Abstract
Macroautophagy is a membrane-trafficking process that delivers cytoplasmic constituents to lysosomes for degradation. The process operates under basal conditions as a mechanism to turnover damaged or misfolded proteins and organelles. As a result, it has a major role in preserving cellular integrity and viability. In addition to this basal function, macroautophagy can also be modulated in response to various forms of cellular stress, and the rate and cargoes of macroautophagy can be tailored to facilitate appropriate cellular responses in particular situations. The macroautophagy machinery is regulated by a group of evolutionarily conserved autophagy-related (ATG) proteins and by several other autophagy regulators, which either have tissue-restricted expression or operate in specific contexts. We report here the characterization of a novel autophagy regulator that we have termed DRAM-3 due to its significant homology to damage-regulated autophagy modulator (DRAM-1). DRAM-3 is expressed in a broad spectrum of normal tissues and tumor cells, but different from DRAM-1, DRAM-3 is not induced by p53 or DNA-damaging agents. Immunofluorescence studies revealed that DRAM-3 localizes to lysosomes/autolysosomes, endosomes and the plasma membrane, but not the endoplasmic reticulum, phagophores, autophagosomes or Golgi, indicating significant overlap with DRAM-1 localization and with organelles associated with macroautophagy. In this regard, we further proceed to show that DRAM-3 expression causes accumulation of autophagosomes under basal conditions and enhances autophagic flux. Reciprocally, CRISPR/Cas9-mediated disruption of DRAM-3 impairs autophagic flux confirming that DRAM-3 is a modulator of macroautophagy. As macroautophagy can be cytoprotective under starvation conditions, we also tested whether DRAM-3 could promote survival on nutrient deprivation. This revealed that DRAM-3 can repress cell death and promote long-term clonogenic survival of cells grown in the absence of glucose. Interestingly, however, this effect is macroautophagy-independent. In summary, these findings constitute the primary characterization of DRAM-3 as a modulator of both macroautophagy and cell survival under starvation conditions.
Similar content being viewed by others
Main
Macroautophagy (hereafter autophagy) is a cellular process that delivers cytoplasmic constituents to lysosomes for degradation.1 Autophagy operates at basal levels in virtually all, if not all, cells. At the initiation of autophagy, membranes termed isolation membranes nucleate in the cytoplasm from a variety of sources.2, 3, 4, 5 Two ubiquitin-like conjugation mechanisms involving evolutionarily conserved autophagy-related (Atg) genes then function together to expand these membranes to form the characteristic organelles of autophagy, the autophagosome.6, 7 During this process, cargoes are recruited to the lumen of the autophagosome via a protein called LC3, which becomes tethered to autophagosome membranes during biogenesis.8 Adapter proteins such as p62/SQSTM1, NBR1 and OPTN then act as ‘bridges’ for cargo recruitment by simultaneously binding LC3, and the ubiquitin moieties on proteins and organelles destined for degradation.9
Following autophagosome formation, a variety of fusion events can occur with other organelles including multi-vesicular bodies and endosomes.10 Ultimately, however, fusion occurs with lysosomes to form new organelles called autolysosomes in which lysosomal acidic hydrolases invoke cargo degradation.10, 11 Under basal conditions, the breakdown products are then recycled into biosynthetic pathways.10, 11 As a result, autophagy is a critical mechanism within cells to remove damaged proteins and organelles, thereby preserving cellular fidelity, homeostasis and ultimately viability of the cell and organism.1, 12
Autophagy can also be modulated by a variety of internal and external cues.13 This can increase the rate of autophagic flux and/or modulate the cargoes that are digested. In this regard, several selective forms of autophagy have been described including mitophagy – the selective digestion of mitochondria.14, 15 The best characterized situation in which autophagy is modulated is in response to starvation conditions.16, 17, 18, 19 This evolutionarily conserved response utilizes autophagy to provide fuel for catabolic pathways to maintain ATP levels during periods of diminished nutrient availability.
To understand the regulation of autophagy, it is important to identify factors that regulate the process in both general and specific situations. For example, we previously identified DRAM-1 (damage-regulated autophagy modulator-1) as an autophagy regulator downstream of the tumor suppressor p53.20, 21 Subsequently, we found that DRAM-1 belongs to a previously undescribed, evolutionarily-conserved protein family.22 To date, however, we have only characterized DRAM-1 and the most related protein in terms of amino-acid sequence that we termed DRAM-2.22 We report here initial characterization of another DRAM-1-related protein that is encoded by TMEM150B and that we have named DRAM-3. This protein localizes to endosomes and autolysosomes/lysosomes, but unlike DRAM-1 is not induced by p53. DRAM-3 does, however, regulate autophagic flux and promotes cell survival in response to nutrient deprivation, but DRAM-3’s effect on cell survival is autophagy-independent.
