Abstract
Caspase-6 (CASP6) has an important role in axonal degeneration during neuronal apoptosis and in the neurodegenerative diseases Alzheimer and Huntington disease. Decreasing CASP6 activity may help to restore neuronal function in these and other diseases such as stroke and ischemia, where increased CASP6 activity has been implicated. The key to finding approaches to decrease CASP6 activity is a deeper understanding of the mechanisms regulating CASP6 activation. We show that CASP6 is posttranslationally palmitoylated by the palmitoyl acyltransferase HIP14 and that the palmitoylation of CASP6 inhibits its activation. Palmitoylation of CASP6 is decreased both in Hip14−/− mice, where HIP14 is absent, and in YAC128 mice, a model of Huntington disease, where HIP14 is dysfunctional and where CASP6 activity is increased. Molecular modeling suggests that palmitoylation of CASP6 may inhibit its activation via steric blockage of the substrate-binding groove and inhibition of CASP6 dimerization, both essential for CASP6 function. Our studies identify palmitoylation as a novel CASP6 modification and as a key regulator of CASP6 activity.
Similar content being viewed by others
Main
Caspase-6 (CASP6) belongs to a family of cysteine proteases that have classically been described as important players in orchestrating the complex process of apoptosis. However, it has recently become apparent that they also have non-apoptotic functions, including regulation of synaptic function and plasticity1, 2 and developmental neurite pruning.3 Recently, CASP6 has been shown to have a critical role in neurological disorders, including Alzheimer and Huntington disease (AD and HD, respectively),4 as well as in axonal degeneration during stroke,5 global brain ischemia,6 and inflammatory pain.7
CASP6 activation is a very early event in AD, occurring prior to clinical diagnosis, and is an important mediator of axonal degeneration.8, 9, 10 Active CASP6 is abundant in neurofibrillary tangles, neuritic plaques, and neuropils in the AD brains.9 CASP6 cleaves the intracellular domain of amyloid precursor protein (APP)11 and the microtubule-associated protein tau,8 which may trigger microtubule destabilization and lead to tangle formation. Interestingly, mutating the CASP6 cleavage site (VEVD) at D664 in APP prevents synaptic loss and other AD phenotypes, including dentate gyral atrophy, memory loss, and learning deficits in AD mice.12, 13, 14, 15
HD is an adult-onset, autosomal-dominant, fatal neurodegenerative disease characterized by motor, cognitive, and psychiatric disturbances and is caused by a CAG repeat expansion in the HTT gene.16, 17 In HD, activation of CASP6 is an early pathogenic event that can be observed prior to onset of motor symptoms both in human HD brain tissue and in the YAC128 HD mouse model, a transgenic model expressing full-length human HTT with 128 CAG repeats that accurately recapitulates features of HD.18 The huntingtin protein (HTT) is a substrate for CASP6 and cleavage at the CASP6 cleavage site (IVLD) at D586 is an important step in the pathogenesis of HD.19 YAC128 mice expressing CASP6-resistant (C6R) mutant HTT, in which the CASP6 cleavage site is mutated to prevent the generation of the toxic N-terminal 586 amino-acid HTT fragment,20 are protected against striatal neurodegeneration, behavioural deficits, and biochemical alterations.18, 21, 22 The implication of CASP6 as a critical target in the pathogenesis of HD has recently been supported by a study showing that a novel CASP6 inhibitor protects presymptomatic HD mice from developing motor dysfunction and other behavioural abnormalities by preventing the cleavage of HTT at D586.23
Furthermore, the genetic ablation of Casp6 in mice is neuroprotective in an ischemic stroke model5 and reducing CASP6 activity can protect against inflammatory pain in a formalin-induced pain model.7 Together, these studies implicate CASP6 as an important player in numerous disorders of the central nervous system, making it an attractive molecular target for the development of neuroprotective therapeutics. To develop therapeutic approaches to modulate CASP6, it is crucial to understand the mechanisms regulating CASP6 activation.
Similar to other caspases, CASP6 is expressed as a zymogen comprised of a prodomain, large (p20), and small (p10) subunits and a linker sequence. The active form of CASP6 is a heterotetramer of two p20 subunits and two p10 subunits generated from the zymogen following cleavage and removal of the prodomain and cleavage on either side of the linker releasing the two subunits.24, 25 This process can occur by an intramolecular auto-activation mechanism where the catalytic residue C163 cleaves the linker domain24, 26 or by cleavage by other caspases, such as CASP1, CASP3, CASP7, or CASP9.5, 27, 28 CASP6 activity can furthermore be regulated at both transcriptional and posttranslational levels, including transcriptional initiation, alternate splicing, and phosphorylation events.29, 30, 31, 32, 33 However, it remains unclear which mechanisms are responsible for the increased CASP6 activation in neurodegenerative diseases, such as HD.
Interestingly, we previously showed that the palmitoyl acyltransferase (PAT) HIP14 (DHHC17) is dysfunctional in the presence of mutant HTT in the YAC128 mice and displays a reduced ability to transfer palmitate to its substrates,34 suggesting that altered palmitoylation of HIP14 substrates could contribute to the pathogenesis of HD. This is further supported by our recent work demonstrating a significant association between palmitoylation and HD in a curated list of palmitoylated proteins35 and by the finding that mice with a targeted disruption of the Hip14 gene display HD-like phenotypes.34 S-palmitoylation (here referred to simply as palmitoylation) is the reversible, posttranslational covalent attachment of palmitic acid to cysteine residues of proteins via a thioester bond.36 Palmitoylation increases the hydrophobicity of proteins and thereby regulates membrane association, subcellular localization,36 and enzyme function and activity.37, 38 As CASP6 activation and reduced palmitoylation of HIP14 substrates are both associated with phenotypes in HD,18, 34 we investigated whether CASP6 is palmitoylated and whether palmitoylation regulates its activation.
Here we show that CASP6 can indeed be palmitoylated and that this posttranslational modification has a significant role in regulating CASP6 activity.
Results
Caspase 6 is palmitoylated by HIP14
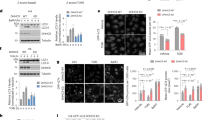
We first sought to determine whether CASP6 is palmitoylated. Immunoprecipitation of CASP6 from transfected COS7 cells followed by Acyl-Biotin Exchange assay (ABE)39 showed that CASP6 is robustly palmitoylated (Figure 1a). To confirm this finding, we treated COS7 cells transfected with CASP6 with 2-bromopalmitate (2-BP), an irreversible inhibitor of PAT activity.40 Treatment of cells with increasing concentrations of 2-BP resulted in a dose-dependent reduction in palmitoylation (Figure 1b), confirming that CASP6 is indeed palmitoylated. Next, to determine whether HIP14 palmitoylates CASP6, we co-transfected COS7 cells with CASP6 and HIP14. As a control for transfection conditions and protein expression, we used an inactive HIP14 mutant with the entire DHHC-cysteine-rich domain deleted (HIP14ΔDHHC).41 In the presence of active HIP14, a significant ~73% increase in CASP6 palmitoylation was observed, indicating that HIP14 is a PAT for CASP6 (Figure 1c). To further confirm that HIP14 is a PAT for CASP6, we assessed the levels of CASP6 palmitoylation in Hip14−/− mice by immunoprecipitation of CASP6 from brain lysates followed by the ABE assay. Compared with wild-type (WT) littermates, palmitoylation of CASP6 was decreased by ~30% in the brains of Hip14−/− mice (Figure 1d). These data confirm that CASP6 is palmitoylated both in vitro and in vivo and that HIP14 modulates the palmitoylation of CASP6.
