Abstract
Tumor-suppressor p53 is frequently mutated in human cancers. Many tumor-associated mutant p53 (mutp53) proteins gain new functions in promoting tumorigenesis, defined as gain-of-function (GOF). The mechanisms for mutp53 GOF are not well understood. Here, we report Pontin, a highly conserved AAA+ ATPase important for various cellular functions, as a new mutp53-binding protein. This Pontin–mutp53 interaction promotes mutp53 GOF in invasion, migration and anchorage-independent growth of tumor cells. The ATPase domain of Pontin is crucial for its promoting effect on mutp53 GOF; blocking the ATPase activity of Pontin by a Pontin-specific ATPase inhibitor or an ATPase-deficient dominant-negative Pontin expression vector greatly diminished mutp53 GOF. Pontin promotes mutp53 GOF through regulation of mutp53 transcriptional activity; knockdown of Pontin abolished the transcriptional regulation of mutp53 toward a group of genes. Furthermore, overexpression of Pontin in tumors is associated with the poor survival in cancer patients, especially those containing mutp53. Our results highlight an important role and mechanism for Pontin, a new mutp53 partner, in promoting mutp53 GOF in tumorigenesis.
Similar content being viewed by others
Main
Tumor-suppressor p53 has a crucial role in tumor prevention.1 p53 function is compromised in many human tumors, mostly due to p53 mutation. The majority of tumor-associated p53 mutations are point mutations in p53 DNA-binding domain. Major categories of p53 mutations include DNA contact mutations (including R248Q and R273H) and conformational mutations (including R175H).2 Many tumor-associated mutant p53 (mutp53) proteins gain new properties to promote tumorigenesis in addition to the loss of tumor-suppressive activity of wild-type p53 (wtp53), which is defined as mutp53 gain-of-function (GOF). The mutp53 GOF includes the abilities to promote proliferation, metastasis, cell transformation and metabolic changes.2, 3, 4, 5 Mutp53 proteins often accumulate to high levels in tumors but not normal tissues, which is critical for mutp53 GOF in tumorigenesis.3, 4 It has been suggested that chaperone protein Hsp90, which is frequently overexpressed in tumors, interacts with mutp53 to prevent its degradation, promoting mutp53 accumulation in tumors.6 Recently, we found that tumor-associated MDM2 isoforms inhibit MDM2-mediated mutp53 degradation and promote mutp53 accumulation in tumors.7
Although the concept of mutp53 GOF is well established, the underlying mechanisms for mutp53 GOF are not well understood. Mechanisms being proposed include protein interaction and the regulation of gene expression and microRNA expression.8 In particular, the interaction of mutp53 with proteins to regulate their functions has been suggested as an important mechanism. For example, mutp53 interacts with transcriptional factors p63, p73, SREBP and VDR to modulate their transcriptional activities.9, 10, 11 Mutp53 forms protein complex with MRE11, and perturbs its function in DNA damage response.12 Furthermore, mutp53 function can be regulated by its interacting proteins. For example, PML interacts with mutp53 and enhances mutp53 transcriptional activity.13 Pin1, which regulates protein conformation, binds to mutp53 and enhances mutp53 GOF.14
Pontin is a highly conserved AAA+ ATPase, which is associated with several protein complexes and involved in multiple biological processes, including chromatin remodeling, transcriptional regulation, cell replication and telomerase activities.15, 16, 17 Some Pontin-binding proteins, including Tip60, c-Myc and β-catenin, have known functions in cancer.16, 18, 19 Pontin is frequently overexpressed in many human cancers, suggesting the potential role of Pontin in tumorigenesis.20, 21 In this study, we identified Pontin as a novel mutp53-binding protein, which enhances mutp53 GOF in tumor cells. Results from this study showed an important role of Pontin in promoting mutp53 GOF and revealed an unidentified mechanism for mutp53 GOF in tumorigenesis.
Results
Pontin is a novel mutp53-interacting protein
Recently, Pontin was reported to bind to the p53 promoter and downregulate wtp53 levels and function in human colorectal cancer RKO cells.22 This finding raised an interesting question whether Pontin regulates mutp53 accumulation and GOF. We first confirmed that Pontin downregulates wtp53 levels in RKO cells; knockdown of Pontin increased both the mRNA and protein levels of wtp53 (Supplementary Figure S1). However, this effect of Pontin on wtp53 appeared to be cell type specific because Pontin did not affect wtp53 at either mRNA or protein level in human colorectal cancer HCT116 p53+/+ cells (Supplementary Figure S1). Further, Pontin did not regulate mutp53 levels in cells. In human breast cancer SK-BR-3 and colorectal cancer HCT116 p53R248W/– cells, which contain a single copy of p53 gene with tumor hotspot mutations R175H and R248W, respectively, and human p53-null lung cancer H1299 cells with stable ectopic expression of mutp53 (R175H), knockdown of Pontin did not clearly affect mutp53 mRNA and protein levels (Supplementary Figure S1).
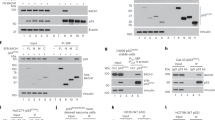
Interestingly, we found that Pontin interacts with mutp53 protein. In H1299 cells transfected with human Pontin-Flag expression vectors together with wtp53 or mutp53 (R175H) expression vectors, Pontin preferentially bound to mutp53 compared with wtp53 as determined by co-immunoprecipitation (co-IP) assays (Figure 1a). The Pontin–mutp53 interaction was also observed in H1299 cells with ectopic expression of different hotspot mutp53 proteins, including R248Q and R273H, in addition to R175H (Figure 1b). The endogenous Pontin–mutp53 interaction was also observed in SK-BR-3 cells (Figure 1c).
Pontin is a novel mutp53-interacting protein in both human cells and mutp53R172H/R172H tumors. (a) Pontin preferentially bound to ectopically expressed mutp53 (R175H) protein compared with wtp53 in human p53-null H1299 cells. H1299 cells transiently transfected with expression vectors of wtp53 or mutp53 (R175H) along with Pontin-Flag expression vectors. Antibodies used for IP: Flag for Pontin-Flag and DO-1 for p53. (b) Pontin interacted with several hotspot mutp53 proteins. H1299 cells were transiently transfected with expression vectors of wtp53 or mutp53 (R175H, R248Q or R273H) along with Pontin-Flag expression vectors. (c) The interaction of endogenous Pontin with mutp53 (R175H) was observed in human SK-BR-3 cells but not SK-BR-3-p53sh cells with stable knockdown of endogenous mutp53 by shRNA. (d) Pontin interacted with mutp53 at its DBD. Left panel: schematic diagram showing the domain structure of mutp53 and the fragments of mutp53 and wtp53 proteins used for co-IP assays. Right panel: H1299 cells transiently transfected with expression vectors of different fragments of mutp53 and wtp53 along with Pontin-Flag expression vectors. Antibodies used for IP: Flag for Pontin-Flag. (e) The interaction of Pontin with mouse mutp53 protein was observed in thymic lymphomas from p53R172H/R172H mice expressing high levels of mutp53 protein but not normal thymic tissues from p53R172H/R172H mice expressing very low levels of mutp53 protein or thymic lymphomas from p53−/− mice. Tumors and normal thymic tissues were subjected to co-IP assays using an anti-p53 (FL-393) antibody
To determine the regions of mutp53 required for the Pontin–mutp53 interaction, expression vectors of different mutp53 domains (Figure 1d, left panel) were co-transfected with Pontin-Flag into H1299 cells. Expression vectors of wtp53 DNA-binding domain (DBD) were included as a control. Pontin interacted with all mutp53 fragments containing DBD, but not with mutp53 fragment lacking DBD or wtp53 DBD (Figure 1d). These results show that Pontin interacts with mutp53 through mutp53 DBD domain.
