID and NSD of the cytoplasmic part of TNFR1 (Figure 6a) and transfected these mutants into HEK 293T cells, respectively. Coimmunoprecipitation experiments revealed that STAT1 could be recruited to wild type and Δ338-455-TNFR1, but not to Δ319-455- and Δ266-455-TNFR1 (Figure 6b). The pull-down assay showed again that only Δ338-455-TNFR1 could directly interact with STAT1 as wild-type TNFR1 did (Figure 6c), indicating the region spanning amino acids 319–337 of TNFR1 for STAT1 binding. Thus we named the region as STAT1 domain (SD). Ectopic expression of Δ338-455-TNFR1 in HEK 293T cells increased STAT1 recruitment only in response to tmTNF-α, but not to sTNF-α (Figure 6d). It is very likely that tmTNF-α stimulates STAT1 binding to SD of TNFR1 and further recruitment of DISC proteins to exert its cytotoxicity.
STAT1 as an adaptor recruited DISC to TNFR1 in tmTNF-α-mediated apoptotic signaling. (a) The scheme of three TNFR1 truncations used in the following experiments. (b) HEK 293T cells were transfected to express wild-type TNFR1, Δ338-455-TNFR1, Δ319-455-TNFR1 or Δ266-455-TNFR1, and then treated with R32W-tmTNF-α for 30 min. Total protein was immunoprecipitated with an anti-TNFR1 antibody and analyzed by immunoblotting. (c) Bacterially expressed different GST fusion proteins and GST alone were incubated with His-tagged STAT1 bound to Ni-NTA resin, respectively, and then western blot was performed using antibodies against TNFR1 or STAT1. (d) HEK 293T cells transfected to express Δ338-455-TNFR1 were treated with 100 ng/ml sTNF-α or R32W-tmTNF-α for 30 min. STAT1 was detected by immunoprecipitation (IP)/western blotting. (e) Western blot of STAT1 phosphorylation in U937 cells treated with 100 ng/ml sTNF-α or R32W-tmTNF-α for 30 min. (f) U3A cells expressing STAT1, S727A-, Y701I- or DM-STAT1 were stimulated with R32W-tmTNF-α for 24 h. The cytotoxicity was detected by an MTT assay. Data represent means±S.D. of at least three independent experiments. ***P<0.001 versus corresponding treatment in the control group. (g) U3A cells expressing STAT1, S727A- or Y701I-STAT1 were treated for 30 min with R32W-tmTNF-α. STAT1 and DISC were visualized by IP/western blotting. For neutralization, R32W-tmTNF-α- expressing cells were treated with anti-TNF-α (Ab) for 30 min prior to addition to the target cells (d–f). All IP/western data are representative of three independent experiments
STAT1 can be phosphorylated at both tyrosine and serine residues (Y701, S727),26, 27 which is associated with its function. Figure 6e showed that tmTNF-α induced phosphorylation of S727 and Y701 of STAT1 in U937 cells, while sTNF-α caused phosphorylation of Y701 only. tmTNF-α-mediated cytotoxicity was significantly increased by reconstitution of U3A cells with wild-type STAT1 or Y701I-STAT1, but not with S727A-STAT1 or DM (Y701I and S727A double mutations)-STAT1 (Figure 6f). Consistently, immunoprecipitation of TNFR1 confirmed that S727A-STAT1 mutant failed to recruit DISC proteins in tmTNF-α-treated cells (Figure 6g). These data imply that serine phosphorylation at 727 of STAT1 is required for tmTNF-α-induced DISC formation and its cytotoxicity.
To investigate whether the transcriptional activity of STAT1 is involved in tmTNF-α signaling, the nuclear translocation of STAT1 in U937 cells was examined by double immunofluorescent staining. tmTNF-α induced colocalization (yellow) of STAT1 (green) and TNFR1 (red) at the plasma membrane, whereas sTNF-α caused colocalization of STAT1 and TNFR1 in the cytoplasm. Both forms of TNF-α did not induce evident localization of STAT1 in the nucleus of U937 cells (Supplementary Figure S3a). In addition, immunoblotting assay also revealed a marked increase of STAT1 in the membrane fraction upon tmTNF-α stimulation and in the cytoplasmic fraction upon sTNF-α stimulation. However, in the nuclear fraction, both forms of TNF-α had no effect (Supplementary Figure S3b). These data suggest that in the signaling pathways of both forms of TNF-α, STAT1 does not function as a transcription factor in the nucleus, but rather localize to the cell membrane functioning as an adaptor for DISC formation to transduce tmTNF-α apoptotic signaling, or to the cytoplasm to transduce sTNF-α signaling.