CASP6 is palmitoylated by HIP14. (a) COS7 cell lysates were subjected to the acyl biotin exchange (ABE) assay, which showed that immunoprecipitated CASP6 was robustly palmitoylated (HAM+ lane). The top panel shows palmitoylation (BMCC-Biotin detected with Streptavidin (str)). No palmitoylation signal was observed in the HAM-negative control lane. Total CASP6 immunoprecipitated is shown in the middle panel (CASP6), and an overlay of the two (str in red and CASP6 in green) is shown in the bottom panel. (b) Treatment with increasing doses of palmitoylation inhibitor 2-BP demonstrates a reduction in CASP6 palmitoylation (Kruskal–Wallis: H=9.09, P=0.011; N=4). (c) In the presence of overexpressed HIP14, the palmitoylation of CASP6 is significantly increased compared with inactive HIP14ΔDHHC as assessed by the ABE assay (Student’s t-test; P=0.0039; N=9). (d) ABE assays were performed on immunoprecipitated CASP6 from the 5-month-old Hip14−/− brains, and CASP6 palmitoylation was found to be significantly reduced by 30% (Student’s t-test: P=0.0016; N=7). The top panel shows the palmitoylation signal (Streptavidin (str)), the middle panel shows total CASP6 immunoprecipitated (CASP6), and the bottom panel shows an overlay of the two (str in red and CASP6 in green). (e) Treatment with increasing dose of 2-BP demonstrates a significant increase in CASP6 activation measured by the level of CASP6 processing (p10 fragment) (one-way analysis of variance: P=0.0009; N=4; Tukey’s post hoc test). (f) HIP14 overexpression significantly reduces CASP6 activity compared with inactive HIP14ΔDHHC (Student’s t-test: P=0.0009; N=3). The cropped representative images in panels (a) or (c) are from the same WBs. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. ns, not significant
Palmitoylation regulates CASP6 activation
In order to determine whether the reduced palmitoylation of CASP6 results in altered function of CASP6, we treated CASP6-transfected COS7 cells with 2-BP and assessed the activation of CASP6. Generation of the p10 CASP6 fragment, which occurs when full-length CASP6 is processed into its active form, can be used as a surrogate marker for CASP6 activation and can be quantified directly by western immunoblotting (WB).19, 31 We detected a 2-BP concentration-dependent increase in the p10 fragment with an increase of ~255% at the highest dose tested (Figure 1e), suggesting that reduced palmitoylation of CASP6 results in increased activity. We next sought to determine whether increasing palmitoylation of CASP6 would correspondingly reduce its activation. We assessed the levels of the p10 fragment in COS7 cells co-transfected with CASP6 and either HIP14 or HIP14ΔDHHC. Compared with inactive HIP14ΔDHHC, a significant decrease in the generation of p10 CASP6 fragment by ~75% was observed in cells transfected with HIP14, confirming that the HIP14-mediated increase in palmitoylation of CASP6 results in a decrease in CASP6 activation (Figure 1f).
CASP6 palmitoylation is decreased in STHdhQ111 cells and in the YAC128 HD mouse brain
As HIP14 is dysfunctional in the presence of mutant HTT in YAC128 mice,34 we assessed the levels of palmitoylation of CASP6 in the brains from YAC128 mice, as well as in the HD cell model STHdhQ111 of striatal origin.42 Indeed, the palmitoylation of CASP6 was significantly decreased by ~15% in the YAC128 brains (Figure 2a). In addition, using the acyl resin-assisted capture (ARAC) of palmitoylated proteins,43 we found a 53% decrease in CASP6 palmitoylation in the HD STHdhQ111 cell line compared with the control STHdhQ7 (Figure 2b). Thus palmitoylation of CASP6 is decreased both in the absence of HIP14 (Hip14−/− mice) and when HIP14 is dysfunctional in the presence of mutant HTT.
Palmitoylation of CASP6 is reduced in HD models. (a) Using ABE assays, we found that palmitoylation of CASP6 from the 9-month-old YAC128 mouse brains is significantly reduced by 15% (Student’s t-test: P=0.0066; N=5), agreeing with the fact that HIP14 is dysfunctional in the presence of mutant HTT in the YAC128 mice. The top panel shows the palmitoylation signal (Streptavidin (str)), the middle panel shows total CASP6 immunoprecipitated (CASP6), and the bottom panel shows an overlay of the two (str in red and CASP6 in green). (b) Using the ARAC assay, we found a significant 53% decrease in CASP6 palmitoylation in STHdhQ111 compared with STHdhQ7 (Student’s t-test: P=0.0004; N=6 and 7, respectively). The top panel shows CASP6 levels in the input samples and in the ARAC purified HAM+ and HAM− samples, and the bottom panel shows GAPDH (glyceraldehyde 3-phosphate dehydrogenase) in the input to show equal loading. Palmitoylation levels are calculated as a ratio of signal from the HAM+ ARAC purified lanes over signal from the corresponding input lane. The cropped representative images in panels (a) or (c) are from the same WBs. (c) Using the MSD assay to measure the cleavage of lamin A, we found a 55% increase in CASP6 activity in STHdhQ111 compared with STHdhQ7 (Student’s t-test: P=0.017; N=3, respectively). (d) FRET (fluorescence resonance energy transfer) analysis showed a 250% increase of the 586 HTT fragment when normalized to total HTT for STHdhQ111 compared with STHdhQ7 (Student’s t-test: P<0.0001; N=8). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001
We have previously demonstrated that CASP6 activity is increased in the cellular models of HD, YAC128 HD mouse brain, and human HD brain.18, 31 Here we assessed the impact of decreased CASP6 palmitoylation on CASP6 activity in the HD STHdhQ111 cell line. Lysates from STHdhQ111 cells show a ~55% higher cleavage activity for lamin A, a highly specific CASP6 substrate,44 compared with control STHdhQ7 (Figure 2c). As cleavage of mutant HTT at the CASP6 cleavage site (IVLD) at D586 has been shown to have a central role in the pathogenesis of HD and that blocking cleavage at this site completely ameliorates the disease pathogenesis,19, 20, 23 we assessed the cleavage of HTT at D586 by FRET and found a ~250% increase in the 586 HTT fragment in STHdhQ111 compared with STHdhQ7 (Figure 2d), suggesting that the increase in CASP6 activity potentiates HTT cleavage at D586 in these cells.
CASP6 is palmitoylated at cysteines 264 and 277
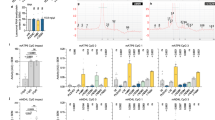
Though palmitoylation is well described for many substrates, a consensus sequence for palmitoylation has yet to be determined.45 However, programs that predict sites of palmitoylation based on a database of known sites of palmitoylation have been developed.46 The palmitoylation prediction algorithm, SeqPalm (http://lishuyan.lzu.edu.cn/seqpalm)46 predicted C277 as the most likely site of palmitoylation in CASP6 and C264 as the next highest predicted site of CASP6 palmitoylation (Table 1). CASP6 has 10 cysteines (Figure 3a) and we generated single mutants of all cysteines to confirm the sites of palmitoylation of CASP6. Of all the 10 single mutations of cysteines to serines, only C264S and C277S showed a reduction in palmitoylation (Figure 3b and data not shown), with each mutation resulting in ~30% decrease in palmitoylation. We next generated the C264/277S double mutant, which showed a ~50% reduction in palmitoylation (Figure 3b). These data confirm C264 and C277 not only as the primary sites of CASP6 palmitoylation but also indicate that other secondary sites of palmitoylation may exist.