Results
DRAM-3 is expressed in multiple tissues and tumor cell lines, but is not induced by a variety of stimuli
To identify DRAM-1-related proteins we performed Blast searches using the DRAM-1 peptide sequence. One protein identified comprises 233 amino acids and has 30% identity and 43% similarity to DRAM-1 (Figure 1a). This protein is encoded by TMEM150B, but, due to its homology to DRAM-1, we decided to rename this gene and its protein product, DRAM-3 (DRAM-Related/Associated Member 3). Analysis of the DRAM-3 peptide sequence revealed that, like DRAM-1,20, 23 DRAM-3 appears to contain a signal peptide and several hydrophobic regions, indicating that DRAM-3 is likely a transmembrane protein located in one or more compartments of the secretory pathway (Figure 1b).
DRAM-3 is expressed in various human tissues and cancer cell lines. (a) Alignment of DRAM-3 and DRAM-1. DRAM-3 shares 30% identity and 43% similarity with DRAM-1 at the amino-acid level. (b) Schematic representation of DRAM-3. DRAM-3 is predicted to encode for a 233-aa protein with six putative transmembrane domains and a predicted N-terminal signal peptide. (c) DRAM-3 mRNA expression varies in different body tissues. qRT-PCR products of the Origene TissueScan Human Normal cDNA Array using Origene primers against DRAM-3 on a 2% agarose gel. (d) Quantification of qRT-PCR from (c). GAPDH was used as a standard. (e) DRAM-3 mRNA expression in a panel of cancer cell lines. Results obtained through qRT-PCR. All values were measured in triplicates and normalized to 18S RNA. The red line denotes the detection limit of the qRT-PCR. (f) DRAM-1 mRNA expression in a panel of cancer cell lines. Results obtained through qRT-PCR. All values were measured in triplicates and normalized to 18S RNA. All values shown were above the detection limit
We next analyzed DRAM-3 expression in a variety of human tissues that revealed DRAM-3 is clearly detectable in some, but not all tissues (Figure 1c). Quantification revealed that the colon and lung had the highest DRAM-3 mRNA levels with comparatively high levels also detectable in the lymph nodes, placenta, duodenum, peripheral blood mononuclear cells and spleen. Other tissues either had very little or undetectable levels of DRAM-3 mRNA (Figure 1d). We also profiled DRAM-3 expression in human tumor cell lines, which again showed expression in some lines, but virtually no expression in others (Figure 1e). DRAM-1 was also selectively expressed, but there did not appear to be any level of co-regulation of the two genes across the lines (Figure 1f).
Our previous studies showed DRAM-1 is markedly induced by p53.20 By contrast, DRAM-2 is not induced in this context.22 We were therefore keen to examine whether DRAM-3 was modulated by p53. We utilized a previously described Saos-2 cell line (null for endogenous p53), which contains a tetracycline-responsive p53 transgene.24 Treatment of these cells with doxycycline (Dox) potently upregulates p53 with maximal expression being reached by 24 h (Figure 2a). DRAM-1 mRNA was also induced over this time frame (Figure 2b),20 but DRAM-3, like DRAM-2, was completely unaffected (Figure 2b).
Regulation of DRAM-3 expression in selected cancer cell lines. (a) Western blot of Saos-2 tet-on p53 cells with and without addition of doxycycline. p53 is induced by doxycycline in a time-dependent manner. (b) DRAM-3 is not induced by p53. In Saos-2 tet-on p53 cells, DRAM-3 mRNA is not induced after addition of doxycyline, while DRAM-1 mRNA is upregulated. Results obtained through qRT-PCR. The data represented are from three independent experiments and are plotted as means; error bars are S.E.M. (c, d) Analysis of DRAM-3 mRNA levels in response to various stimuli in p53-null Saos-2 cells (c) or p53 wild-type U2OS cells (d). Results obtained through qRT-PCR. Values shown are relative to control, which was normalized to 1. The data represented are from three independent experiments and are plotted as means; error bars are S.E.M. (e, f) Analysis of DRAM-1 mRNA levels in response to various stimuli in p53-null Saos-2 cells (e) and p53 wild-type U2OS cells (f). Results obtained through qRT-PCR. Values shown are relative to control, which was normalized to 1. The data represented are from three independent experiments and are plotted as means; error bars are S.E.M. (c–f) No glucose medium contained 10% dialyzed FBS
Although unaffected by p53, we considered that DRAM-3 expression may be modulated by factors that elicit an autophagic or cellular stress response. We therefore treated U2OS, HepG2, A549 and LNCaP cells (wild-type for endogenous p53) and Saos-2 and H1299 cells (p53-null) with, where indicated, cisplatin, etoposide, actinomycin D, TNFα and IFN-γ. Saos-2 and U2OS cells were also incubated in either Earle’s Balanced Salt Solution (EBSS, a medium lacking amino acids and serum) or regular medium lacking serum or glucose. DNA-damaging agents have been reported to upregulate DRAM-1 in cells containing wild-type p53,20 but no induction of DRAM-3 was observed in response to etoposide, actinomycin D or cisplatin in any of the cells tested whether wild type or null for p53 (Figure 2c and d and Supplementary Figure 1). Similarly, DRAM-3 was not induced in response to TNFα or IFN-γ, nor was any effect observed on incubation in starvation conditions (Figure 2c and d). The effect of these treatments on DRAM-1 mRNA levels is shown for comparison (Figure 2e and f). We conclude that DRAM-3 levels vary widely between different tissues, but, at least in the cells tested, DRAM-3 is not markedly up- or downregulated by DNA damage, nutrient deprivation or by exposure to TNFα and IFN-γ.