Discussion
In this study, we found that tmTNF-α and sTNF-α employ different signal cascades to induce apoptosis via TNFR1. In contrast to sTNF-α signaling that activates both the NF-κB pro-survival pathway and apoptotic pathway, tmTNF-α induces the binding of STAT1 to TNFR1, activating apoptotic pathway but not NF-κB pathway.
A large body of evidence has revealed the role of internalization of TNFR1 and CD95 in pro-apoptotic signaling.8, 18, 28, 29, 30 The internalization of these death receptors is important for the compartmentalization of multiple signaling molecules and decides cell death or survival. Internalization of CD95 is required for DISC recruitment and apoptosis induced by either soluble or transmembrane ligand. Here, we show that the two forms of TNF-α differ in the requirement of TNFR1 internalization for the induction of apoptosis. Namely, the internalization of TNFR1 is only necessary for sTNF-α-, but not for tmTNF-α-mediated apoptosis, because tmTNF-α induces DISC formation at the plasma membrane without TNFR1 internalization, and blocking the internalization of TNFR1 by MDC or transfection to express Y236A-TNFR1 has no effect on tmTNF-α-mediated cytotoxicity and apoptosis. It has been reported that targeting TNFR1 endocytosis by adenovirus E3-14.7 K protects infected cells from apoptosis induced by sTNF-α, CD95 and TRAIL.28, 31, 32 Thus, tmTNF-α-mediated apoptosis may be beneficial for anti-virus strategies, as a selective blockage of tmTNF-α processing into sTNF-α may prevent immune evasion by the pathogen.
The DD is necessary for TNFR1 to mediate the functions of sTNF-α. However, our results uncovered that deletion of DD of TNFR1 does not affect tmTNF-α-induced DISC formation and its cytotoxic activity. In addition, both neutral SMase (sphingomyelinase) and acidic SMase are involved in sTNF-α-mediated signal transduction.33, 34, 35 Consistently, our results showed that addition of S1P that prevents ceramide-induced apoptosis23 blocks sTNF-α-induced cell death. In contrast, tmTNF-α-mediated cytotoxicity is unaffected either by expression of ΔNSD-TNFR1 or by addition of S1P, suggesting that nSMase and ceramide are not involved in tmTNF-α-mediated cytotoxicity. These data indicate that tmTNF-α might require different domain of TNFR1 for its functions.
The DD of TNFR1 binds TRADD in sTNF-α signaling, but is not required for tmTNF-α signaling, indicating that other molecule than TRADD might bind to TNFR1 in response to tmTNF-α. Our results uncovered that STAT1 is essential for tmTNF-α apoptotic signaling and cytotoxicity, as STAT1-deficiency in U3A cells or silence of STAT1 expression in HT1080 cells significantly reduces the DISC formation and cytotoxicity induced by tmTNF-α. The defective tmTNF-α signaling can be restored by reconstitution of U3A cells with STAT1. Consistently, overexpression of tmTNF-α markedly suppresses the growth of STAT1-containing tumors, but not of STAT1-deficient tumors.
Although STAT1 is involved in the signaling of two forms of TNF-α, we found that STAT1 binds to TNFR1 at the plasma membrane in tmTNF-α signaling, whereas colocalization of STAT1 and TNFR1 in the cytoplasm suggests binding of STAT1 to TNFR1–TRADD after internalization in sTNF-α signaling. STAT1 is known to be able to bind both TNFR1 and TRADD, but not FADD.24 We identified that STAT1 directly interacts with SD, a region from 319 to 337, of TNFR1 upon tmTNF-α stimulation. It is likely that STAT1 binds to TNFR1 and further recruits TRADD in a DD-independent manner to form DISC in tmTNF-α signaling (Figure 7).