CASP6 is palmitoylated at cysteines 264 and 277. (a) Schematic of the cysteines in CASP6 that are potential palmitoylation sites and internal cleavage sites at the prodomain (D23) and at the linker (D179 and D193). (b) CASP6 palmitoylation is reduced when cysteines 264 and 277 are mutated to serines (Kruskal–Wallis: H=20.30, P=0.0001, N=8). The top panel shows the palmitoylation signal (Streptavidin (str)), the middle panel shows total CASP6 immunoprecipitated (CASP6), and the bottom panel shows an overlay of the two (str in red and CASP6 in green). (c) Increased CASP6 p10 fragment is observed by WB (DDK antibody) in cells transfected with palmitoylation-deficient CASP6 C264/277S (Student’s t-test: P=0.008; N=3). (d) Cleavage of lamin A is significantly increased in the presence of palmitoylation-deficient CASP6 (Student’s t-test: P=0.00027; N=3). (e) Cleavage of HTT 1212 128Q 4C measured by FRET (fluorescence resonance energy transfer) is increased by 12% in the presence of palmitoylation-deficient CASP6 (Student’s t-test: P=0.011; N=9). WB shows the normal expression levels of the two transgenes using actin as a loading control. (f) The CASP6 p20 fragment is increased by 108% and 79% in cells transfected with CASP6 C264/277S compared with WT CASP6 measured by HD91 and LE22, respectively (HD91 antibody: Student’s t-test: P=0.0001; N=6, and LE22 activity probe: Student’s t-test: P=0.0018; N=6). (g) An increase in LE22 binding by 134 and 27% is observed for full-length CASP6 and CASP6 without the prodomain (ΔCASP6), respectively, for CASP6 C264/277S compared with WT CASP6 (CASP6: Student’s t-test: P=0.0001; N=6, and ΔCASP6: Student’s t-test: P=0.0016; N=6). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001
Palmitoylation-deficient mutant display increased CASP6 activation
To directly confirm that the loss of palmitoylation of CASP6 resulted in its increased activation, we assessed activation of the C264/277S CASP6 mutant by quantifying the p10 CASP6 fragment by WB. A significant increase in the p10 fragment was detected in the C264/277S palmitoylation-deficient CASP6 mutant compared with WT CASP6 (Figure 3c), suggesting that reduced palmitoylation results in increased processing and activation of CASP6. To further validate this finding, we used the cleavage of lamin A as a direct measure of CASP6 activity.31 Indeed, a significant ~140% increase in lamin A cleavage was observed by WB in lysates containing the palmitoylation-deficient C264/277S mutant when compared with WT CASP6 (Figure 3d), confirming that reduced palmitoylation results in increased CASP6 activity.
To determine whether loss of CASP6 palmitoylation affects mutant HTT cleavage by CASP6, we used an in vitro FRET assay that uses 128Q HTT amino acids 1–1212 with the four non-CASP6 caspase cleavage sites mutated (D513A, D530A D552A, and D589A) (1212 128Q 4C HTT).47 This allows easy detection of the mutant HTT 586 fragment without any other cleavage events occurring. We co-transfected 1212 128Q 4C HTT with WT CASP6 or mutant CASP6 C264/277S and found a significant ~12% increase in cleavage of mutant HTT and generation of the 586 HTT fragment by palmitoylation-deficient CASP6 compared with WT (Figure 3e). These findings taken together demonstrate that the loss of CASP6 palmitoylation promotes increased cleavage of critical substrates involved in apoptosis and the pathogenesis of HD.
To more directly assess the effects of CASP6 palmitoylation on substrate access to the catalytic site, we used the activity-based probe, LE22, that binds the active site cysteine, C163, and efficiently labels the processed active form of CASP6 and, to a lesser extent, full-length CASP6.48 Lysates from CASP6 or CASP6 C264/277S-transfected COS7 cells were incubated with LE22 and assessed by WB. Similar to the increase in p10 fragment, we found a significant increase by ~108 and ~79% for the p20 fragment detected by a CASP6-specific antibody, HD91, or LE22, respectively (Figure 3f), confirming that reduced CASP6 palmitoylation promotes its processing and activation. Interestingly, when we evaluated the LE22 labelling of the unprocessed full-length CASP6 and the activation intermediate CASP6 lacking the prodomain (ΔCASP6), we found an increase in LE22 labelling by ~134 and ~27%, respectively, of the palmitoylation-deficient CASP6 mutant compared with WT CASP6 (Figure 3g), suggesting that reduced CASP6 palmitoylation may promote a conformation where the active site is more accessible to the LE22 activity probe.
CASP6 palmitoylation is predicted to block substrate binding and dimerization
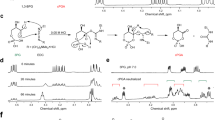
To further determine how palmitoylation of CASP6 might affect its processing and activation (Figure 4a), we utilized computational molecular modelling based on the published crystal structures of CASP6.24, 25, 49, 50, 51, 52, 53, 54, 55 Generation of the model of CASP6 required the construction of coordinates of loop 2 (L2), which forms the substrate-binding groove (active site) and is not resolved in any of the several crystal structures available, presumably owing to high flexibility. The structure of CASP6 has a six-stranded β-sheet flanked by five α-helices and two small β-strands.24, 25 The loops (L1–L4) protrude from the central β-sheet forming the active site.24, 25
CASP6 modelling suggests that palmitoylation inhibits CASP6 activation by blocking the substrate-binding site and dimerization. (a) Schematic representation of the cleavage sites in CASP6. Surface and cartoon representation of the structure of unmodified WT CASP6 (b) and CASP6 with C264 palmitoylated (turquoise stick) (c). Interactions between L2 and L4 are highlighted (dashed lines). Loops L2, L3, and L4 are coloured in magenta, blue and orange, respectively, with rest of the protein in grey. The cleavage site 190TEVD193 is highlighted in green. (d) Distance probability distributions in conformations that are sampled during the MD simulations with representative models (e) illustrating the distance between the active site cysteine C163, dyad histidine H121, and the linker site aspartate D193. (f) Superposition of CASP6 palmitoylated at C277 (turquoise stick), with the dimeric CASP6 structure, highlighting the steric clash. The palmitoylated CASP6 subunit of the dimer is shown in grey and the other CASP6 subunit is shown as a transparent purple surface. (g) Representative microscopic images from COS7 cells transfected with WT CASP6 or mutant CASP6 C264/277S show no difference in localization when comparing the ratios of nuclear and cytoplasmic intensities. CASP6 is visualized by anti-FLAG antibody and the nucleus is stained with DAPI (4,6-diamidino-2-phenylindole). The two images are merged on the right panel to visualize CASP6 distribution (Student’s t-test: P=0.54; N=90 WT CASP6 cells and 88 CASP6 C264/277S cells from two separate experiments with two transfections per experiment)
During the molecular dynamics (MD) simulations, the CASP6 structure was very stable and remained close to its crystallographic state with the loops showing high flexibility, as expected (Figure 4b). C264 is located at the middle of L4 and is well exposed. Upon palmitoylation of C264, increased flexibility of loops L2, L3, and L4 was observed during the simulations, bringing L2 into close contact with L4, occluding active site access (Figure 4c). Several intramolecular interactions between loops L2 and L4 stabilize this closed conformation (Figure 4c). In contrast, no such interactions are observed during the simulations of non-palmitoylated CASP6 (Figure 4b). In non-palmitoylated CASP6, the C163/H121 catalytic dyad, which initiates the nucleophilic attack on D193 to initiate self-cleavage, is intact, whereas upon palmitoylation of C264, the catalytic dyad expands (the distance between C163 and H121 increases from 5.5 to ~7.5 Å, the distance between C163 and D193 increases from 4.0 to ~5.1 Å, and the distance between H121 and D193 increases from 4.5 to ~6.5 Å; Figures 4d and e). This increased separation would prevent C163 from carrying out a nucleophilic attack on D193 and the subsequent initiation of self-cleavage and activation of CASP6. Finally, the other site of palmitoylation, C277, is located at the start of the β-strand and the dimer interface. Palmitoylation of C277 will result in the long palmitoyl chain sterically impeding the approach of the other molecule of CASP6, thus preventing dimerization and activation (Figure 4f).