DRAM-3 localizes to various compartments within the cell
DRAM-1 and DRAM-2 localize to lysosomes.20, 22 In addition, DRAM-1 is also found in early endosomes, late endosomes and autophagosomes with smaller isoforms of DRAM-1 showing partial localization in the peroxisomes and ER.25 We therefore tested whether DRAM-3 occupies similar or different subcellular compartments when compared with DRAM-1 and DRAM-2. This revealed that DRAM-3, like DRAM-1, was detectable in regions of the cell stained with antibodies against LAMP2, LC3B (punctate stain) and the endosomal marker, EEA1 (Figure 3a–c). In addition and in contrast to DRAM -1 and DRAM-2, DRAM-3 was also found in the plasma membrane with a concentrated localization at actin-rich focal adhesions (Figure 3d). Localization in the plasma membrane and endosomes was also confirmed by live-cell imaging (Figure 3e and f). By contrast, DRAM-3 was not found in phagophores, the endoplasmic reticulum or Golgi (Figure 3g and h and Supplementary Figure 2a).
DRAM-3 localizes to a variety of cellular compartments (a–d). Exogenous DRAM-3 partially localizes within close proximity to the autophagosome marker LC3B (a), the early endosome marker EEA1 (b), the lysosome marker LAMP2 (c) and actin-coated pits (d). Actin staining was performed using Phalloidin-red. DRAM-3 was visualized with an α-Myc-Tag antibody. Immunofluorescence analysis was performed on a Zeiss 710 confocal microscope with × 63 objective. (e) DRAM-3 containing vesicles closely localize to Rab5-containing vesicles in live cells. GFP-tagged DRAM-3 and RFP-tagged early endosome marker Rab5 were co-transfected into Saos-2 cells via calcium phosphate transfection. Live-cell imaging was performed after a minimum of 2 days on an Andor spinning disc confocal system. (f) DRAM-3 containing vesicles partially localize close to Rab7-containing vesicles in live cells. GFP-tagged DRAM-3 and RFP-tagged late endosome marker Rab7 were co-transfected into Saos-2 cells via calcium phosphate transfection. Live-cell imaging was performed after a minimum of 2 days on an Andor spinning disc confocal system. (g) Exogenous DRAM-3 does not colocalize with Golgi marker GM130. Immunofluorescence analysis was performed on an Olympus FV1000 confocal microscope with × 60 objective. (h) Exogenous DRAM-3 does not colocalize with endoplasmic reticulum marker Calnexin. Immunofluorescence analysis was performed on an Olympus FV1000 confocal microscope with × 60 objective
Because LC3B is connected to autophagy, we explored the relationship between DRAM-3 and LC3B more closely. Quantification of LC3B colocalized with DRAM-3 revealed that ~20% of LC3B is in close proximity to DRAM-3 (Supplementary Figure 2b). By contrast, <5% of DRAM-3 can be found in close proximity to LC3B (Supplementary Figure 2b). In addition, because LC3B marks both autophagosomes and autolysosomes, we examined the localization of DRAM-3-associated LC3B. Interestingly, cellular regions that stain positive for DRAM-3 and LC3B also appear positive for LAMP2 (Supplementary Figure 2c), indicating that when DRAM-3 associates with LC3B it does so in autolysosomes. Taken together, these findings identify common and also divergent localization sites for DRAM-3 when compared with DRAM-1 and DRAM-2.
DRAM-3 promotes autophagy, but does not affect cell viability in replete nutrient conditions
DRAM-1 can contribute to p53’s cell death response,20 but when expressed alone it has no detrimental impact on cell viability.20 To test whether DRAM-3 affects cell death, we generated tetracycline-regulated DRAM-3-inducible cells. In these cells, a small, but reproducible increase in cells with sub-G1 DNA was observed 48 h after DRAM-3 induction (Figure 4a), but this effect was not statistically significant (Figure 4a). In line with a lack of apoptosis, DRAM-3 expression does not induce any detectable increase in the cleaved form of caspase 3 (Supplementary Figure 3a). Annexin V/Propidium iodide (PI) staining also showed that DRAM-3 has no impact on apoptosis (Annexin V-positive, PI-negative cells), although a small increase in cells that were Annexin V positive and PI positive indicates that DRAM-3 may cause a small amount of necrosis (Supplementary Figure 3b).