To investigate whether Pontin interacts with mouse mutp53, p53R172H/R172H mice with knock-in of mutp53 R172H (equivalent to human mutp53 R175H) and p53−/− mice, which mainly develop thymic lymphomas, were used. Thymic lymphomas and normal thymus from p53R172H/R172H mice, and thymic lymphomas from p53−/− mice were subjected to co-IP assays using an anti-p53 antibody. Consistent with previous reports,3, 4 mutp53 protein accumulation was observed in thymic lymphomas but not normal tissues from p53R172H/R172H mice. The Pontin–mutp53 interaction was observed in thymic lymphomas from p53R172H/R172H mice but not normal thymus from p53R172H/R172H mice or thymic lymphomas from p53−/− mice (Figure 1e). Together, these results show that Pontin is a novel mutp53-binding protein, and this interaction is conserved in both human tumor cells and mouse tissues.
Loss of the Pontin–mutp53 complex compromises mutp53 GOF
To investigate whether the Pontin–mutp53 interaction contributes to mutp53 GOF, endogenous Pontin was knocked down and its impact upon mutp53 GOF was evaluated in p53-null H1299 and human osteosarcoma Saos2 cells with stable ectopic expression of mutp53 R175H (H1299-R175H and Saos2-R175H), respectively. Promoting tumor cell metastasis is an important mutp53 GOF.3, 4 Migration and invasion are critical steps for tumor metastasis. As shown in Figures 2a and b and Supplementary Figures S2a and b, H1299-R175H and Saos2-R175H cells displayed increased migration and invasion abilities compared with H1299-Con and Saos2-Con cells transfected with control vectors as determined by transwell assays. Notably, knockdown of Pontin by siRNA decreased the migration and invasion abilities of H1299-R175H and Saos2-R175H but not H1299-Con and Saos2-Con cells (Figures 2a and b and Supplementary Figures S2a and b). Similarly, in SK-BR-3 cells containing endogenous mutp53 R175H, Pontin knockdown significantly decreased the migration and invasion abilities of SK-BR-3 but not SK-BR-3-p53sh cells with stable mutp53 knockdown (Figures 2c and d). The Pontin knockdown was confirmed at both mRNA and protein levels by Taqman real-time PCR and western blot assays, respectively (Supplementary Figures S2c and S3). Similar results were obtained by using two different siRNA oligos against Pontin. In addition to mutp53 R175H, Pontin knockdown decreased the migration ability of H1299 cells with stable ectopic expression of mutp53 R248Q and R273H, respectively (Supplementary Figure S4). This effect of Pontin knockdown on mutp53 GOF in migration and invasion was not because of the changes in cell cycle distribution, cell viability or apoptosis as determined in H1299-Con and H1299-R175H cells with or without Pontin knockdown (Supplementary Figure S5). The effect of Pontin knockdown on mutp53 GOF in migration was further confirmed by scratch motility assays. Motility of H1299-R175H cells was faster than that of H1299-Con cells (Figure 2e). Notably, Pontin knockdown significantly reduced the motility of H1299-R175H but not H1299-Con cells (Figure 2e). The effect of Pontin on mutp53 GOF in metastasis was further investigated in vivo. H1299-Con and H1299-R175H cells with or without knockdown of Pontin were injected into nude mice via the tail vein and lung metastasis was analyzed. As shown in Figure 2f, mutp53-promoted tumor lung metastasis; H1299-R175H cells formed significantly higher number of lung metastatic tumors compared with H1299-Con cells. Notably, Pontin knockdown greatly reduced lung tumor formation by H1299-R175H cells, and this effect was largely reduced in H1299-Con cells.
Knockdown of Pontin decreases mutp53 GOF in migration, invasion and anchorage-independent growth. (a) Knockdown of Pontin by siRNA inhibited migration of H1299-R175H but not H1299-Con cells as determined by transwell assays. Left panel: representative images from a portion of field; right panel: quantification of average number of migrated cells/field (× 200 magnification). Two different siRNA oligos were used and very similar results were obtained. For the sake of clarity, in (a– e), results from one siRNA were presented. (b) Knockdown of Pontin decreased invasion of H1299-R175H cells but not H1299-Con cells as determined by transwell assays in chambers coated with matrigel. (c and d) Pontin knockdown reduced the migration (c) and invasion (d) abilities of SK-BR-3-Consh cells but not SK-BR-3-p53sh cells. SK-BR-3 cells stably transfected with control shRNA (Consh) or shRNA against p53 (p53sh) were transiently transfected with control siRNA (Con) or siRNA against Pontin. (e) Pontin knockdown reduced the motility of H1299-R175H but not H1299-Con cells as determined by scratch motility assays. (f) Pontin knockdown significantly reduced lung tumors formed by H1299-R175H cells but not H1299-Con cells. H1299-Con and H1299-R175H cells with or without knockdown of Pontin were injected into nude mice via the tail vein. The number and area of lung tumors were determined after 6 weeks (n=8 per group). Left panel: representative H&E images of lung sections. Black arrows indicate tumors. Scale bar: 100 μm. (g) Pontin knockdown inhibited anchorage-independent cell growth on soft agar of H1299-mutp53 (R175H, R248Q, and R273H) cells but not H1299-Con cells. H1299-mutp53 and H1299-Con cells were transfected with shRNA against Pontin or control shRNA before assays. Left panel: representative images; right panel: relative colony numbers of cells on soft agar. For a– g, data are presented as mean±S.D. (n=3 for a– e, g; n=8 for f)
Mutp53 promotes the anchorage-independent growth of tumor cells.5 H1299-R175H, H1299-R248Q and H1299-R273H cells displayed increased anchorage-independent cell growth on soft agar compared with H1299-Con cells (Figure 2g). Notably, Pontin knockdown by shRNA significantly reduced the anchorage-independent growth of H1299-R175H, H1299-R248Q and H1299-R273H but not H1299-Con cells (Figure 2g). Together, these results show that Pontin knockdown decreases mutp53 GOF in migration, invasion, metastasis and anchorage-independent cell growth of tumor cells.