tmTNF-α-induced STAT1-dependent apoptotic signaling via TNFR1. sTNF-α induces binding of TRADD to the DD of TNFR1. TNFR1-bound TRADD further recruits TRAF2 and cIAP1/2 at the plasma membrane to activate NF-κB pathway. However, after TNFR1 internalization, STAT1 binds to TRADD and is phosphorylated at tyrosine 701, which excludes the binding of TRAF2 and cIAP1 to TRADD and attenuates NF-κB activation. Meanwhile, TRADD recruits FADD and caspase 8 to form DISC in the cytoplasm to mediate apoptosis. In contrast, tmTNF-α induces binding of STAT1 to the SD of TNFR1 and STAT1 serine phosphorylation at 727. The S727 phosphorylated STAT1 further recruits TRADD to form DISC at the plasma membrane to trigger apoptosis, but not NF-κB activation
More importantly, our data indicate that STAT1 transduces different signaling pathways via TNFR1 upon stimulation with tmTNF-α and sTNF-α. Our data reveal that STAT1 is required for tmTNF-α-induced apoptotic signaling but not for NF-κB activation, as RIP1 and cIAP1, two members of complex activating NF-κB, were undetectable in the TNFR1 signaling complex. In sTNF-α signaling, TNFR1-bound TRADD recruits RIP1-containing complex I proteins on the plasma membrane to induce NF-κB activation, STAT1 competitively suppresses binding of TRADD to RIP and TRAF2, inhibiting NF-κB activation without disrupting TRADD-FADD interaction.24 However, in response to tmTNF-α, TRADD is recruited to TNFR1-bound STAT1 in a DD-independent manner, which may prevent TRADD binding to RIP1 and subsequent NF-κB activation. Therefore, inhibition of NF-κB by PDTC only enhances sTNF-α-, but not tmTNF-α-mediated cytotoxicity. This difference may explain why tmTNF-α is able to kill tumor cells that are resistant to sTNF-α-mediated cytotoxicity.
sTNF-α-induced phosphorylation of Y701-STAT1 is necessary for its recruitment by TRADD and reduction of NF-κB activation.24 However, ectopic expression of S727A- but not Y701I-STAT1 disrupts tmTNF-α-induced DISC formation and cytotoxicity in U3A cells, indicating the serine phosphorylation of STAT1 is essential for tmTNF-α signaling, although both S727 and Y701 are phosphorylated by tmTNF-α stimulation. Interestingly, the loss of S727 phosphorylation prevents recruitment of TRADD, but not of the STAT1 mutant to TNFR1, suggesting that S727 phosphorylation is required for the interaction between STAT1 and TRADD. Although phosphorylation of STAT1 at both Y701 and S727 contributes to its transcriptional activity,26 in consistent to the other reports, we could not detect any nuclear translocation of STAT1 in response to either form of TNF-α, suggesting STAT1 acts as an adaptor in TNF-α signaling.
In summary, we revealed that tmTNF-α induces STAT1-dependent DISC formation and apoptosis through TNFR1. Although both forms of TNF-α are able to induce apoptosis via TNFR1, sTNF-α signaling activates NF-κB that reduces the sensitivity of tumor cells to apoptosis. Therefore, tmTNF-α may be more efficient for the treatment of tumors in comparison with sTNF-α.
Materials and methods
Cell culture
U937, U3A, HT1080 cells, HEK 293T, COS-7 and NIH3T3 stably transfected with TNF-α 7 were cultured in RPMI-1640 or DMEM medium (GIBCO, Grand Island, NY, USA) supplemented with 10% pyrogen-free FCS and incubated at 37 °C in 5% CO2.
Mutation, plasmid construction, siRNAs and transfection
The cDNA of full length of human TNFR1 was generated by RT-PCR from THP1 cells and cloned into pTriEx4.0His-N1 vector at XhoI and BamHI site. Different mutants of TNFR1 cDNA encoding Y236A-TNFR1, a point mutation by replacing tyrosine with alanine at 236, and thus losing its ability to internalize,8 ΔNSD-TNFR1, a deletion of NSD domain from 338 to 348, ΔDD-TNFR1, lacking the DD from 356 to 455, Δ338-455-TNFR1, Δ319-455-TNFR1 and Δ266-455-TNFR1, lacking C-terminal fragment from 338, 319 or 266 to 455, respectively, were generated by recombinant PCR and inserted into pTriEx4.0His-N1 vector at XhoI and BamHI site. Furthermore, a mutant of tmTNF-α cDNA encoding R32W-tmTNF-α (selectively binds to TNFR1)19, 20 was generated by recombinant PCR and inserted into pIRES2-EGFP vector at XhoI and BamHI site. All the constructs were verified by DNA sequencing (TSINGKE Biological Technology, Beijing, China) and the sequences of primers used are listed in Supplementary Table S1. The plasmids pcDNA3.1 containing wild-type STAT1, Y701I-STAT1, S727A-STAT1 or DM (Y701I and S727A double mutations)-STAT1 were kindly gifted by Dr. Guanxin Shen (Department of Immunology, Tongji Medical College, Huazhong University of Science and Technology).
siRNA against FADD, STAT1 and TNFR2, and a scrambled control siRNA were designed and synthesized by RiboBio (Guangzhou, China). The sequences used for siRNA targeting were FADD 5′-GGAACTCAGACGCATCTAC-3′, STAT1 5′-CCTAAAGGAACTGGATATA-3′ and TNFR2 5′-GGCTCAGAGAATACTATGA-3′.