DRAM-3 does not affect apoptotic cell death or cell growth processes. (a) DRAM-3 overexpression does not induce apoptotic cell death. Saos-2 Tet-On DRAM-3 cells were treated with doxycycline for 48 h and analyzed for Sub-G1 content via FACS (a reliable measure of apoptotic death). The data represented are from three independent experiments and are plotted as means. Error bars represent S.E.M. Statistical analysis was undertaken with an unpaired t-test. (b) DRAM-3 expression is strongly induced after doxycycline treatment of Saos-2 Tet-On DRAM-3 cells. Cells were induced with doxycycline for 24, 48 and 72 h, and western blot analysis was performed to ensure DRAM-3 expression. The doxycycline-inducible construct is fused to a Myc-His tag, and an α-Myc-tag antibody was used to detect the exogenous protein. (c) Cells stably overexpressing DRAM-3-Myc-His were generated via ecotropic viral infection of Saos-2 EcoR cells. The extent of DRAM-3 overexpression was measured in triplicates by qRT-PCR, and DRAM-3 measurements were normalized to 18S RNA. Data represent means. Error bars are S.D. (d) Saos-2 pWZL DRAM-3 cells express DRAM-3 protein. DRAM-3 protein was visualized via western blot, using an α-Myc-tag antibody. (e) Saos-2 cells stably overexpressing DRAM-3 do not show proliferation defects. DRAM-3 overexpressing and control cells were counted in triplicates on the specified days, and a growth curve was generated for the course of 6 days. (f) Infection of Saos-2 cells with a retroviral construct (pWZL) expressing DRAM-3 does not impair clonogenic survival under standard growth conditions. Saos-2 pWZL and pWZL DRAM-3 cells were seeded at low density 1 week after selection
It was difficult to know whether DRAM-3 expression has a long-term effect on cell viability using this system because DRAM-3 expression becomes attenuated over time (Figure 4b). Induction of DRAM-3 in two waves (1 week of Dox, followed by 1 week without Dox and then addition of Dox again) resulted in lower DRAM-3 levels at the second round of induction, indicating potentially that some DRAM-3-expressing cells (possibly those with the highest DRAM-3 levels) had died during the first wave of induction (Supplementary Figure 3c).
To test the long-term effects of DRAM-3 on cell death, we infected Saos-2 cells with a retrovirus expressing Myc-tagged DRAM-3 or an ‘empty’ retroviral construct as a control (Figure 4c and d). This revealed that retroviral expression of DRAM-3 had no impact on cell growth in the short-term nor did it affect the clonogenic potential of cells in the long term (Figure 4e and f). Therefore, whereas high levels of DRAM-3 may affect cell viability, lower levels do not affect growth or viability when expressed alone in replete medium.
The best characterized effect of DRAM-1 is on autophagy20 and so we tested whether DRAM-3 is also an autophagy modulator. First, we analyzed LC3B distribution in cells expressing DRAM-3. In cells with low levels of autophagy, the majority of LC3B is diffused in the cytoplasm in a form termed LC3-I. On autophagosome formation, LC3-I is cleaved and becomes conjugated to phosphatidylethanolamine to give the form of LC3 termed LC3-II. In this form, LC3B integrates into the autophagosome membrane via its lipid moiety.8 As a result, the appearance of LC3B puncta by immunofluorescence can be used as a measure of autophagosome levels and an indicator of changes in autophagic flux. In replete growth medium, DRAM-3-expressing cells had significantly more LC3B-positive puncta than control cells (Figure 5a and b). Western blotting for LC3-II also showed increased levels of LC3-II in DRAM-3-expressing cells when compared with controls (Figure 5c and Supplementary Figure 4a–c). Under starvation conditions, however, where autophagy is markedly induced, the difference between DRAM-3-expressing and control cells was no longer apparent (Figure 5a–c).