The C-terminus of Pontin is required for the Pontin–mutp53 interaction and mutp53 GOF
Pontin has following three distinct domains. Domain I (1–120 aa and 296–365 aa) contains Walker A and Walker B motifs that are responsible for binding and hydrolysis of ATP, respectively. Domain II (121–295 aa) is important for DNA/RNA binding. Domain III (368–456 aa) contains 4 α-helices and is important for ATP binding.23 To determine the regions of Pontin required for the Pontin–mutp53 interaction, expression vectors of different domains of Pontin (Figure 3a) were co-transfected with mutp53 (R175H) expression vectors into H1299 cells for co-IP assays. As shown in Figure 3b, mutp53 interacted with the Pontin fragments containing the C-terminus, including aa 292–456 and Δaa 125–291, which both contain Walker B motif in the C-terminus of domain I and domain III. Mutp53 did not interact with either the N terminus of Pontin (aa 1–124), which contains Walker A motif in domain I, or the Pontin domain II (aa 191–295). These results show that Pontin interacts with mutp53 through its C-terminus.
The C-terminus of Pontin is required for Pontin–mutp53 interaction and promotes mutp53 GOF. (a) Schematic diagram showing the domain structure of Pontin and the fragments of Pontin protein used for co-IP assays. (b) Pontin fragments containing the C-terminus of Pontin bound to mutp53 (R175H) protein as determined by co-IP assays. H1299 cells transiently transfected with expression vectors of different fragments of Pontin-Flag protein along with mutp53 (R175H) expression vectors. Antibodies used for IP: Flag for Pontin fragment-Flag. (c and d) Ectopic expression of Pontin fragments containing the C-terminus of Pontin abolished the inhibitory effect of Pontin knockdown on mutp53 GOF in migration (c) and invasion (d). H1299-Con and H1299-R175H cells with knockdown of Pontin by siRNA were transfected with expression vectors of siRNA resistant Pontin-Flag fragments or full-length Pontin-Flag (FL-Pontin-R). The migration and invasion abilities of cells were determined by transwell assays in chambers coated without or with matrigel, respectively. For (c and d), data are presented as mean±S.D. (n=3). The knockdown of endogenous Pontin as well as the expression of Pontin fragments and FL-Pontin-R were determined by western blot assays using anti-Pontin (for endogenous Pontin and FL-Pontin-R) and anti-Flag (for Pontin-Flag fragments and FL-Pontin-R) antibodies (c, lower panel). For (c and d), data are presented as mean±S.D. (n=3)
To determine the regions of Pontin protein required for mutp53 GOF, the effects of different Pontin fragments on mutp53 GOF in migration and invasion were examined. H1299-Con and H1299-R175H cells were first transfected with siRNA to knockdown endogenous Pontin, and then transfected with the vectors expressing different Pontin fragments or the full-length siRNA-resistant Pontin (FL-Pontin-R) that are all resistant to Pontin siRNAs. Knockdown of endogenous Pontin significantly inhibited migration and invasion of H1299-R175H but not H1299-Con cells, which was fully reversed by the expression of FL-Pontin-R (Figures 3c and d). Interestingly, expression of the Pontin fragments containing the C-terminus (aa 292–456 and Δaa 125–291, respectively), which are the fragments that interact with mutp53, largely reversed the inhibitory effect of Pontin knockdown on mutp53 GOF in migration and invasion (Figures 3c and d). In contrast, expression of the Pontin fragments aa 1–124 and aa 121–295, which do not interact with mutp53, had no obvious effect on migration and invasion of H1299-R175H cells with knockdown of endogenous Pontin. These results suggest that the C-terminus of Pontin is required for Pontin’s function in promoting mutp53 GOF.
The ATPase activity in Walker B motif is required for Pontin to promote mutp53 GOF
As both Pontin fragments (aa 292–456 and Δaa 125–291) that interact with mutp53 and promote mutp53 GOF contain the conserved Walker B motif, which is responsible for the ATPase activity, we investigated whether the ATPase activity is required for Pontin to promote mutp53 GOF. A single missense mutation in aa 302 in Walker B motif (DEVH to NEVH) was reported to create an ATPase-deficient dominant-negative mutant Pontin-D302N.24 Pontin-D302N expression vectors were co-transfected with mutp53-R175H into H1299 cells. As shown in Figure 4a, Pontin-D302N interacted with mutp53 to a similar degree compared with wild-type Pontin. Interestingly, Pontin-D302N greatly decreased the migration and invasion abilities of H1299-R175H but not H1299-Con cells, which is similar to the effect of Pontin knockdown (Figures 4b and c). Consistently, blocking Pontin’s ATPase activity by Rottlerin, a Pontin-specific ATPase inhibitor,25 significantly decreased the migration ability of H1299-R175H but not H1299-Con cells (Figure 4d). To confirm that the inhibitory effect of Rottlerin on the migration ability of mutp53 was due to its inhibition of Pontin’s ATPase activity but not nonspecific effects of Rottlerin, cells were simultaneously treated with Rottlerin and transfected with Pontin siRNA. Rottlerin treatment or Pontin knockdown decreased the migration ability of H1299-R175H cells to a similar extent (Figure 4e). Notably, combined treatments of Rottlerin and Pontin knockdown did not exhibit an additive or synergistic effect in H1299-R175H cells (Figure 4e). Furthermore, blocking Pontin’s ATPase activity by Pontin-D302N significantly reduced the anchorage-independent cell growth in H1299-R175H but not H1299-Con cells (Figure 4f). These results show that the ATPase activity of Walker B motif is important for Pontin’s function in promoting mutp53 GOF.