All the plasmids and siRNAs were transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The cells were harvested at 48 h after transfection.
TNF-α bioassay
Target cells were seeded in a 96-well microtiter plate and incubated overnight. 100 ng/ml sTNF-α (300-01 A; PeproTech, Rocky, Hill, NJ, USA) or 4% paraformaldehyde fixed tmTNF-α or its mutant expressing cells were added to target cells at an effector and target ratio of 10:1 and incubated for 24 h. For the neutralization of TNF-α, the fixed effector cells were pre-treated with a specific antibody (#7321; Cell Signaling Technology, Danvers, MA, USA) for 30 min before addition to the target cells. Cell viability was then measured by staining for 4 h with glucose-PBS containing 0.45 mg/ml MTT (Sigma Chemical Co., St. Louis, MO, USA), followed by cell lysis with 0.1 ml DMSO. The OD value at 570 nm was measured on a microplate reader (Tecan, Melbourne, Victoria, Australia). The cytotoxicity of TNF-α was calculated by the following formula: cytotoxicity (%)=(1–ODsample/ODcontrol) × 100%.
Flow cytometry
For the detection of expression of TNFR2, TNFR1 and its mutants, or tmTNF-α and its mutant, cells were incubated for 1 h at 4 °C with primary antibodies against TNFR1, TNFR2 or TNF-α listed in Supplementary Table S2, followed by the secondary antibodies conjugated with PE (Jackson Biotech, West Chester, PA, USA). The expression of molecules was analyzed on a FACS Calibur 440 E flow cytometer (Becton Dickinson,San Jose, CA, USA).
For the detection of TNF-α-mediated apoptosis, U937 cells after stimulation were stained with FITC-conjugated annexin V and propidium iodide (PI) for 10 min according to the manufacturer's instructions (KeyGEN, Nanjing, China). The percentage of apoptotic (annexin V-positive) cells was determined by flow cytometry. Apoptosis (%)=percentage of Annexin V-positive cells+percentage of both Annexin V and PI positive cells.
Confocal microscopy
After stimulation with two forms of TNF-α, cells were washed with PBS, permeabilized for 10 min in 0.1% Triton X-100/PBS and blocked with 5% BSA in PBS for 1 h at room temperature (RT). The cells were then incubated overnight at 4 °C with primary antibodies, including anti-TNFR1, anti-EEA, anti-FADD, anti-caspase 8 or anti-STAT1, listed in Supplementary Table S2. After washing with PBS, the appropriate secondary antibodies conjugated with FITC, PE or DyLight 488 (Thermo Scientific, Waltham, MA, USA), were applied. The nucleus was counterstained with DAPI for 5 min. The localization of interested molecules in cells was visualized under a confocal microscope FU5000 (Olympus, Tokyo, Japan).
Isolation of total, cytoplasmic, nuclear and membrane protein
Cytoplasmic and membrane protein was isolated with a commercial kit, ProteoJET Membrane Protein Extraction Kit, according to the manufacturer's instructions (Fermentas, Burlington, Ontario, Canada). Briefly, after stimulation and washes, cells were resuspended in 1.5 ml of ice-cold Cell Permeabilization Buffer and incubated for 10 min at 4 °C with continuously rocking. After centrifugation at 16 000 × g for 15 min at 4 °C, cytoplasmic protein in the supernatant was collected. The cell debris pellet was mixed with 1 ml of ice-cold Membrane Protein Extraction Buffer and incubated for 30 min at 4 °C in the thermomixer with shaking at 1400 r.p.m. The membrane protein in the supernatant was obtained by centrifugation at 16 000 × g for 15 min at 4 °C.
For total protein extraction, cells were lysed in pre-cold buffer A (10 mM HEPES, pH 7.8, 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 5 μg/ml aprotinin and 5 μg/ml leupeptin) and incubated on ice for 20 min. After centrifugation at 12 000 × g for 20 min at 4 ˚C, the total protein was collected.