CASP6 localization is not affected by reduced palmitoylation
Palmitoylation has been shown to regulate protein membrane association and subcellular localization,36 therefore we assessed whether loss of palmitoylation at cysteines 264 and 277 affects CASP6 localization. We transfected COS7 cells with WT CASP6 or mutant CASP6 C264/277S and evaluated their localization by confocal microscopy. We observed a diffuse distribution across the cytoplasm and nucleus for both proteins and found no difference for the ratio of nuclear to cytoplasmic intensity between CASP6 C264/277S and WT CASP6 (Figure 4g). This finding supports our results from the molecular modelling studies, suggesting that loss of palmitoylation regulates CASP6 activity primarily by inducing a conformational change rather than by altering its localization.
Discussion
Our study provides a novel mechanism for the regulation and activation of CASP6, which may be particularly relevant in HD, where HIP14 is dysfunctional.34, 56 Taken together, our data show that CASP6 is palmitoylated by the PAT HIP14 and that cysteines 264 and 277 are the main sites of palmitoylation. Most importantly, we demonstrate that reduced palmitoylation of CASP6 results in its increased activation and that increasing CASP6 palmitoylation reduces its activation. We also show that the increased CASP6 activity owing to reduced palmitoylation leads to cleavage of HTT and generation of the N-terminal 586 amino acid mutant HTT fragment, which has been shown in three recent studies to have a central role in the pathogenesis of HD. First, preventing cleavage at the CASP6 cleavage site in mutant HTT can completely prevent the HD phenotype.18 Second, the HD-like symptoms in a transgenic mouse model expressing the 586 mutant HTT fragment progress faster than full-length HTT models.20 Finally, reducing CASP6 activity pharmacologically protects against the HD phenotype by preventing the cleavage of HTT at D586.23
When we mutated all 10 cysteines in CASP6, we only saw a reduction in palmitoylation when either C264 or C277 were mutated. However, when changing both sites simultaneously, we did not observe a complete loss of palmitoylation, which suggests that other cysteines may be palmitoylated as a compensatory mechanism.
In both the YAC128 and STHdhQ111 models of HD, we found significant decreases in CASP6 palmitoylation, which is consistent with previous findings demonstrating both dysfunctional HIP1434 and increased CASP6 activation in the presence of mutant HTT.18 The reduction in CASP6 palmitoylation is less when comparing whole YAC128 mouse brain to the striatal precursor cell line STHdhQ111, which may be a result of increased impact of mutant HTT in the striatal-derived cell line or differentially regulated CASP6 palmitoylation between different neuronal and glial cell types.57
Interestingly, we did not observe a complete loss of CASP6 palmitoylation in the brains from Hip14−/− mice, and it is thus possible that other PATs might contribute to CASP6 palmitoylation. This notion is supported by similar findings for HTT, which can be palmitoylated by both HIP14 and HIP14L (DHHC13).58 Thus the observed smaller loss of CASP6 palmitoylation in vivo could be a result of a compensation by PATs that are expressed and active in vivo but not in vitro.
CASP6 can be activated by three different mechanisms in Mammalian cells: First, as a result of CASP6 auto-activation; second, as a result of cleavage by the effector CASP3 and CASP7 of the full-length CASP6; and third, by cleavage of the 23 amino acid prodomain by initiator caspases followed by auto-activation.8 Once activated, the localization and proximity to its substrates could add an additional layer of complexity to the regulation of substrate cleavage by CASP6. However, when we investigated whether palmitoylation affected CASP6 localization, we did not find any differences between WT CASP6 and mutant CASP6 C264/277S, suggesting that the conformational changes found by molecular modelling is the main contributing factor to altered CASP6 activity.
We propose two distinct mechanisms for the regulation of CASP6 activation: First, palmitoylation at C264 negatively regulates the auto-activation of CASP6 by altering the conformation of the catalytic site, as well as sterically hindering access to the substrate-binding groove thus reducing its activity; and second, palmitoylation at C277 impedes dimerization of the heterotetramer CASP6 complex.
Our modelling studies show that the C163/H121 catalytic dyad of CASP6, which is essential for self-cleavage, is intact in the non-palmitoylated state. When C264 is palmitoylated, the distance between the residues of the catalyatic dyad expands, preventing the initiation of self-cleavage and activation of CASP6 via nucleophilic attack of C163 on D193. As substrate binding and optimal positioning of the catalyatic dyad is necessary for caspase proteolytic activity, it is possible that palmitoylation of C264 locks the protein in an inhibited state by promoting interactions between loops that occlude the substrate-binding groove. This finding is strongly supported by our LE22 labelling data showing increased access to the active site for the palmitoylation-deficient CASP6 mutant. Palmitoylation at C264 may be particularly relevant for limiting auto-activation, as well as restricting substrate access, but may not prevent activation arising directly from cleavage by other effector caspases, which does not require an accessible substrate-binding groove in CASP6.
Our modelling studies also suggest that palmitoylation at C277 may sterically interfere with the dimerization of CASP6, which is an essential step for its activation, and may work independent of effector caspases. Although our predictions suggest that the two sites of palmitoylation may regulate CASP6 activity through different mechanisms, it remains unclear whether these sites of palmitoylation work independently or together and whether they may regulate other CASP6 posttranslational modifications.
Interestingly, phosphorylation of CASP6 at S257 also inhibits its activation. Crystal structure studies of the phospho-mimetic S257D suggested that phosphorylation of S257 inactivates CASP6 through a steric clash with the proline side chain of residue P201 in the L2’ loop,49 causing reorganization of the L2 loop into an inactive position.59 Indeed, removal of the clashing proline side chain of P201 (P201G) reversed the S257 phosphorylation-mediated inhibition of CASP6 activity, suggesting that steric hindrance was the underlying mechanism of inhibition.59
This is similar to our proposition that palmitoylation-mediated inhibition of CASP6 activation may result from steric hindrance. Interestingly, protein palmitoylation has been shown to regulate phosphorylation of several proteins.60, 61 However, it remains to be investigated whether CASP6 phosphorylation and palmitoylation regulate each other and whether they work synergistically or independently to regulate CASP6 activation. Thus further studies on the interaction between phosphorylation and palmitoylation of CASP6 are needed.
Here we demonstrated that CASP6 activation is decreased by HIP14-mediated palmitoylation, a novel mechanism for the regulation of this protein with a critical role in the pathogenesis of both HD and AD. Future studies will be critical to explore the potential of modulating the activity of HIP14 in in vivo models of both diseases to reduce CASP6 activity and associated toxicity.
Materials and methods
CASP6 and palmitoylation mutants
Human CASP6 plasmid with a DDK (FLAG) tag was purchased from Origene (Rockville, MD, USA) and used for all in vitro studies. Potential palmitoylation site cysteine mutants were generated using a PCR-based protocol as previously described.62 HIP14-GFP and HIP14ΔDHHC-GFP were previously described.41, 63
Cell culture
The COS7 cells were cultured at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum, 1 × Penicillin/ Streptomycin (Gibco), and 2 mM L-glutamine (Gibco) (complete DMEM). COS7 cells were transfected using Fugene X-tremeGENE (Roche, Mississauga, ON, Canada) and incubated for 24 h. For the 2-BP experiments, the cells were transfected for 16 h and subsequently treated with 15 or 50 μM 2-BP for 8 h. The immortalized striatal neuronal cell lines STHdhQ7 and STHdhQ111 were cultured at 33 °C in complete DMEM supplemented with 0.5 mg/ml active G418 (Gibco). Cells were grown in a humidified atmosphere containing 5% CO2 and harvested by scraping the cells in media.