The ATPase activity in Walker B motif is required for Pontin to promote mutp53 GOF in migration, invasion and anchorage-independent cell growth. (a) The ATPase-deficient dominant-negative mutant Pontin-D302N bound to mutp53 protein. H1299 cells were co-transfected with expression vectors of mutp53 (R175H) along with wild-type Pontin or Pontin-D302N for co-IP assays. (b and c) Ectopic expression of Pontin-D302N largely blocked mutp53 GOF in migration (b) and invasion (c) as determined by transwell assays. H1299-Con and H1299-R175H cells were transfected with expression vectors of Pontin-D302N or control vectors. Cells were also transfected with siRNA against Pontin or control siRNA to compare the effect of Pontin-D302N with knockdown of endogenous Pontin on mutp53 GOF in migration and invasion. (d) Rottlerin, a Pontin-specific small molecular ATPase inhibitor, greatly compromised mutp53 GOF in migration. H1299-Con and H1299-R175H cells were treated with Rottlerin (0.5 μM) for 48 h, and their migration abilities were determined by transwell assays. (e) Combined treatments of Rottlerin and Pontin knockdown did not have an additive or synergistic effect on downregulation of mutp53 GOF. H1299-Con and H1299-R175H cells were subjected to following treatments: transfection with siRNA against Pontin, treatment with Rottlerin or Pontin knockdown by siRNA followed by Rottlerin treatment as indicated. The migration abilities of cells were determined by transwell assays. (f) Ectopic expression of Pontin-D302N inhibited anchorage-independent cell growth on soft agar of H1299-R175H cells but not H1299-Con cells. H1299-R175H and H1299-Con cells were transfected with control or Pontin-D302N expression vectors. For (b– f), data are presented as mean±S.D. (n=3)
Pontin promotes mutp53 GOF through regulation of mutp53 transcriptional activity
Pontin is associated with several chromatin-remodeling complexes, including Ino80, TIP60/NuA4 and SWR1 complexes, and has an important role in chromatin-remodeling and transcriptional regulation.15, 16 Mutp53 can transcriptionally regulate a group of genes involved in tumorigenesis.8 Here, we investigated whether Pontin promotes mutp53 GOF through regulating mutp53 transcriptional activity. First, immunofluorescence (IF) staining assays in H1299-R175H and SK-BR-3 cells showed that both Pontin and mutp53 proteins were predominantly localized in the nucleus. Furthermore, Pontin was largely colocalized with mutp53 in the nucleus (Figure 5a). The transcriptional regulation ability of mutp53 was then determined in SK-BR-3-Consh cells transduced with control shRNA and SK-BR-3-p53sh cells transfected with control siRNA or Pontin siRNA by Affymetrix microarray GeneChip analysis. The expression data were analyzed using Affymetrix Microarray Suite version 5.0 (MAS 5.0, Santa Clara, CA, USA). Change of expression levels for each gene by mutp53 was calculated as expression levels in SK-BR-3-Consh compared with SK-BR-3-p53sh cells. As shown in Figure 5b, the expression levels of a group of genes were significantly altered (P<0.01) following mutp53 knockdown in SK-BR-3 cells; knockdown of mutp53 resulted in decreased expression (>2-fold) of 67 genes and increased expression (>2-fold) of 31 genes. A more detailed list of genes regulated by mutp53 is shown in Supplementary Table S1. Notably, Pontin knockdown reduced the transcriptional regulation of mutp53 toward a group of genes (Figure 5b). These genes that require Pontin for their transcriptional regulation by mutp53 (n=40 for genes upregulated by mutp53; n=12 for genes downregulated by mutp53) were involved in many important signaling pathways, including gap junction signaling, IGF-1 signaling, EGF signaling etc., as analyzed by ingenuity pathway analysis (IPA; Figure 5c). The dysregulation of these pathways has an important role in tumorigenesis and metastasis.26, 27, 28 The requirement of Pontin in mutp53 function in transcriptional regulation was confirmed in a group of represented genes, including ADRB1, receptor (TNFRSF)-interacting serine-threonine kinase 1 (RIPK1), inositol 1,4,5-triphosphate receptor, type I (ITPR1), P2RX5, stathmin 1 (STMN1) and SH3BGR by real-time PCR (Figure 5d). The gene products of ADRB1, RIPK1 belong to the gap junction signaling, which is important for cell motility.29, 30, 31 STMN1, which has a role in cell migration, was reported to be upregulated by mutp53.32 It is possible that some of these genes mediate the promoting effect of Pontin on mutp53 GOF. Mutp53 often transcriptionally regulates genes through interacting with other transcription factors, including SREBP, NF-Y, VDR, NF-κB, and binding to the binding sites of their regulated genes.8, 11, 33, 34 Using the Champion ChiP Transcription Factor Search (SABiosciences, Valencia, CA, USA) to analyze the promoter and regulatory region of above-mentioned genes, we identified a SREBP-binding site in the intron 1 of the STMN1 gene, and NF-κB-binding sites in the intron 1 of the RIPK1 and ITPR1 genes. Using chromatin IP (ChIP) assays, significant binding of mutp53 and Pontin was observed in the vicinity of the SREBP-binding site of the STMN1 gene and the NF-κB-binding sites of the RIPK1 and ITPR1 genes in SK-BR-3 cells (Figure 5e). Pontin knockdown resulted in the loss of Pontin interaction with these regions, and largely reduced mutp53 interaction with these regions (Figure 5e). STMN1 was reported to promote cell migration.35 We found that STMN1 partially mediates mutp53 GOF in migration in a Pontin-dependent manner (Figure 5f). Knockdown of STMN1 by siRNA reduced the migration ability of H1299-R175H but not H1299-Con cells (Figure 5f). Notably, the inhibitory effect of STMN1 knockdown on mutp53 GOF in migration was largely disappeared in H1299-R175H cells with knockdown of Pontin (Figure 5f). These results suggest that the Pontin–mutp53 interaction regulates mutp53 transcriptional activity, which in turn promotes mutp53 GOF.
The interaction of Pontin with mutp53 is crucial for the transcriptional regulation of mutp53 toward a group of genes. (a) Pontin was predominantly colocalized with mutp53 in the nucleus in H1299-R175H and SK-BR-3 cells as determined by IF staining. Anti-p53 and anti-Pontin antibodies were used for IF. Nuclei were stained with DAPI. Scale bar: 10 μM. (b) Knockdown of Pontin in SK-BR-3 cells reduced the transcriptional activity of mutp53 toward a subset of genes. Heatmap represents the change of gene expression levels by mutp53 obtained from microarray data of SK-BR-3-Consh and SK-BR-3-p53sh cells with or without knockdown of Pontin. The color scale represents the fold change of gene expression levels by mutp53 (R175H). The genes that were upregulated are presented in red and genes that were downregulated are presented in green. (c) Blue bars represent top scoring pathways regulated by mutp53 that are significantly changed in SK-BR-3 cells with or without knockdown of Pontin. Data were analyzed by using of IPA. (d) Validation of microarray data by real-time PCR. The expression levels of a list of genes, including ADRB1, PIPK1, ITPR1, P2RX5, STMN1 and SH3BGR were determined by real-time PCR in SK-BR-3-Consh and SK-BR-3-p53sh cells with or without knockdown of Pontin by siRNA. All expression levels were normalized to actin. (e) Mutp53 and Pontin bound to the DNA regions in the vicinity of the SREBP-binding site in the STMN1 gene and the NF-κB-binding sites in the RIPK1 and ITPR1 genes in SK-BR-3 cells but not SK-BR-3 cells with knockdown of Pontin. Upper panel: the putative SREBP-binding site in the STMN1 gene and the NF-κB-binding sites in the RIPK1 and ITPR1 genes. Lower panel: SK-BR-3 cells were subjected to ChIP assays using antibodies against p53, Pontin and mouse IgG antibodies, respectively. A region of the coding sequence of the STMN1 gene was included for ChIP assays as a negative control. (f) STMN1 mediated Pontin’s promoting effect on mutp53 GOF in migration. Knockdown of STMN1 by siRNA reduced the migration ability of H1299-R175H cells but not H1299-Con cells. This effect of STMN1 knockdown on mutp53 GOF in migration was largely disappeared in H1299-R175H cells with knockdown of Pontin. Upper panel: representative images from a portion of field; middle panel: quantification of average number of migrated cells/field (200 × magnification). The knockdown of endogenous STMN1 and Pontin was determined by western blot assays (lower panel). For (d– f), data are presented as mean±S.D. (n=3)
High Pontin expression in tumors correlates with poor prognosis, especially for tumors carrying mutp53
Pontin is frequently overexpressed in many human cancers, including breast, lung and colorectal cancers.20, 21 Here, we investigated the prognostic value of Pontin by using the KM Plotter, an online tool that incorporates microarray data with overall survival information of 1926 lung and 1115 breast cancer patients.36 Patients were divided into two groups by auto-selection of best cutoff. There was a significant association of high Pontin mRNA expression levels with poor prognosis in both lung (HR=1.62, P=8.3e-14) and breast (HR=1.61, P=0.00066) cancer patients (Figures 6a and b), which suggests the significant prognostic value of Pontin expression levels for lung and breast cancer patients. Pontin promotes mutp53 transcriptional activity toward a group of genes, including STMN1 and RIPK1. Notably, the prognostic value of Pontin together with STMN1 and RIPK1 was stronger than that of Pontin alone (Figures 6c and d). These results suggest that regulation of mutp53 transcriptional activity is an important mechanism by which Pontin promotes tumor progression.