For nuclear protein extraction, a commercial kit was used (Boster, Wuhan, China). Briefly, cells were resuspended in ice-cold Cytoplasmic Extraction Reagent and incubated for 30 min at 4 °C with continuously rocking. After centrifugation at 16 000 × g for 10 min at 4 °C, the cell debris pellet was mixed with ice-cold Nuclear Extraction Reagent and incubated for 30 min at 4 °C. The nuclear protein was collected after centrifugation at 16 000 × g for 10 min at 4 °C.
Western blot analysis
Fifty micrograms of total, membranous, cytoplasmic or nuclear protein or immunoprecipitated protein were loaded and separated on 12% SDS-polyacrylamide gels by electrophoresis. The separated protein was then transferred to PVDF membranes (Millipore, Billerica, MA, USA) using a semi-dry transfer system (BioRad Laboratories, Hercules, CA, USA). These membranes were blocked for 2 h at RT with 5% non-fat dry milk in PBS-Tween 20 (0.05%), and then probed overnight at 4 °C with primary antibodies, including anti-TNFR1, anti-STAT1, anti-β-actin, anti-laminB1, anti-caveolin, anti-His, anti-GAPDH, anti-cIAP1, anti-IκB, αnti-TRAF2, anti-FADD, anti-TRADD, anti-RIP-1, anti-p-Y701-STAT1, anti-p-S727-STAT1, anti-caspase 8 or anti-caspase 3, listed in Supplementary Table S2. After washes, the membranes were incubated for 1 h with corresponding horseradish peroxidase-conjugated secondary antibodies, and then visualized using SuperSignal West Pico Chemiluminescence Substrate (Thermo Scientific, Waltham, MA, USA).
Immunoprecipitation
The total, membranous or cytoplasmic protein was extracted after stimulation, and then subjected to immunoprecipitation using a monoclonal antibody specific to TNFR1 or to His (Supplementary Table S2) at 4 °C for 2 h. Subsequently, the immune complexes were incubated with protein A sepharose (sc-2001; Santa Cruz Biotechnology, Dallas, TX, USA) in rotation at 4 °C overnight. The immunoprecipitated molecules were analyzed by immunoblotting.
Pull-down assay
His-tagged STAT1 was obtained using pET28a-His-STAT1 for transformation and expression in E. coli strain Rosetta (DE3). His-STAT1 was purified and then immobilized on Ni-NTA-resin. His-STAT1 on NI-NTA-resin was incubated with bacterially expressed GST-TNFR1, GST-Δ319-455-TNFR1, GST-Δ338-455-TNFR1 or GST proteins overnight at 4 °C with continuous rocking. After washing with PBS, the resin-bound proteins were eluted and then analyzed by western blot.
Animal model
The animal study was approved by the Animal Care and Use Committee of Huazhong University of Science and Technology. Five-week-old male BALB/c nude mice (Beijing HFK Bioscience Co., ITD, Beijing, China) were housed in specific pathogen-free environment in 12-h light–dark cycles and allowed ad libitum access to food and water. 5 × 106 HT1080 or U3A cells were subcutaneously injected into forelimb armpit of nude mice. One hundred micrograms of plasmid containing R32W-tmTNF-α or empty vector were injected into the tumor site at day 5 after tumor cell inoculation. The volume of the tumors was measured every 3 days and it was calculated by assuming the following equation (ref. 7): length × width2 × π/6.
Immunohistochemistry and terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling staining
Paraffin sections of tumor tissues were deparaffinized in xylene and rehydrated in graded ethanol, and then incubated for 60 min at RT with a house made antibody specific to tmTNF-α without crossreaction to sTNF-α.36 After a 30-min incubation with biotin-conjugated secondary antibody against IgG (Beijing Zhongshan Biotechnology, Beijing, China), the sections were then incubated with peroxidase-labeled streptavidin for 20 min. After washing, color was developed by incubation with its substrate diaminobenzidine for 5 min and the expression of tmTNF-α was observed under a microscope. To evaluate apoptosis in tumors, the terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay was performed with a commercial kit (Boster, Wuhan, China) according to the manufacturer's protocol. Nuclei were counterstained with DAPI.