DRAM-3 overexpression affects autophagic processes. (a) The levels of LC3B puncta are increased in cells stably overexpressing DRAM-3. Saos-2 cells were treated with control or starvation media (EBSS) for 2 h and examined by immunofluorescence analysis. Endogenous autophagosomes were visualized with LC3B-specific antibody on a Zeiss 710 confocal microscope with × 63 objective. (b) Quantification of (a). Immunofluorescence stacks were overlayed with maximum intensity, and the area of LC3B puncta relative to the cell area was measured using Image J. Quantification of 12 cells per condition. A Student’s t-test was performed as statistical analysis, and the bars in the figure represent mean values; errors are S.D. NSP>0.05; **P<0.01. (c) DRAM-3 overexpression increases LC3-II levels under normal growth conditions. Saos-2 cells stably overexpressing DRAM-3 were subjected to 1 or 2 h EBSS starvation or DMEM supplemented with 10% FBS and Glutamine. (d) DRAM-3 overexpression increases autophagic flux. Saos-2 cells stably overexpressing DRAM-3 were subjected to 5 μM chloroquine treatment (CQ) for 2 h. (e) DRAM-3 overexpression accelerates mitophagy. Saos-2 Eco cells containing DRAM-3 or control (pWZL) constructs and overexpressing Parkin-YFP were treated with 12.5 μM CCCP two times during a 24 h period and then analyzed for quantity of mitochondria via western blotting, using an α-COX IV antibody. (f) DRAM-3 overexpression modulates autophagic flux in different cell lines. MDA-MB-231 cells stably overexpressing DRAM-3 were subjected to 1 or 2 h EBSS starvation or DMEM supplemented with 10% FBS and glutamine. (g) DRAM-3 overexpression increases autophagic flux in different cell lines. MDA-MB-231 cells stably overexpressing DRAM-3 were subjected to 5 μM chloroquine treatement (CQ) for 2 h
Increased LC3B puncta or LC3-II levels can indicate enhanced autophagic flux, but, as most cells have a basal autophagic rate, an increase can also indicate a block or retardation in autophagosome turnover. To discern between these two possibilities, we examined LC3B accumulation following treatment with the lysosomotropic agent, chloroquine, to block autophagosome turnover. Cells with higher rates of flux accumulate more LC3-II upon chloroquine treatment than those with lower flux. However, if cells already have blocked flux, treatment with chloroquine causes no further increase in the levels of LC3-II. In the context of DRAM-3 expression, we found that chloroquine caused a greater accumulation of LC3-II when compared with control (Figure 5d), indicating that DRAM-3 promotes autophagic flux. Interestingly, the autophagy adaptor protein p62/SQSTM1, which is often used as a measure of flux,26 was not increased by chloroquine treatment (Figure 5d), nor was it affected by incubation in EBSS (Figure 5c). This indicates that, although useful in some scenarios, p62/SQSTM1 is not actively degraded by autophagy in these cells under these conditions and therefore cannot be utilized to measure autophagic flux.
To address this issue further we analyzed autophagic flux by ‘enhanced-mitophagy’.27 To do this, we infected DRAM-3-expressing and control cells with a retrovirus-expressing Parkin. Treatment of Parkin-expressing cells with the mitochondrial uncoupling agent CCCP causes autophagy-dependent loss of mitochondria, with the rate of loss being dependent on the autophagic capacity and rate in the cell. Using this system we found that depletion of the mitochondrial protein COX IV was greater in DRAM-3-expressing cells when compared with controls (Figure 5e), confirming that DRAM-3 promotes autophagic flux.
We next tested whether DRAM-3 affects autophagy in another line. As Saos-2 have relatively high DRAM-3 levels, we decided to examine MDA-MB-231 cells in which endogenous DRAM-3 levels are comparatively low (Figure 1e). In contrast to Saos-2 cells, DRAM-3 expression in MDA-MB-231 did not affect basal LC3-II levels, indicating that this effect may be cell-type specific (Figure 5f). On starvation, however, there was less LC3-I and LC3-II in DRAM-3-expressing cells (Figure 5f), indicating that DRAM-3 was promoting autophagic flux. To examine this further, we treated with chloroquine, which caused a greater accumulation of LC3-II in DRAM-3-expressing MDA-MB-231 cells when compared with controls (Figure 5g), showing in line with what was observed in Saos-2 (Figure 5d and e) that DRAM-3 promotes autophagic flux.
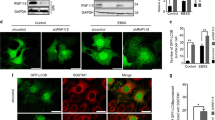
As DRAM-3 overexpression enhances autophagic flux, we were keen to ascertain whether loss of endogenous DRAM-3 could also affect autophagic flux. To do this we utilized CRISPR/Cas9 technology to introduce mutations in the dram-3 gene, which disrupt DRAM-3 expression. In the absence of an effective DRAM-3 antibody, we performed two tests to confirm CRISPR/Cas9 targeting of the dram-3 locus. First, we sequenced DNA from DRAM-3-targeted cells in the region targeted by the CRISPR guide RNA. This revealed major disruption of dram-3 DNA sequence in cells targeted with a DRAM-3 guide RNA when compared with those targeted with a GFP guide RNA (Figure 6a). To test whether the DRAM-3 CRISPR/Cas9 could also disrupt protein expression, we transfected the two cell pools with a plasmid expressing DRAM-3-GFP. Expression of DRAM-3-GFP was clearly detectable in control cells, but expression was lost in cells expressing guide RNAs targeting DRAM-3 or GFP, indicating effective targeting of dram-3 and gfp sequences in these two cell pools (Figure 6b).