High levels of Pontin are associated with poor prognosis, especially in patients with mutp53 in tumors. (a and b). Kaplan–Meier curves indicating the overall survival of 1926 lung cancer patients (a) and 1115 breast cancer patients (b) with different expression levels of Pontin. (c and d) Kaplan–Meier curves indicating the overall survival of 1926 lung cancer patients (c) and 1115 breast cancer patients (d) with different expression levels of Pontin, STMN1 and RIPK1. In (a– d), the overall survival information and expression levels of Pontin, STMN1 and RIPK1 of patients were obtained from the KM plotter (2015 version for lung cancer and 2014 version for breast cancer). The Affymetrix IDs used for analysis are: 201614_s_at for Pontin; 217714_x_at for STMN1 and 209941_at for RIPK1. Tumors with high levels of Pontin are associated with poor survival in both lung and breast cancer patients. Tumors with high levels of Pontin, STMN1 and RIPK1 are associated with poorer survival in both lung and breast cancer patients. (e) Kaplan–Meier curves indicating the overall survival of breast patients with different expression levels of Pontin in tumors containing mutp53 or wtp53. High levels of Pontin are associated with poor prognosis in breast cancer patients containing mutp53 in tumors but not in patients containing wtp53 in tumors. The survival information, expression levels of Pontin and p53 mutation status were obtained from the public available database (GSE3494). Left panel: total patients in the cohort; middle panel: patients carrying mutp53 in tumors; right panel: patients carrying wtp53 in tumors
To further investigate whether the promoting effect of Pontin on mutp53 GOF contributes to the poor survival associated with high Pontin expression levels, a cohort of 236 breast cancer patients (GSE3494) from public available database with known p53 mutation status and survival information was used.37 Consistent with the results obtained from the KM plotter, there was a significant association of high Pontin mRNA levels with poor prognosis in these patients (n=234, HR=1.69, P=0.047; Figure 6e, left panel). The prognostic value of Pontin mRNA levels was further compared between patients carrying wtp53 or mutp53 in tumors. Although Pontin levels showed no significant effect on prognosis in patients with wtp53 (n=181, HR=1.27, P=0.35), high Pontin mRNA levels were significantly associated with poor prognosis in patients with mutp53 (n=53, HR=2.43, P=0.034; Figure 6e, middle and right panels). These results show that high Pontin expression levels are associated with poor prognosis, especially for tumors carrying mutp53.
Discussion
Ample evidence has demonstrated that mutp53 can promote tumorigenesis through GOF mechanism.8 However, its underlying mechanisms are not well understood. In this study, Pontin was identified as a new mutp53-binding protein. Pontin preferentially binds to mutp53, through its C-terminus. Pontin is an essential component of several protein complexes that regulate transcription, telomerase activities, chromatin remodeling, metabolism and DNA repair.15, 16, 17 Pontin is frequently overexpressed in tumors. Some of the Pontin-interacting proteins, including c-Myc and β-catenin, have pivotal roles in tumorigenesis, suggesting an important role of Pontin in tumorigenesis.18, 19 However, the mechanism of Pontin in tumorigenesis is poorly understood. This study shows that Pontin promotes mutp53 GOF in tumorigenesis, including increased migration, invasion, metastasis and anchorage-independent cell growth. Our results established an important role of Pontin in promoting mutp53 GOF, and also provided a new mechanism for Pontin in tumorigenesis.
As an ATPase, Pontin has Walker A and Walker B motifs that are involved in ATP binding and ATP hydrolysis, respectively. The ATPase activity of Pontin is important for many of its functions. Results in this study show that Pontin-D302N, an ATPase-deficient dominant-negative mutant, binds to mutp53 to a similar extent compared with wild-type Pontin, indicating that the ATPase activity is not required for the Pontin–mutp53 interaction. Notably, Pontin-D302N, which competes with endogenous wild-type Pontin to interact with mutp53, greatly inhibits mutp53 GOF. Similarly, Rottlerin, a Pontin-specific ATPase inhibitor, greatly inhibits mutp53 GOF. Furthermore, although knockdown of endogenous Pontin inhibits mutp53 GOF, ectopic expression of the Pontin fragments containing the Walker B motif restores mutp53 GOF in cells with knockdown of endogenous Pontin. These results show that its ATPase activity is required for Pontin to regulate mutp53 GOF.
Pontin is a component of several chromatin-remodeling complexes, including Ino80 complex, TIP60/NuA4 complex and SWR1 complex, and is essential for their chromatic-remodeling activities and transcriptional regulation.15, 16 It is possible that the Pontin–mutp53 interaction recruits mutp53 to specific DNA-binding sites or chromatin landscapes, and facilitates mutp53 to transcriptionally regulate a group of genes directly or together with other transcription factors. Results from IF assays show that Pontin is predominantly localized in the nucleus and largely colocalized with mutp53. Knockdown of Pontin abolishes mutp53’s transcriptional activity toward a group of genes. Some genes that require Pontin for their transcriptional regulation by mutp53, including STMN1, ADRB1, ITPR1, RIPK1, CtBP2, TPD52 and TOX3, have been suggested to have roles in tumorigenesis, especially for migration and metastasis.30, 31, 38, 39, 40, 41 Results from ChIP assays show that mutp53 interacts with some target genes at their regulatory regions in a Pontin-dependent manner. It remains unclear why Pontin selectively regulates mutp53 transcriptional activity toward certain genes. Future studies should further increase our understanding on the role and mechanisms of Pontin in regulating mutp53 GOF.