Palmitoylation assays
COS7 cells overexpressing CASP6 alone, CASP6 with HIP14 or HIP14ΔDHHC, and CASP6 palmitoylation mutants were harvested, washed, snap frozen, and stored at −80 °C. ABE assays were performed as previously described.39 Cell pellets were resuspended in 500 μl Lysis Buffer (150 mM NaCl, 50 mM Tris, 5 mM EDTA, 1% Triton X-100 pH 7.4, 1 × Roche Complete protease inhibitor tablets, 5 μM zVAD (Calbiochem, San Diego, CA, USA; Caspase Inhibitor I)) with 100 mM N-ethylmaleimide (NEM; Sigma, St. Louise, MO, USA), to block free cysteines, sonicated, and cleared by centrifugation for 10 min at 14 000 r.p.m. Cleared lysates were incubated with protein-G DynaBeads (ThermoFisher, Waltham, MA, USA) and anti-DDK (Origene, Rockville, MD, USA, 4 μl per sample) antibody directed to the CASP6 DDK tag overnight to immunoprecipitate CASP6. The beads were then washed with Lysis buffer, divided into two portions, and one was treated with 1 M hydroxylamine (HAM) at pH 7.0 (HAM+) and the other with Lysis Buffer (HAM−) for 1 h at room temperature to remove palmitate. Following HAM treatment, the beads were washed and then treated with EZ-Link BMCC-Biotin (ThermoFisher) to label the previously palmitoylated cysteines with biotin.39 Samples were eluted from beads using 1 × LDS sample buffer (ThermoFisher) with 100 mM dithiothreitol (DTT) and run on 10% SDS-PAGE gels. Palmitoylation was detected using Alexa Fluor 680-conjugated Streptavidin (ThermoFisher, 1 : 5000), and total CASP6 immunoprecipitated was detected with anti-Flag antibody (rabbit, Sigma, 1 : 1000) and IRDye 800 goat anti-mouse antibody (Rockland, Limerick, PA, USA, 1 : 5000). All antibody incubations were performed in 5% bovine serum albumin in PBS with 0.1% Tween-20. The fluorescence was scanned and quantified using the Odyssey Infrared Imaging System (Li-COR Bioscience, Lincoln, NE, USA). Palmitoylation was calculated as a ratio of signal from Streptavidin over signal from the Flag antibody.
For palmitoylation assays from brain samples, the brains were harvested and immediately snap frozen in liquid nitrogen and stored at −80 °C. Frozen brains were homogenized in 4 ml lysis buffer (same as above) with 100 mM NEM. Endogenous CASP6 was immunoprecipitated using an in-house antibody against CASP6 (HD88, 20 μl/IP) and ABE assays were performed as described above. Samples were eluted from beads using 1 × LDS sample buffer (ThermoFisher) with 100 mM DTT and run on 10% SDS-PAGE gels. Palmitoylation was detected using Alexa Fluor 680-conjugated Streptavidin (ThermoFisher, 1 : 5000) and total CASP6 immunoprecipitated was detected using HD88 (rabbit, in-house, 1 : 250) and IRDye 800 goat anti-rabbit antibody (Rockland, 1 : 5000). All primary antibody incubations were performed in 5% milk in TBS with 0.1% Tween-20, and all secondary incubations were performed in 5% bovine serum albumin in PBS with 0.1% Tween-20. The fluorescence was scanned and quantified using the Odyssey Infrared Imaging System (Li-COR Bioscience). Palmitoylation was calculated as a ratio of signal from Streptavidin over signal from the Flag antibody. The HD88 was generated as previously described64 by immunizing a rabbit with a synthetic peptide GFYKSREVFDPAEQ corresponding to the N-terminus of mouse Casp6. The antibody was purified by protein A and peptide affinity chromatography.
For palmitoylation assays from STHdhQ7 and STHdhQ111 cells, a modified ARAC assay was used.43 Cells were harvested and immediately frozen and stored at −80 °C. Samples were resuspended in 1 ml RIPA Buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.5% sodium deoxycholate, 1% Igepal CA-630, 0.1% SDS, 1 × Roche Complete protease inhibitor tablets, 5 μM ZVAD (Calbiochem, Caspase Inhibitor I)) with 100 mM NEM, sonicated, cleared by centrifugation at 14 000 r.p.m. for 10 min, and lysates were incubated overnight at 4 degrees with rotation to block free cysteines. The samples were then subjected to a chloroform–methanol precipitation to remove excess NEM, and protein concentrations were determined by the DC Protein Assay Kit (Bio-Rad, Mississauga, ON, Canada). A total of 50 μg (20%) of the total protein was saved at this point as input protein for each sample. Also, 250 μg of each sample was incubated with 1 M hydroxylamine at pH 7.0 (HAM+) or with lysis buffer (HAM−) for 4 h at room temperature in the presence of 200 μl (50/50 bead/buffer) slurry of Thiopropyl sepharose beads (GE Healthcare, Mississauga, ON, Canada) to simultaneously cleave palmitate groups and capture palmitoyl proteins. Proteins were captured on the Thiopropyl Sepharose beads by the formation of a disulphide bond between the beads and the newly free cysteine residues allowing the capture of the entire palmitoyl proteome. Beads were then washed and proteins eluted using 150 mM fresh DTT and run on 8% BIS-Tris SDS-PAGE gels along with the 20% input of each sample. Blots were probed with HD88 (rabbit, in-house, 1 : 250) and IRDye 800 goat anti-rabbit antibody (Rockland, 1 : 5000) to detect CASP6 in the input and ARAC-purified HAM+ and HAM− samples. All antibody incubations were performed in 5% BSA in PBS with 0.1% Tween-20. The fluorescence was scanned and quantified using the Odyssey Infrared Imaging System (Li-COR Bioscience). Palmitoylation was calculated as a ratio of ARAC HAM+ signal over input signal.
Western immunoblotting
Cell pellets were lysed in lysis buffer including protease inhibitors (50 mM Tris pH 8.0, 150 mM NaCl, 1% Igepal/NP40, 40 mM β-glycerophosphate, 10 mM NaF, 1 mM sodium vanadate, 1 mM PMSF, 5 μM zVAD, and 1 × Roche Complete). Protein concentration was measured by the DC Protein Assay Kit (Bio-Rad) and lysates were separated on 4–12% Bis-Tris Gels (Invitrogen, Waltham, MA, USA). For LE22 labelling, cell pellets were lysed in complete lysis buffer without caspase inhibitor zVAD. Aliquots corresponding to 50 μg protein were diluted to 20 μl and final concentrations of 0.5 μM LE22 and 0.5 mM DTT were added.48 All subsequent steps were performed in the dark. Samples were incubated for 2 h at 37 °C, mixed with SDS sample buffer, and separated on 12% Bis-Tris Gels (Invitrogen). Following transfer to PVDF Immobilon-FL membranes, proteins of interest were detected using the following antibodies. CASP6 was detected with anti-DDK antibody (1:1000; Origene, Rockville, MD, USA) detecting the DDK tag, or HD91 (1:1000; in-house64). For the quantification of CASP6 levels, both the full-length and ΔCASP6 bands were estimated together except for the LE22 experiment, where they were estimated separately. Lamin A was detected with anti-Lamin A/C (1:1000; Cell Signaling, Danvers, MA,USA) and anti-cleaved lamin A (1:1000; Cell Signaling) antibodies. Secondary antibodies goat-anti-mouse or goat-anti-rabbit conjugated to either Alexa Fluor 680 or 800 IR dye (1 : 5000 for both; ThermoFisher or Rockland, respectively) were used for detection of primary antibodies using the LiCor Odyssey Infrared Imaging System.