Statistical analysis
Statistical analysis was performed with GraphPad Prism V6 software using one- or two-way (where appropriate) ANOVA followed by Bonferroni post-test. Data are represented as mean±S.D. Statistical significance in the figures is indicated as follows: *P<0.05, **P<0.01, ***P<0.001.
References
Kriegler M, Perez C, DeFay K, Albert I, Lu SD . A novel form of TNF/cachectin is a cell surface cytotoxic transmembrane protein: ramifications for the complex physiology of TNF. Cell 1988; 53: 45–53.
Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 1997; 385: 729–733.
Grell M, Douni E, Wajant H, Lohden M, Clauss M, Maxeiner B et al. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 1995; 83: 793–802.
Ardestani S, Li B, Deskins DL, Wu H, Massion PP, Young PP . Membrane versus soluble isoforms of TNF-alpha exert opposing effects on tumor growth and survival of tumor-associated myeloid cells. Cancer Res 2013; 73: 3938–3950.
Zhang H, Yan D, Shi X, Liang H, Pang Y, Qin N et al. Transmembrane TNF-alpha mediates 'forward' and 'reverse' signaling, inducing cell death or survival via the NF-kappaB pathway in Raji Burkitt lymphoma cells. J Leuk Biol 2008; 84: 789–797.
Shi W, Li, Gong FL, Xiong P, Xu Y . Comparison of the cytocidal effect induced by transmembrane and secreted TNF-alpha. Chin J Microbiol Immunol 1998; 18: 499–504.
Li Q, Li L, Li Z, Gong F, Feng W, Jiang X et al. Antitumor effects of the fibroblasts transfected TNF-alpha gene and its mutants. J Huazhong Univ Sci Technol Med Sci 2002; 22: 92–95.
Schneider-Brachert W, Tchikov V, Neumeyer J, Jakob M, Winoto-Morbach S, Held-Feindt J et al. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity 2004; 21: 415–428.
Adam D, Wiegmann K, Adam-Klages S, Ruff A, Kronke M . A novel cytoplasmic domain of the p55 tumor necrosis factor receptor initiates the neutral sphingomyelinase pathway. J Biol Chem 1996; 271: 14617–14622.
Tartaglia LA, Ayres TM, Wong GH, Goeddel DV . A novel domain within the 55 kd TNF receptor signals cell death. Cell 1993; 74: 845–853.
Brakebusch C, Nophar Y, Kemper O, Engelmann H, Wallach D . Cytoplasmic truncation of the p55 tumour necrosis factor (TNF) receptor abolishes signalling, but not induced shedding of the receptor. EMBO J 1992; 11: 943–950.
Micheau O, Tschopp J . Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 2003; 114: 181–190.
Van Herreweghe F, Festjens N, Declercq W, Vandenabeele P . Tumor necrosis factor-mediated cell death: to break or to burst, that's the question. Cell Mol Life Sci 2010; 67: 1567–1579.
Neumeyer J, Hallas C, Merkel O, Winoto-Morbach S, Jakob M, Thon L et al. TNF-receptor I defective in internalization allows for cell death through activation of neutral sphingomyelinase. Exp Cell Res 2006; 312: 2142–2153.
Wajant H, Pfizenmaier K, Scheurich P . Tumor necrosis factor signaling. Cell Death Differ 2003; 10: 45–65.
Chen G, Goeddel DV . TNF-R1 signaling: a beautiful pathway. Science 2002; 296: 1634–1635.
Schutze S, Tchikov V, Schneider-Brachert W . Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat Rev Mol Cell Biol 2008; 9: 655–662.
Schutze S, Machleidt T, Adam D, Schwandner R, Wiegmann K, Kruse ML et al. Inhibition of receptor internalization by monodansylcadaverine selectively blocks p55 tumor necrosis factor receptor death domain signaling. J Biol Chem 1999; 274: 10203–10212.
Van Ostade X, Vandenabeele P, Everaerdt B, Loetscher H, Gentz R, Brockhaus M et al. Human TNF mutants with selective activity on the p55 receptor. Nature 1993; 361: 266–269.
Loetscher H, Stueber D, Banner D, Mackay F, Lesslauer W . Human tumor necrosis factor alpha (TNF alpha) mutants with exclusive specificity for the 55-kDa or 75-kDa TNF receptors. J Biol Chem 1993; 268: 26350–26357.
Spiegel S, Cuvillier O, Edsall LC, Kohama T, Menzeleev R, Olah Z et al. Sphingosine-1-phosphate in cell growth and cell death. Ann NY Acad Sci 1998; 845: 11–18.