Pontin overexpression has been observed in many human cancers.21 Results from this study and previous reports show that Pontin overexpression in tumors is strongly associated with poor prognosis.21 Interestingly, Pontin expression levels have a much stronger prognostic value in breast cancers containing mutp53 than those without mutp53. This observation suggests that Pontin has an important role in mutp53-promoted tumorigenesis, and highlights the biological significance of Pontin overexpression in tumors, especially those containing mutp53.
In summary, this study established Pontin as a new mutp53-binding protein, and demonstrated that Pontin promotes mutp53 GOF in tumorigenesis in an ATPase-dependent manner. Results from this study raise interesting and intriguing possibilities to block mutp53 GOF in tumors through targeting Pontin or its ATPase activity.
Materials and Methods
Cell culture, mouse strains, constructs and cell treatments
Human lung cancer H1299, breast cancer SK-BR-3, osteosarcoma Saos2, colorectal cancer RKO p53+/+ and HCT116 p53+/+ cell lines were obtained from ATCC (Manassas, VA, USA). Human HCT116 p53R248W/- cells were gifts from Dr. Bert Vogelstein at Johns Hopkins University. Stable cell lines expressing mutp53 R175H, R248Q and R273H were established as previously described.7 SK-BR-3-p53sh cells with knockdown of endogenous mutp53 were established by stable transduction of retroviral shRNA vectors against p53 into SK-BR-3 cells. p53−/− mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA) and p53R172H/R172H mice were gifts from Dr. Gigi Lozano at MD Anderson Cancer Center. Expression vectors of Flag-tagged Pontin and Pontin fragments were gifts from Dr. Steve Artandi at Stanford University. Expression vectors of Flag-tagged ATPase-deficient dominant-negative mutant Pontin-D302N were gifts from Dr. Michael Cole at Dartmouth College. Site-directed mutagenesis was performed to produce the expression vectors of Pontin and Pontin fragments resistant to siRNA by using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). The primers used for site-directed mutagenesis are: F: 5′-CCAAGATGTGACCTTGCACGATTTAGACGTGGCTAATGCGCGGCCC-3′; R: 5′-GGGCCGCGCATTAGCCACGTCTAAATCGTGCAAGGTCACATCTTGG-3′. The mutant sites are shown in bold.
Expression vectors of mutp53 fragments containing aa 1–363 and aa 43–363 were obtained by using site-directed mutagenesis to introduce R175H mutation into expression vectors of wtp53 fragments containing aa 1–363 and aa 43–363, respectively, which were generous gifts from Dr. Xinbin Chen at University of California, Davis. Expression vectors of mutp53 and wtp53 DBD were constructed by amplifying the fragments from pRC-mutp53-R175H and pRC-wtp53 vectors, respectively. Two-step overlap extension PCR was used to obtain mutp53 fragments without DBD. Primer sequences are listed in Supplementary Table S2. The final PCR products were inserted into pCMV-Flag vectors.
For siRNA knockdown, two siRNA oligos against Pontin and STMN1 were purchased from Integrated DNA Technologies (Coralville, IA, USA). siRNA targeting Pontin: siRNA-1: 5′-GCAUGACUUGGAUGUGGCUAAUGC-3′; siRNA-2: 5′-GGACCUCCUGGAACUGGCAAGACAG-3′. siRNA targeting STMN1: siRNA-1: 5′-AGAACCGAGAGGCACAAAUGGCUGC-3′; siRNA-2: 5′-GCAGAAGAAAGACGCAAGUCCCATG-3′. Transfection of cells with expression vectors or siRNA was performed by using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA). Retroviral shRNA vectors against Pontin (Thermo Scientific, Grand Island, NY, USA) were transduced into cells. For Rottlerin treatment, cells were treated with Rottlerin (Calbiochem, Billerica, MA, USA; 0.5 μM) for 48 h.
IP assays
IP assays were performed as previously described.7 In brief, 1 mg protein lysates were used for IP using anti-p53 (DO-1 for human cells and FL-393 for mouse tissues, Santa Cruz, Dallas, TX, USA), anti-Pontin (Sigma, St. Louis, MO, USA) and anti-Flag antibodies (Sigma) to pull-down p53, Pontin and Pontin-Flag proteins, respectively.
Quantitative real-time PCR
Total RNA was prepared from cells by using an RNeasy kit (Qiagen, Valencia, CA, USA) and treated with DNase I to remove residual genomic DNA. Real-time PCR was performed in triplicate with Taqman PCR Mix (Applied Biosystems, Grand Island, NY, USA) in the ABI Step-One Plus system. All primers were purchased from Applied Biosystems. The expression of genes was normalized with the β-actin gene.
Western blot assays
Standard western blot assays were used to analyze the levels of protein. Anti-p53 (FL-393) (Santa Cruz), anti-Pontin (Sigma), anti-Flag (Sigma), anti-STMN1 (Cell Signaling, Billerica, MA, USA) and anti-β-actin (Sigma) antibodies were used to determine the levels of p53, Pontin, Flag-tagged Pontin proteins (including full-length and fragments of Pontin), STMN1 and β-actin, respectively.
Cell migration and invasion assays
The transwell system (BD Biosciences, San Jose, CA, USA) was used for cell migration and invasion assays as previously described.7, 42 In brief, cells in FBS-free medium were seeded into upper chambers coated with or without matrigel (BD Biosciences) for invasion and migration assays, respectively. The lower chamber was filled with medium supplemented with 10% FBS. For SK-BR-3 cells, the lower chamber was filled with 1:1 mix of medium supplemented with 10% FBS and NIH 3T3 cell-conditioned medium. Cells on the lower surface of upper chambers were counted after culturing at 37 °C for 24 h (NIH, Bethesda, MD, USA).
Scratch motility assays
Cells were scratched with 2 μl pipette tips and then cultured in FBS-free medium. Scratched wound was monitored and pictures were taken at the indicated time points. The distances between the two edges of the scratched wound were measured by using the ImageJ software (NIH, Bethesda, MD, USA).
In vivo lung metastasis analysis
For lung metastasis analysis, H1299-Con and H1299-R175H cells with or without knockdown of Pontin (1 × 106 cells in 0.1 ml PBS) were injected into nude mice via the tail vein (n=8 mice per group). Mice were killed at 6 weeks after the inoculation. The number of lung tumor nodules were counted under a dissecting microscope and confirmed by histopathological analysis. The areas of tumor nodules were quantified in eight representative images taken at × 10 magnification by using the ImageJ software. Animal protocols were approved by the IACUC committee of Rutgers University.
Anchorage-independent growth assays
Anchorage-independent growth assays were performed as previously described.5, 43 In brief, cells were seeded in six-well culture plates coated with media containing 0.6% agarose, and cultured in media containing 0.3% agarose. Each experiment was repeated at least three times with triplicate wells.