FRET assay for CASP6 activity
The N-terminal 1212 amino acids of HTT with 128Q and the 4C mutations (D513A, D530A D552A, and D589A) were transiently transfected and overexpressed together with WT CASP6 or mutant CASP6 C264/277S in COS7 cells for 24 h.47 Cell pellets from COS7 cells or Q111 and Q7 cells were lysed with SDP+ lysis buffer65 and diluted in SDP+ lysis buffer to a final protein concentration of 2 μg/μl.66, 67 The detection of the 586 HTT fragment was performed using a combination of the monoclonal BKP1 antibody raised against the HTT N-terminus and an in-house 586 neo-epitope antibody raised against the C-terminus of 586 cleaved HTT.47 The level of 586 HTT fragment was normalized to total HTT measures with the combination of BKP1 and 2166 raised against exon 2. BKP1 was labelled with terbium and 2166 and Neo-586 were labelled with D2 (Cisbio Bioassays, Bedford, MA, USA) fluorescent tags. Antibody pairs were mixed in 100 mM Tris (pH 7.4), 150 mM NaCl, 0.1% BSA, 0.05% Tween-20 at 1 ng/μl BKP1-terbium and 10 ng/μl Neo-586-HTT-D2. In all, 2 μl of antibody master mix was added to each sample in a white 384-well plate and incubated overnight at 4 °C. For total HTT (BKP1/2166) and 586 HTT fragment, 20 and 200 μg protein was used, respectively. The plate was measured with a xenon lamp Victor Plate Reader (Perkin Elmer, Waltham, MA, USA) after excitation at 340 nm. The relative HTT concentration is represented by the 665/615-nm ratio.66
Mesoscale for cleavage lamin A
Cells were lysed in MCB buffer (50 mM HEPES, pH 7.4, 100 nM NaCl, 0.1% CHAPS, 2 nM EDTA, 1% NP-40, 10% Glycerol, 1 × Roche Complete) and adjusted to 2 μg/μl in 10 μl. In all, 100 ng lamin A (Abcam, Atlanta, GA, USA) in 10 μl of caspase cleavage buffer with fresh DTT was added to the lysate and incubated for 3 h at 37 °C. Also, 5 μl of sample was spotted onto Mesoscale ELISA plates and processed as previously described.44 First, the samples were incubated for 60 min at room temperature and then the wells were blocked with 5% BSA in PBS for another 60 min. The wells were washed three times with 150 μl PBS-T (PBS with 0.05% Tween-20) and incubated for 60 min with 25 μl antibody mix (cleaved lamin A antibody (Cell Signaling 2036, 1:100) and goat anti-mouse sulpho-tag secondary antibody (MSD Technology, 1:500) in 1% BSA/PBS) added to each well. The wells were washed three times with 150 μl PBS-T and 150 μl 2 × reading reagent (MSD Technology, Rockville, MD, USA) was added. The plates were read on the Mesoscale platform electrochemiluminescence reader (MSD Technology) according to the manufacturer’s instructions.
CASP6 molecular modelling
The initial model of CASP6 was developed using the crystal structure 4IYR.pdb.55 This is the structure of CASP6 zymogen with structural information missing from residues 173–187. These missing regions were built using the program Modeller (version 9.12; San Francisco, CA, USA, https://salilab.org/modeller/).68 Several models were generated and the models with the best physicochemical properties (decided by DOPE score within modeller and visual inspection) were refined further using all atom MD simulations.
Three starting models of CASP6 were prepared for the studies using the program Amber1169 (University of California, San Francisco, http://ambermd.org): CASP6 with no modifications, CASP6 with palmitoyl group attached covalently at C264, and CASP6 with palmitoyl group attached covalently at C277. Hydrogen atoms were added to the structures using the Xleap module of the Amber11 package.69 The net charges on all the systems were neutralized by the addition of counter ions. The neutralized systems were solvated with TIP3P70 water molecules to form a truncated octahedral box ensuring a 10-Å shell between the solute atoms and the edges of the box. MD simulations were carried out with the Sander module of the AMBER11 package in combination with the parm03 force field.71 Force field parameters for palmityolated cysteine were taken as previous described.72 All the systems were first subjected to 100 steps of energy minimization. This was followed by MD simulations, for which the protein was initially harmonically restrained (25 kcal/mol/Å2) to the energy minimized coordinates, and the system was heated up to 300 K in steps of 100 K followed by gradual removal of the positional restraints and a 1-ns unrestrained equilibration at 300 K. All production runs were carried out in explicit solvent at 300 K. During all the simulations, the long-range electrostatic interactions were treated with the particle mesh Ewald method using a real space cutoff distance of 9 Å.73 The settle algorithm was used to constrain bond vibrations involving hydrogen atoms,74 which allowed a time step of 2 fs during the simulations. For each system, three independent MD simulations (assigning different initial velocities) were carried out for 100 ns, and conformations were saved every 10 ps. Simulation trajectories were visualized using VMD75 and figures were generated using Pymol (https://www.pymol.org/).76
Confocal microscopy
Immunofluorescence experiments were performed as previously described.77 COS7 cells were grown on glass coverslips and transfected with WT or C264/277S CASP6. Twenty-four hours posttransfection, cells were washed and fixed in 4% paraformaldehyde/PBS for 15 min. Cells were permeabilized with 0.1% Triton/1% paraformaldehyde/PBS for 1 min and blocked in 0.2% Gelatin/PBS for 15 min. Coverslips were incubated for 1 h with primary antibody (Rabbit anti-FLAG, 1 : 400; Sigma-Aldrich, St. Louise, MO, USA) diluted in 0.2% Gelatin/PBS. Cells were washed and incubated in the same way with secondary antibody (Alexa Fluor 594 Goat anti-Rabbit, 1 : 400; Invitrogen). Washed coverslips were mounted on microscope slides with one drop of ProLong Gold Antifade Reagent with DAPI (Invitrogen). Images were captured using a Leica SP8 confocal microscope at 40 times magnification and Leica Application Suite X software (Leica Microsystems Inc., Concord, ON, CA). All steps were performed at room temperature. Images were analysed using the ImageJ software (Andrej Sali, Bethesda, MD, USA; https://imagej.nih.gov/ij/). The nucleus was outlined manually using the DAPI stain and the intensity of FLAG staining within the nucleus was calculated. Three representative regions of interest were then drawn in the cytoplasm and the average FLAG staining intensity of the three was calculated. The ratio of nuclear FLAG intensity to cytoplasmic FLAG intensity was calculated to give the relative CASP6 nuclear localization.
Statistical analysis
Data were graphed using GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA) as the average value±S.E. from at least three independent experiments unless otherwise indicated. Data were analysed by Student’s t-test, ANOVA, or nonparametric tests as indicated. Analyses for P-values were calculated with GraphPad Prism 5.0. Differences were considered statistically significant when P⩽0.05. P-values are illustrated with asterisks (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001).
References
D'Amelio M, Cavallucci V, Middei S, Marchetti C, Pacioni S, Ferri A et al. Caspase-3 triggers early synaptic dysfunction in a mouse model of Alzheimer's disease. Nat Neurosci 2011; 14: 69–76.
Li Z, Jo J, Jia J-M, Lo S-C, Whitcomb DJ, Jiao S et al. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell 2010; 141: 859–871.
Park KJ, Grosso CA, Aubert I, Kaplan DR, Miller FD . p75NTR-dependent, myelin-mediated axonal degeneration regulates neural connectivity in the adult brain. Nat Neurosci 2010; 13: 559–566.
Graham RK, Ehrnhoefer DE, Hayden MR . Caspase-6 and neurodegeneration. Trends Neurosci 2011; 34: 646–656.
Akpan N, Serrano-Saiz E, Zacharia BE, Otten ML, Ducruet AF, Snipas SJ et al. Intranasal delivery of caspase-9 inhibitor reduces caspase-6-dependent axon/neuron loss and improves neurological function after stroke. J Neurosci 2011; 31: 8894–8904.