dram-3 disruption does not affect the number or localization of phagophores, autophagosomes and lysosomes. (a) lentiCRISPR disruption of DRAM-3 in Saos-2 cells. Genomic DNA from Saos-2 cells infected with lentivirus containing control (GFP) or DRAM-3-targeting constructs was sequenced for CRISPR-induced DNA frameshifts. (b) lentiCRISPR cells efficiently inactivate expression of exogenous DRAM-3-GFP. Saos-2 cells containing lentiCRISPR contructs targeting GFP or DRAM-3 were transfected with 10 μg DRAM-3-GFP plasmid and collected 2 days after transfection for western blot analysis. (c) Analysis of autophagic responses on starvation in DRAM-3-knockout cells. Saos-2 DRAM-3 disrupted and control cells were subjected to EBSS starvation for 1–2 h. (d) Autophagic flux is reduced on DRAM-3 disruption. Saos-2 DRAM-3 disrupted and control cells were subjected to +/− addition of 5 μM chloroquine (an inhibitor of autophagic flux). (e) The appearance of cell structures associated with autophagy (phagophores, autophagosomes and autolysosomes/lysosomes) do not change on DRAM-3 disruption. Saos-2 DRAM-3 disrupted and control cells were grown on coverslips and stained for immunofluorescence analysis with LC3B together with WIPI2 or LAMP2 antibodies. Imaging was performed on a Zeiss LSM710 microscope with × 63 lens
In contrast to DRAM-3-overexpressing cells, there was no difference in basal LC3-II levels between wild type and DRAM-3-knockout cells (Figure 6c). However, incubating cells in chloroquine, as outlined above and as previously described,26 caused greater LC3-II accumulation in control cells (GFP) than in cells with DRAM-3 disruption (Figure 6d and Supplementary Figure 4d–f). This is reciprocal to what was observed on retroviral overexpression of DRAM-3 and confirms that DRAM-3 is a modulator of autophagic flux. Interestingly, although this result was seen in several independent experiments, in some cases no effect was observed. This indicates perhaps that DRAM-3 has a role that is dependent on the cellular nutrient status as would be the case for the majority of autophagic responses.
To test whether DRAM-3 loss has an effect on autophagy by affecting the number or localization of structures associated with autophagy – phagophores, autophagosomes and autolysosomes/lysosomes – we stained wild type (Crispr GFP) and DRAM-3 Crispr cells with antibodies against WIPI2, LC3B and LAMP2. No differences, however, were seen between the two cell lines (Figure 6e). In addition, because the endocytic pathway can affect autophagic flux, we also examined endosomes, but staining for EEA1 looked similar in wild type and DRAM-3-expressing cells (Supplementary Figure 5a). This does not, however, provide proof that endocytosis is not affected and so we examined uptake of bovine serum albumin (BSA) over time, but again no difference was evident between wild-type and DRAM-3-expressing cells (Supplementary Figure 5b and c). Collectively, this shows DRAM-3 does not affect endocytosis and that differences in endosomes or endocytic rate cannot explain the effects of DRAM-3 on autophagy.
DRAM-3 promotes autophagy-independent survival on glucose deprivation
As we found DRAM-3 is a regulator of autophagic flux, we re-visited the possibility that DRAM-3 may affect cell viability, but this time under starvation conditions where autophagy can be critical for cell survival.16, 17, 18, 19 This indeed revealed that DRAM-3-expressing Saos-2 and MDA-MB-231 cells had enhanced clonogenic potential when grown in low serum without glucose (Figure 7a and b and Supplementary Figure 6a). This effect appeared cell death-related, as a difference in propidium iodide exclusion (a measure of cell membrane permeability and by association, cell death) was also observed between DRAM-3-expressing and control cells (Figure 7c). By contrast, no major differences in cell-cycle stage or sub-G1 DNA content (a measure of apoptotic cell death) were evident in either replete medium or starvation conditions (Figure 7d). Further analysis again showed DRAM-3’s effect on cell death was not apoptotic, as the extent of death was not significantly affected by treatment with the pan-caspase inhibitor zVAD-fmk (Supplementary Figure 6b). Interestingly, we also noted that this difference in cell survival may not be related to the ability to supply energy in the absence of glucose, as the difference was also evident when cells were incubated in pyruvate, an alternative source of fuel (Figure 7e). Analysis of cells under these conditions by Annexin V/PI staining again showed that the difference in cell death was not apoptotic (Annexin V-positive, PI-negative) and was more likely down to differences in necrosis (Annexin V-positive, PI-positive) (Supplementary Figure 6c).