Olivera A, Kohama T, Edsall L, Nava V, Cuvillier O, Poulton S et al. Sphingosine kinase expression increases intracellular sphingosine-1-phosphate and promotes cell growth and survival. J Cell Biol 1999; 147: 545–558.
Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind S et al. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 1996; 381: 800–803.
Wang Y, Wu TR, Cai S, Welte T, Chin YE . Stat1 as a component of tumor necrosis factor alpha receptor 1-TRADD signaling complex to inhibit NF-kappaB activation. Mol Cell Biol 2000; 20: 4505–4512.
Kim HS, Lee MS . STAT1 as a key modulator of cell death. Cell Signal 2007; 19: 454–465.
Shuai K, Stark GR, Kerr IM, Darnell JE Jr . A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 1993; 261: 1744–1746.
Decker T, Kovarik P . Serine phosphorylation of STATs. Oncogene 2000; 19: 2628–2637.
Schneider-Brachert W, Tchikov V, Merkel O, Jakob M, Hallas C, Kruse ML et al. Inhibition of TNF receptor 1 internalization by adenovirus 14.7 K as a novel immune escape mechanism. J Clin Invest 2006; 116: 2901–2913.
Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J 1995; 14: 5579–5588.
Lee KH, Feig C, Tchikov V, Schickel R, Hallas C, Schutze S et al. The role of receptor internalization in CD95 signaling. EMBO J 2006; 25: 1009–1023.
Tollefson AE, Toth K, Doronin K, Kuppuswamy M, Doronina OA, Lichtenstein DL et al. Inhibition of TRAIL-induced apoptosis and forced internalization of TRAIL receptor 1 by adenovirus proteins. J Virol 2001; 75: 8875–8887.
Chen P, Tian J, Kovesdi I, Bruder JT . Interaction of the adenovirus 14.7-kDa protein with FLICE inhibits Fas ligand-induced apoptosis. J Biol Chem 1998; 273: 5815–5820.
Dressler KA, Mathias S, Kolesnick RN . Tumor necrosis factor-alpha activates the sphingomyelin signal transduction pathway in a cell-free system. Science 1992; 255: 1715–1718.
Chatterjee S . Neutral sphingomyelinase action stimulates signal transduction of tumor necrosis factor-alpha in the synthesis of cholesteryl esters in human fibroblasts. J Biol Chem 1994; 269: 879–882.
Wiegmann K, Schutze S, Machleidt T, Witte D, Kronke M . Functional dichotomy of neutral and acidic sphingomyelinases in tumor necrosis factor signaling. Cell 1994; 78: 1005–1015.
Yu M, Zhou X, Niu L, Lin G, Huang J, Zhou W et al. Targeting transmembrane TNF-alpha suppresses breast cancer growth. Cancer Res 2013; 73: 4061–4074.
Acknowledgements
We thank Dr. Fred Schape for careful review and correction of the manuscript. This study was supported by grants from the National Natural Science Foundation of China (30872294, 81272520 and 30872376).
Author contributions
YJ, MY, XH and LH performed experiments and analyzed the results. KY, HB and ZZ conducted some experiments. BY and XY, the senior authors, contributed to the analysis of the results and provided important insight to this study. ZL and JW designed experiments and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Edited by A Ashkenazi
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Jiang, Y., Yu, M., Hu, X. et al. STAT1 mediates transmembrane TNF-alpha-induced formation of death-inducing signaling complex and apoptotic signaling via TNFR1. Cell Death Differ 24, 660–671 (2017). https://doi.org/10.1038/cdd.2016.162
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.162
This article is cited by
-
Low-level laser selectively inhibiting colorectal cancer cell metabolic activity and inducing apoptosis for delaying the development of intestinal cancer
Photochemical & Photobiological Sciences (2023)
-
Supplementation of polyunsaturated fatty acids (PUFAs) and aerobic exercise improve functioning, morphology, and redox balance in prostate obese rats
Scientific Reports (2021)
-
TNF-α synergises with IFN-γ to induce caspase-8-JAK1/2-STAT1-dependent death of intestinal epithelial cells
Cell Death & Disease (2021)
-
Deciphering insights of novel recombinant tmTNFα in cell growth inhibition
Molecular Biology Reports (2020)
-
The mitochondrial negative regulator MCJ modulates the interplay between microbiota and the host during ulcerative colitis
Scientific Reports (2020)