Ding Z-M, Wu B, Zhang W-Q, Lu X-J, Lin Y-C, Geng Y-J et al. Neuroprotective effects of ischemic preconditioning and postconditioning on global brain ischemia in rats through the same effect on inhibition of apoptosis. Int J Mol Sci 2012; 13: 6089–6101.
Berta T, Park C-K, Xu Z-Z, Xie R-G, Liu T, Lü N et al. Extracellular caspase-6 drives murine inflammatory pain via microglial TNF-α secretion. J Clin Invest 2014; 124: 1173–1186.
LeBlanc AC . Caspase-6 as a novel early target in the treatment of Alzheimer's disease. Eur J Neurosci 2013; 37: 2005–2018.
Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC . Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer's disease. Am J Pathol 2010; 165: 523–531.
Albrecht S, Bourdeau M, Bennett D, Mufson EJ, Bhattacharjee M, LeBlanc AC . Activation of caspase-6 in aging and mild cognitive impairment. Am J Pathol 2007; 170: 1200–1209.
Nikolaev A, McLaughlin T, O’Leary DDM, Tessier-Lavigne M . APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 2009; 457: 981–989.
Banwait S, Galvan V, Zhang J, Gorostiza OF, Ataie M, Huang W et al. C-terminal cleavage of the amyloid-beta protein precursor at Asp664: a switch associated with Alzheimer's disease. J Alzheimers Dis 2008; 13: 1–16.
Galvan V, Gorostiza OF, Banwait S, Ataie M, Logvinova AV, Sitaraman S et al. Reversal of Alzheimer's-like pathology and behavior in human APP transgenic mice by mutation of Asp664. Proc Natl Acad Sci 2006; 103: 7130–7135.
Galvan V, Zhang J, Gorostiza OF, Banwait S, Huang W, Ataie M et al. Long-term prevention of Alzheimer's disease-like behavioral deficits in PDAPP mice carrying a mutation in Asp664. Behav Brain Res 2008; 191: 246–255.
Zhang J, Gorostiza OF, Tang H, Bredesen DE, Galvan V . Reversal of learning deficits in hAPP transgenic mice carrying a mutation at Asp664: a role for early experience. Behav Brain Res 2010; 206: 202–207.
Group T . A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington‘s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell 1993; 72: 971–983.
Zuccato C, Valenza M, Cattaneo E . Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev 2010; 90: 905–981.
Graham RK, Deng Y, Carroll J, Vaid K, Cowan C, Pouladi MA et al. Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J Neurosci 2010; 30: 15019–15029.
Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 2006; 125: 1179–1191.
Waldron-Roby E, Ratovitski T, Wang X, Jiang M, Watkin E, Arbez N et al. Transgenic mouse model expressing the caspase 6 fragment of mutant huntingtin. J Neurosci 2012; 32: 183–193.
Pouladi MA, Graham RK, Karasinska JM, Xie Y, Santos RD, Petersén A et al. Prevention of depressive behaviour in the YAC128 mouse model of Huntington disease by mutation at residue 586 of huntingtin. Brain 2008; 132: 919–932.
Metzler M, Gan L, Mazarei G, Graham RK, Liu L, Bissada N et al. Phosphorylation of huntingtin at Ser421 in YAC128 neurons is associated with protection of YAC128 neurons from NMDA-mediated excitotoxicity and is modulated by PP1 and PP2A. J Neurosci 2010; 30: 14318–14329.
Aharony I, Ehrnhoefer DE, Shruster A, Qiu X, Franciosi S, Hayden MR et al. A Huntingtin-based peptide inhibitor of caspase-6 provides protection from mutant Huntingtin-induced motor and behavioral deficits. Hum Mol Genet 2015; 24: 2604–2614.
Wang X-J, Cao Q, Liu X, Wang K-T, Mi W, Zhang Y et al. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep 2010; 11: 841–847.
Baumgartner R, Meder G, Briand C, Decock A, D'arcy A, Hassiepen U et al. The crystal structure of caspase-6, a selective effector of axonal degeneration. Biochem J 2009; 423: 429–439.
Klaiman G, Champagne N, LeBlanc AC . Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochem Biophys Acta 2009; 1793: 592–601.
Kaushal V, Dye R, Pakavathkumar P, Foveau B, Flores J, Hyman B et al. Neuronal NLRP1 inflammasome activation of Caspase-1 coordinately regulates inflammatory interleukin-1-beta production and axonal degeneration-associated Caspase-6 activation. Cell Death Differ 2015; 22: 1676–1686.
Inoue S, Browne G, Melino G, Cohen GM . Ordering of caspases in cells undergoing apoptosis by the intrinsic pathway. Cell Death Differ 2009; 16: 1053–1061.
Yang C, Kaushal V, Haun RS, Seth R, Shah SV, Kaushal GP . Transcriptional activation of caspase-6 and -7 genes by cisplatin-induced p53 and its functional significance in cisplatin nephrotoxicity. Cell Death Differ 2007; 15: 530–544.
MacLachlan TK, El-Deiry WS . Apoptotic threshold is lowered by p53 transactivation of caspase-6. Proc Natl Acad Sci USA 2002; 99: 9492–9497.
Ehrnhoefer DE, Skotte NH, Ladha S, Nguyen YTN, Qiu X, Deng Y et al. p53 increases caspase-6 expression and activation in muscle tissue expressing mutant huntingtin. Hum Mol Genet 2013; 23: 717–729.
Lee AW, Champagne N, Wang X, Su XD, Goodyer C, LeBlanc AC . Alternatively spliced Caspase-6B isoform inhibits the activation of Caspase-6A. J Biol Chem 2010; 285: 31974–31984.
Suzuki A, Kusakai G-I, Kishimoto A, Shimojo Y, Miyamoto S, Ogura T et al. Regulation of caspase-6 and FLIP by the AMPK family member ARK5. Oncogene 2004; 23: 7067–7075.
Singaraja RR, Huang K, Sanders SS, Milnerwood AJ, Hines R, Lerch JP et al. Altered palmitoylation and neuropathological deficits in mice lacking HIP14. Hum Mol Genet 2011; 20: 3899–3909.
Sanders SS, Martin DDO, Butland SL, Lavallée-Adam M, Calzolari D, Kay C et al. Curation of the Mammalian palmitoylome indicates a pivotal role for palmitoylation in diseases and disorders of the nervous system and cancers. PLoS Comput Biol 2015; 11: e1004405.
Huang K, El-Husseini A . Modulation of neuronal protein trafficking and function by palmitoylation. Curr Opin Neurobiol 2005; 15: 527–535.
Vetrivel KS, Meckler X, Chen Y, Nguyen PD, Seidah NG, Vassar R et al. Alzheimer disease Abeta production in the absence of S-palmitoylation-dependent targeting of BACE1 to lipid rafts. J Biol Chem 2009; 284: 3793–3803.
Cheng H, Vetrivel KS, Drisdel RC, Meckler X, Gong P, Leem JY et al. S-palmitoylation of gamma-secretase subunits nicastrin and APH-1. J Biol Chem 2009; 284: 1373–1384.
Drisdel RC, Green WN . Labeling and quantifying sites of protein palmitoylation. Biotech 2004; 36: 276–285.
Resh MD . Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods 2006; 40: 191–197.
Huang K, Sanders SS, Kang R, Carroll JB, Sutton L, Wan J et al. Wild-type HTT modulates the enzymatic activity of the neuronal palmitoyl transferase HIP14. Hum Mol Genet 2011; 20: 3356–3365.
Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet 2000; 9: 2799–2809.
Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS et al. Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res 2011; 52: 393–398.
Ehrnhoefer DE, Skotte NH, Savill J, Nguyen YTN, Ladha S, Cao L-P et al. A quantitative method for the specific assessment of Caspase-6 activity in cell culture. PLoS One 2011; 6: e27680.