DRAM-3 promotes survival after glucose starvation. (a) DRAM-3 overexpression improves clonogenic survival after glucose starvation. Saos-2 cells stably overexpressing DRAM-3 or a control construct (pWZL) were subjected to DMEM no glucose media +2% dialyzed FBS. After 24 h the starvation media was exchanged with growth media and cells were left recovering for 2–3 days before they were stained with Giemsa. (b) Quantification of (a). Three experiments were quantified for the percentage of the area occupied by cells. A paired Student’s t-test was performed as statistical analysis, and the bars in the figure represent mean values; errors are S.E.M. **P<0.01. (c) DRAM-3 overexpression prevents cell death after glucose starvation. Saos-2 cells stably overexpressing DRAM-3 or a control construct (pWZL) were subjected to DMEM no glucose media +2% dialyzed FBS. After 24 h the cells were collected and incubated with propidium iodide (PI; 5 μg/ml) for 30 min and then analyzed for PI uptake on a FACS Calibur. Results from five experiments measured in triplicates, displayed as mean; error bars are S.E.M. Statistical analysis was performed using a Student’s t-test. *P<0.05 (d) Cell death after glucose starvation does not involve DNA fragmentation in Saos-2 cells. Saos-2 cells stably overexpressing DRAM-3 or a control construct (pWZL) were subjected to DMEM no glucose media +2% dialyzed FBS. After 24 h the cells were collected, fixed and subjected to cell cycle profile analysis. The sub-G1 fraction indicates DNA fragmentation – a marker of apoptosis. Results from three experiments measured in triplicates. (e) Addition of 10% dialyzed serum and sodium pyruvate does not affect the effect of DRAM-3 on glucose starvation-induced cell death. Saos-2 cells stably overexpressing DRAM-3 or a control construct (pWZL) were subjected to DMEM no glucose media +10% dialyzed FBS +0.11 g/l sodium pyruvate. After 48 h the cells were collected and incubated with Propidium Iodide (5 μg/ml) for 30 min and then analyzed for PI uptake on a FACS Calibur. Results from three experiments measured in triplicates, displayed as mean; error bars are S.E.M. Statistical analysis was performed using a Student’s t-test. **P<0.01 (f) Addition of chloroquine to glucose starvation medium blocks autophagic flux. Ten micromolars of chloroquine was added to full DMEM or glucose-free medium supplemented with 2% dialyzed FBS to block the turnover of autophagosomes. Protein samples were collected after 24 h and processed for western blotting. (g) Autophagy block with chloroquine does not abolish the effect of DRAM-3 overexpression under glucose-starved conditions. Cells were collected for PI exclusion FACS analysis after 24 h. The data displayed are from four independent experiments and are presented as mean; error bars are S.E.M. A Student's t-test was performed for statistical analysis. *P<0.05; NSP>0.05
We also tested whether DRAM-3 loss would decrease survival under glucose-deprived conditions. This revealed, however, that loss of DRAM-3 had no impact on cell viability (Supplementary Figure 6d). A result of this kind, however, is not without precedent. For example, XIAP overexpression represses death in multiple situations and yet XIAP-deficient mice are perfectly viable,28 indicating that XIAP loss does not lead to excessive cell death in these animals. It may be that the cells adapt to DRAM-3 loss to survive and/or we have not yet found a condition where endogenous DRAM-3 is critical. Alternatively, because DRAM-3 belongs to a family of proteins, another DRAM protein may compensate for DRAM-3 loss in this scenario.
Finally, we examined whether the differences we had observed in cell death under starvation conditions were related to DRAM-3’s effects on autophagy. LC3-II accumulated in both DRAM-3-expressing and control cells when incubated in low serum without glucose (Figure 7f and Supplementary Figure 7), and to test whether this was biologically significant, we repeated PI exclusion assays as described in Figure 7c, but this time in the presence of chloroquine. If autophagy was responsible for the increased survival of DRAM-3-expressing cells, this should be eliminated by chloroquine. We found, however, that DRAM-3’s ability to repress cell death in low serum with no glucose was unaffected by treatment with chloroquine, indicating that DRAM-3 mediates this effect in an autophagy-independent manner (Figure 7g). This effect was confirmed by analysis of Annexin V/PI staining (Supplementary Figure 6e).
Discussion
We describe here the primary characterization of DRAM-3, a new regulator of autophagy. DRAM-3 shares significant sequence homology with DRAM-1 and localizes to some similar subcellular compartments. Different from DRAM-1, however, DRAM-3 is not induced by p53 or DNA damage. DRAM-3 is also not induced by other autophagy-associated stimuli including culture in EBSS, low glucose, low serum or by treatment with IFN-γ or TNF.17, 18, 19, 29, 30 It remains possible that DRAM-3 could be induced by other DRAM-1-inducing stimuli, for example, the p53 family member, p73.31 Equally, dram-3 may be a gene that is not overtly modulated and yet its function is still important for basal autophagy. In this regard, it is notable that DRAM-3 is readily detectable in multiple tissues and cells, whereas DRAM-1 is virtually undetectable in many tissues in the absence of DNA-damage. Interestingly, we have also examined the levels of dram-3 gene dosage using the cBioPortal database, which revealed that dram-3 is frequently amplified in various tumor types (Supplementary Figure 8). We have no evidence, however, that dram-3 is the driving force for gene amplification in this region and this may instead be connected to a nearby gene to which dram-3 is linked.
As outlined above, one parallel between DRAM-1 and DRAM-3 is subcellular localization. Both proteins localize to endosomes, autolysosomes and lysosomes, and it is plausible that the two proteins perform similar or linked functions at these locations. However, the question remains whether they regulate autophagy in a redundant or in a concerted manner when DRAM-1 is induced by DNA damage, and further studies in this area are undoubtedly worthwhile.