Cell viability and cellular apoptosis assays
Cell viability and cellular apoptosis were determined as previously described.44 The Vi-CELL cell counter (Beckman Coulter, Brea, CA, USA) was used to determine cell viability. Apoptosis was measured by staining cells using Annexin V staining followed by the analysis in a flow cytometry (Beckman Coulter).
Cell cycle analysis
Cell cycle distribution was analyzed as previously described.45 In brief, cells were trypsinized, stained with propidium iodide (50 μg/ml) and then analyzed in a flow cytometry.
IF staining assays
IF staining was performed as previously described.7 In brief, cells grown on slides were fixed with 4% paraformaldehyde, treated with 0.5% TritonX-100 and then stained with anti-p53 (FL-393) and anti-Pontin antibodies to detect p53 and Pontin, respectively. Slides were then incubated with Alexa Fluor 555 Goat Anti-Rabbit IgG (H+L) and Alexa Fluor 488 Goat Anti-Mouse IgG (H+L) (Invitrogen). Nuclei were stained with DAPI (Vector Labs, Burlingame, CA, USA).
Microarray analysis
SK-BR-3-Consh and SK-BR-3-p53sh cells with or without knockdown of Pontin were analyzed for genome-wide expression changes by Affymetrix GeneChip. Total RNA was reverse transcribed and hybridized to an Affymetrix GeneChip expression array (HGU133+2 microarrays) by Functions Genomics Core Facility of Rutgers Cancer Institute of New Jersey. Microarray data were deposited with GEO Accession Number: GSE58140. Data were analyzed by using Microarray Suite version 5.0 (MAS 5.0). Change of expression levels for each gene by mutp53 was calculated as expression levels in SK-BR-3-Consh cells compared with SK-BR-3-p53sh cells with mutp53 knockdown. Heatmap of genes with significant expression change was visualized by using Gene-E software (http://www.broadinstitute.org/cancer/software/GENE-E/index.html). Functional pathway and network analysis of genes with significant expression changes were performed by using the Ingenuity pathways analysis (IPA) software (Ingenuity Systems, http://www.ingenuity.com).
ChIP assays
ChIP assays were performed with an Upstate ChIP assay kit (Billerica, MA, USA) in accordance with the instructions of the manufacturer. SK-BR-3 cells were subjected to ChIP assays with antibodies against p53, Pontin and IgG. DNA fragments pulled-down by antibodies were recovered and subjected to real-time PCR. The primer set that amplifies the region containing the potential SREBP-binding site in the STMN1 gene is: F: 5′-GAGAATGGGGAGCTGGTTCG-3′ and R: 5′-GACCACACTCTGAGCACCAA-3′. The primer sets that amplify the regions containing the potential NF-κB-binding sites in the RIPK1 gene and the ITPR1 gene are: F: 5′-ATCCGAGCACTCTTACCCATTCTA-3′, R: 5′-GTCACCTCTTCGCAAACTCCTG-3′ and F: 5′-CAGAAGTTTTTGCCCCGCTC-3′, R: 5′-GGAGCGCAAGAGAGGGATAC-3′, respectively. The primer set that amplifies the region of the coding sequence of the STMN1 gene is: F: 5′-GTCTCCACTCTGTTCCTGCC-3′, R: 5′-AGGATCCTCTTGGCCAGACT-3′.
Database of cancer patients
The KM plotter online survival database (http://www.kmplot.com) can assess the effect of 22 277 genes on overall survival in 1926 lung cancer and 1115 breast cancer patients.36 Another cohort of the breast cancer patients with known p53 mutation status and survival information was obtained from a publically available database (GSE3494).37 Genome-wide expression levels of all tumor specimens were assessed on Affymetrix HGU133A and B arrays. p53 mutation status was identified by cDNA sequence analysis of exons 2–11 of the p53 gene.37 To analyze the prognostic value of Pontin expression levels, the patients were split into two groups by auto-selection of best cutoff in the KM plotter database and according to the median expression level of Pontin mRNA in the breast cancer database (GSE3494). The Affymetrix IDs used for analysis are: 201614_s_at for Pontin; 217714_x_at for STMN1 and 209941_at for RIPK1.
Statistical analysis
The data were expressed as mean±S.D. Statistical analysis of the results was performed by using the GraphPad Prism software (La Jolla, CA, USA). Kaplan–Meier statistics and log-rank (one tail) test were performed to estimate the significance of differences in overall survival of patients among different groups. All other P-values were obtained using Student's t-test or χ2 test.
Abbreviations
- mutp53:
-
mutant p53
- wtp53:
-
wild-type p53
- GOF:
-
gain-of-function
- DBD:
-
DNA-binding domain
- FL-Pontin-R:
-
full-length siRNA-resistant Pontin
- IP:
-
immunoprecipitation
- IF:
-
immunofluorescence
- ChIP:
-
chromatin immunoprecipitation
- IPA:
-
ingenuity pathway analysis
- ITPR1:
-
Inositol 1,4,5-triphosphate receptor, type I
- RIPK1:
-
receptor (TNFRSF)-interacting serine-threonine kinase 1
- STMN1:
-
stathmin 1
References
Levine AJ, Oren M . The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9: 749–758.
Muller PA, Vousden KH . p53 mutations in cancer. Nat Cell Biol 2013; 15: 2–8.
Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004; 119: 861–872.
Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004; 119: 847–860.
Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun 2013; 4: 2935.
Muller P, Hrstka R, Coomber D, Lane DP, Vojtesek B . Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene 2008; 27: 3371–3383.
Zheng T, Wang J, Zhao Y, Zhang C, Lin M, Wang X et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun 2013; 4: 2996.
Freed-Pastor WA, Prives C . Mutant p53: one name, many proteins. Genes Dev 2012; 26: 1268–1286.
Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C . A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol 2001; 21: 1874–1887.
Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 2012; 148: 244–258.
Stambolsky P, Tabach Y, Fontemaggi G, Weisz L, Maor-Aloni R, Siegfried Z et al. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell 2010; 17: 273–285.
Song H, Hollstein M, Xu Y . p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol 2007; 9: 573–580.
Haupt S, di Agostino S, Mizrahi I, Alsheich-Bartok O, Voorhoeve M, Damalas A et al. Promyelocytic leukemia protein is required for gain of function by mutant p53. Cancer Res 2009; 69: 4818–4826.
Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 2011; 20: 79–91.
Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem 2005; 280: 41207–41212.
Jha S, Shibata E, Dutta A . Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol 2008; 28: 2690–2700.
Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE . Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 2008; 132: 945–957.
Wood MA, McMahon SB, Cole MD . An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell 2000; 5: 321–330.
Feng Y, Lee N, Fearon ER . TIP49 regulates beta-catenin-mediated neoplastic transformation and T-cell factor target gene induction via effects on chromatin remodeling. Cancer Res 2003; 63: 8726–8734.