Resh MD . Membrane targeting of lipid modified signal transduction proteins. Subcell Biochem 2004; 37: 217–232.
Li S, Li J, Ning L, Wang S, Niu Y, Jin N et al. In silico identification of protein s-palmitoylation sites and their involvement in human inherited disease. J Chem Inf Model 2015; 55: 2015–2025.
Warby SC, Doty CN, Graham RK, Carroll JB, Yang YZ, Singaraja RR et al. Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum Mol Genet 2008; 17: 2390–2404.
Edgington LE, van Raam BJ, Verdoes M, Wierschem C, Salvesen GS, Bogyo M . An optimized activity-based probe for the study of caspase-6 activation. Chem Biol 2012; 19: 340–352.
Velázquez-Delgado EM, Hardy JA . Phosphorylation Regulates assembly of the Caspase-6 substrate-binding groove. Structure 2012; 20: 742–751.
Stanger K, Steffek M, Zhou L, Pozniak CD, Quan C, Franke Y et al. Allosteric peptides bind a caspase zymogen and mediate caspase tetramerization. Nat Chem Biol 2012; 8: 655–660.
Heise CE, Murray J, Augustyn KE, Bravo B, Chugha P, Cohen F et al. Mechanistic and structural understanding of uncompetitive inhibitors of Caspase-6. PLoS One 2012; 7: e50864.
Müller I, MBAC Lamers, Ritchie AJ, Dominguez C, Munoz-Sanjuan I, Kiselyov A . Structure of human caspase-6 in complex with Z-VAD-FMK: new peptide binding mode observed for the non-canonical caspase conformation. Bioorg Med Chem Lett 2011; 21: 5244–5247.
Müller I, MBAC Lamers, Ritchie AJ, Park H, Dominguez C, Munoz-Sanjuan I et al. A new apo-caspase-6 crystal form reveals the active conformation of the apoenzyme. J Mol Biol 2011; 410: 307–315.
Vaidya S, Velázquez-Delgado EM, Abbruzzese G, Hardy JA . Substrate-induced conformational changes occur in all cleaved forms of caspase-6. J Mol Biol 2011; 406: 75–91.
Cao Q, Wang XJ, Li LF, Su XD . The regulatory mechanism of the caspase 6 pro-domain revealed by crystal structure and biochemical assays. Acta Cryst 2014; D70: 58–67.
Yanai A, Huang K, Kang R, Singaraja RR, Arstikaitis P, Gan L et al. Palmitoylation of huntingtin by HIP14is essential for its trafficking and function. Nat Neurosci 2006; 9: 824–831.
Tabrizi SJ, Langbehn DR, Leavitt BR, Roos RA, Dürr A, Craufurd D et al. Biological and clinical manifestations of Huntington's disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol 2009; 8: 791–801.
Sutton LM, Sanders SS, Butland SL, Singaraja RR, Franciosi S, Southwell AL et al. Hip14l-deficient mice develop neuropathological and behavioural features of Huntington disease. Hum Mol Genet 2013; 22: 452–465.
Dagbay K, Eron SJ, Serrano BP, Velázquez-Delgado EM, Zhao Y, Lin D et al. A multipronged approach for compiling a global map of allosteric regulation in the apoptotic caspases. Meth Enzymol 2014; 544: 215–249.
Shipston MJ . Ion channel regulation by protein palmitoylation. J Biol Chem 2011; 286: 8709–8716.
Salaun C, Greaves J, Chamberlain LH . The intracellular dynamic of protein palmitoylation. J Cell Biol 2010; 191: 1229–1238.
Singaraja RR, Kang MH, Vaid K, Sanders SS, Vilas GL, Arstikaitis P et al. Palmitoylation of ATP-binding cassette transporter A1 is essential for its trafficking and function. Circ Res 2009; 105: 138–147.
Huang K, Sanders S, Singaraja R, Orban P, Cijsouw T, Arstikaitis P et al. Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J 2009; 23: 2605–2615.
Wong BKY, Ehrnhoefer DE, Graham RK, Martin DDO, Ladha S, Uribe V et al. Neurobiol Dis 2015; 76: 24–36.
Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, Skotte N et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther 2011; 19: 2178–2185.
Graham RK, Deng Y, Pouladi MA, Vaid K, Ehrnhoefer D, Southwell AL et al. Caspase-6-resistant mutant huntingtin does not rescue the toxic effects of caspase-cleavable mutant huntingtin in vivo. J Huntingtons Dis 2012; 1: 243–260.
Weiss A, Abramowski D, Bibel M, Bodner R, Chopra V, DiFiglia M et al. Single-step detection of mutant huntingtin in animal and human tissues: a bioassay for Huntington’s disease. Anal Biochem 2009; 395: 8–15.
Schroeder T, Barandun J, Flütsch A, Briand C, Mittl PRE, Grütter MG . Specific inhibition of Caspase-3 by a competitive DARPin: molecular mimicry between native and designed inhibitors. Structure 2013; 21: 277–289.
Case DA, Betz RM, Botello-Smith W, Cerutti DS, Cheatham, III, TE, Darden TA et al. (2009). AMBER 2009, University of California, San Francisco..
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML . Comparison of simple potential functions for simulating liquid water. J Chem Phys 1983; 79: 926.
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 2003; 24: 1999–2012.
Khoury GA, Thompson JP, Smadbeck J, Kieslich CA, Floudas CA . Forcefield_PTM: ab initio charge and AMBER forcefield parameters for frequently occurring post-translational modifications. J Chem Theory Comput 2013; 9: 5653–5674.
Darden T, York D, Pedersen L . Particle mesh Ewald: an Nlog(N) method for Ewald sums in large systems. J Chem Phys 1993; 98: 10089.
S M, PA K . Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 1992; 13: 952–962.
Humphrey W, Dalke A, Schulten K . VMD: visual molecular dynamics. J Mol Graph 1996; 14: 33–8–27–8.
De Lano W The PyMOL Molecular Graphics System.
Butland SL, Sanders SS, Schmidt ME, Riechers SP, Lin DTS, Martin DDO et al. The palmitoyl acyltransferase HIP14 shares a high proportion of interactors with huntingtin: implications for a role in the pathogenesis of Huntington's disease. Hum Mol Genet 2014; 23: 4142–4160.
Acknowledgements
We thank Dr Matthew Bogyo and Dr Laura E Edgington who kindly provided the caspase-activity probe LE22 and Dr Luke Chamberlain who provided a protocol for the ARAC assay. NHS was support by a Canadian Institutes of Health Research (CIHR) postdoctoral fellowship award. RRS was supported by the Michael Smith Foundation for Health Research (MSFHR). SSS was supported by CIHR and MSFHR doctoral fellowship awards. MRH is a University Killam professor and holds a Canada Research Chair in human genetics and molecular medicine. This work was funded by a grant from CIHR (GPG-102165).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by H Ichijo
Rights and permissions
About this article
Cite this article
Skotte, N., Sanders, S., Singaraja, R. et al. Palmitoylation of caspase-6 by HIP14 regulates its activation. Cell Death Differ 24, 433–444 (2017). https://doi.org/10.1038/cdd.2016.139
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.139
This article is cited by
-
Protein lipidation in health and disease: molecular basis, physiological function and pathological implication
Signal Transduction and Targeted Therapy (2024)
-
How does caspases regulation play role in cell decisions? apoptosis and beyond
Molecular and Cellular Biochemistry (2023)
-
Enhanced cerebral branched-chain amino acid metabolism in R6/2 mouse model of Huntington’s disease
Cellular and Molecular Life Sciences (2019)
-
The Contribution of Necroptosis in Neurodegenerative Diseases
Neurochemical Research (2017)