Lacombe J, Mange A, Jarlier M, Bascoul-Mollevi C, Rouanet P, Lamy PJ et al. Identification and validation of new autoantibodies for the diagnosis of DCIS and node negative early-stage breast cancers. Int J Cancer 2013; 132: 1105–1113.
Lauscher JC, Elezkurtaj S, Dullat S, Lipka S, Grone J, Buhr HJ et al. Increased Pontin expression is a potential predictor for outcome in sporadic colorectal carcinoma. Oncol Rep 2012; 28: 1619–1624.
Taniue K, Oda T, Hayashi T, Okuno M, Akiyama T . A member of the ETS family, EHF, and the ATPase RUVBL1 inhibit p53-mediated apoptosis. EMBO Rep 2011; 12: 682–689.
Tucker PA, Sallai L . The AAA+ superfamily—a myriad of motions. Curr Opin Struct Biol 2007; 17: 641–652.
Mezard C, Davies AA, Stasiak A, West SC . Biochemical properties of RuvBD113N: a mutation in helicase motif II of the RuvB hexamer affects DNA binding and ATPase activities. J Mol Biol 1997; 271: 704–717.
Elkaim J, Castroviejo M, Bennani D, Taouji S, Allain N, Laguerre M et al. First identification of small-molecule inhibitors of Pontin by combining virtual screening and enzymatic assay. Biochem J 2012; 443: 549–559.
Cronier L, Crespin S, Strale PO, Defamie N, Mesnil M . Gap junctions and cancer: new functions for an old story. Antioxid Redox Signal 2009; 11: 323–338.
Feng Z, Levine AJ . The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol 2010; 20: 427–434.
Tomas A, Futter CE, Eden ER . EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol 2014; 24: 26–34.
Xia Y, Gong KZ, Xu M, Zhang YY, Guo JH, Song Y et al. Regulation of gap-junction protein connexin 43 by beta-adrenergic receptor stimulation in rat cardiomyocytes. Acta Pharmacologica Sinica 2009; 30: 928–934.
Guo K, Ma Q, Wang L, Hu H, Li J, Zhang D et al. Norepinephrine-induced invasion by pancreatic cancer cells is inhibited by propranolol. Oncol Rep 2009; 22: 825–830.
Bist P, Leow SC, Phua QH, Shu S, Zhuang Q, Loh WT et al. Annexin-1 interacts with NEMO and RIP1 to constitutively activate IKK complex and NF-kappaB: implication in breast cancer metastasis. Oncogene 2011; 30: 3174–3185.
Masciarelli S, Fontemaggi G, Di Agostino S, Donzelli S, Carcarino E, Strano S et al. Gain-of-function mutant p53 downregulates miR-223 contributing to chemoresistance of cultured tumor cells. Oncogene 2014; 33: 1601–1608.
Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 2006; 10: 191–202.
Cooks T, Pateras IS, Tarcic O, Solomon H, Schetter AJ, Wilder S et al. Mutant p53 prolongs NF-kappaB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 2013; 23: 634–646.
Byrne FL, Yang L, Phillips PA, Hansford LM, Fletcher JI, Ormandy CJ et al. RNAi-mediated stathmin suppression reduces lung metastasis in an orthotopic neuroblastoma mouse model. Oncogene 2014; 33: 882–890.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123: 725–731.
Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA 2005; 102: 13550–13555.
Messai Y, Noman MZ, Hasmim M, Janji B, Tittarelli A, Boutet M et al. ITPR1 protects renal cancer cells against natural killer cells by inducing autophagy. Cancer Res 2014; 74: 6820–6832.
Chen YW, Paliwal S, Draheim K, Grossman SR, Lewis BC . p19Arf inhibits the invasion of hepatocellular carcinoma cells by binding to C-terminal binding protein. Cancer Res 2008; 68: 476–482.
Goto Y, Nishikawa R, Kojima S, Chiyomaru T, Enokida H, Inoguchi S et al. Tumour-suppressive microRNA-224 inhibits cancer cell migration and invasion via targeting oncogenic TPD52 in prostate cancer. FEBS Lett 2014; 588: 1973–1982.
Shan J, Dsouza SP, Bakhru S, Al-Azwani EK, Ascierto ML, Sastry KS et al. TNRC9 downregulates BRCA1 expression and promotes breast cancer aggressiveness. Cancer Res 2013; 73: 2840–2849.
Li X, Yang Q, Yu H, Wu L, Zhao Y, Zhang C et al. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget 2014; 5: 788–801.
Liu J, Zhang C, Lin M, Zhu W, Liang Y, Hong X et al. Glutaminase 2 negatively regulates the PI3K/AKT signaling and shows tumor suppression activity in human hepatocellular carcinoma. Oncotarget 2014; 5: 2635–2647.
Yu H, Yue X, Zhao Y, Li X, Wu L, Zhang C et al. LIF negatively regulates tumour-suppressor p53 through Stat3/ID1/MDM2 in colorectal cancers. Nat Commun 2014; 5: 5218.
Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS et al. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell 2010; 38: 689–699.
Acknowledgements
We thank Dr. Arnold Levine for helpful discussion, Emmanuel Zachariah for technical assistance in microarray experiments, Arthur I Robert for technical assistance in flow cytometry analysis, Dr. Hao Wu for assistance in histopathological analysis of mouse lung tumors. WH is supported by the grants from NIH (1R01CA160558), DOD (W81XWH-10-1-0435), the Ellison Foundation, the New Jersey Health foundation and the New Investigator Award of CINJ. ZF is supported by the grants from NIH (1R01CA143204) and NJCCR. This research was supported in part by Functional Genomics Shared Resource of CINJ (P30CA072720) and Flow Cytometry Shared Resource of CINJ (P30CA072720) and NIH Shared Instrumentation grant (1S10RR025468-01).
Author Contributions
ZF and WH designed experiments. YZ, CZ, XY, XL, JL and HY performed experiments. YZ, VB, QY, ZF and WH analyzed data. YZ, ZF and WH wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Oren
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhao, Y., Zhang, C., Yue, X. et al. Pontin, a new mutant p53-binding protein, promotes gain-of-function of mutant p53. Cell Death Differ 22, 1824–1836 (2015). https://doi.org/10.1038/cdd.2015.33
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2015.33
This article is cited by
-
Δ133p53β isoform pro-invasive activity is regulated through an aggregation-dependent mechanism in cancer cells
Nature Communications (2021)
-
Non-canonical roles of canonical telomere binding proteins in cancers
Cellular and Molecular Life Sciences (2021)
-
Impact of RUNX2 on drug-resistant human pancreatic cancer cells with p53 mutations
BMC Cancer (2018)
-
Parkin targets HIF-1α for ubiquitination and degradation to inhibit breast tumor progression
Nature Communications (2017)
-
A novel mutant p53 binding partner BAG5 stabilizes mutant p53 and promotes mutant p53 GOFs in tumorigenesis
Cell Discovery (2016)