Perhaps the most surprising finding from our study was the fact that while DRAM-3 can positively modulate autophagy, its ability to promote survival under starvation conditions is autophagy-independent. This function of DRAM-3 may be associated with its localization to the plasma membrane, a region not currently considered to be involved in autophagy beyond its participation as a source of membrane for autophagosome biogenesis.4 DRAM-3 may have some role in extracellular matrix engulfment or growth factor receptor recycling, both of which can modulate cell survival. Interestingly, DRAM-3’s localization at the plasma membrane is not uniform with particularly focused localization at focal adhesions. As focal adhesions can also signal cell survival via integrins,32 this may also be an area of focus for subsequent studies on DRAM-3 function.
In summary, to the best of our knowledge, this is the first study detailing DRAM-3 as a modulator of autophagy and, independently, cell survival under starvation conditions. Although many questions remain regarding the function of DRAM-3, due to the intense current interest in autophagy, nutrient sensing and cellular adaptation to starvation conditions, we hope that the findings we present here act as a starting point and catalyst for further investigation into the mechanism of action and functional consequences of this interesting new protein.
Materials and Methods
Cell culture, transfections and infections
All cells (except for HL-60, Raji and Jurkat cell lines) were grown in DMEM high glucose supplemented with 10% FBS, 0.292 mg/ml Glutamine and Penicillin (100 Units/ml)/Streptomycin (100 μg/ml) (all from Life Technologies, Carlsbad, CA, USA). Suspension cells (HL-60, Raji and Jurkat) were grown in RPMI (Life Technologies) supplemented with the reagents mentioned above.
For starvations, EBSS (Sigma, St. Louis, MO, USA) or DMEM no glucose (Gibco, Life Technologies) supplemented with 2% or 10% dialyzed FBS (Gibco) and sodium pyruvate (Life Technologies), as indicated, were used. Where indicated, late-stage autophagy inhibitor chloroquine (Sigma) or mitophagy uncoupler CCCP (Sigma) was used.
Cells were transfected using calcium phosphate precipitates as previously described.33 Tet-on p53 Saos-2 cells24 were induced with 1 μg/ml doxycycline (Sigma).
Ecotropic and amphotrophic retroviral infections with the following constructs were performed in Saos-2, Saos-2 EcoR and MDA-MB-231 EcoR cells as previously described:34 pWZL Hygro, LZRS-Parkin-wt (a kind gift from Stephen Tait), pWZL DRAM-3 Hygro and pLZRS Parkin-YFP Zeo (a kind gift from Stephen Tait) characterized in Tait et al.35 were carried out according to a previously described protocol.34 After infection, Saos-2 cells were selected with 100 μg/ml Hygromycin B (Roche Diagnostics, Burgess Hill, UK) for 7 days or 200 μg/ml Zeocin (Invitrogen, Life Technologies) for 10 days, as appropriate.
Saos-2 DRAM-3 KO cells were generated using a lentiviral CRISPR construct.36 Lentiviral infections were performed as previously described,37 and the infected cells were selected with 1 μg/ml Puromycin (Sigma) for 5 days.
qRT-PCR
RNA was collected and purified according to the manufacturer’s protocol of the Qiagen RNeasy Mini Kit (Qiagen, Venlo, Netherlands), including an optional on-column DNase digestion. cDNA was produced using the DyNAmo SYBR Green qPCR Kit (Life Technologies), and 1 μg RNA was used for the reverse transcription. For human tissue analysis, we purchaed a TissueScan Human Normal cDNA array from Origene (Rockville, MD, USA). qPCR was performed on a Chromo4 RT-PCR detector (Bio-Rad, Hercules, CA, USA). Samples were measured in triplicates, and results were normalized to 18S RNA control.
Primers used were as follows: 18S: fw: 5′-GTA ACC CGT TGA ACC CCA TT-3′, rev: 5′-CCA TCC AAT CGG TAG TAG CG-3′ DRAM-3 (Origene): fw: 5′-GTC TGC ACC ATC CTC ATT GTG G-3′, rev: 5′-CGG AGA AGT CAA CGG CTA AGA G-3′ DRAM-1 full length: fw: 5′-GCC ACA TAC GGA TGG TCA TCT CTG-3′, rev: 5′-GTG ACA CTC TGG AAA TCT TGG GAT-3′.
Clonogenic assays
For clonogenic assays, cells were seeded in 10 -cm dishes or six-well plates and left attached for 24 h before treatment. For starvation assays, cells were washed four times with PBS and incubated with starvation media. Media was exchanged to normal growth media at the indicated time points and cells were left growing for further 3–7 days.
Cell staining was performed using Giemsa-modified stain (Sigma). In short, cells were washed with PBS once and then incubated with Giemsa stain for 15 min. The stained cells were then washed with 10% methanol four times and left drying overnight.
Western blotting
Protein extraction was performed as previously described.38 Protein lysates were separated by SDS–PAGE and blotted onto PVDF membranes. Western blot analysis was performed according to standard techniques. Antibodies used were as follows: α-actin (Abcam, Cambridge, UK; ab8227), α-LC3B (Cell Signaling, Danvers, MA, USA; #2775), α-Myc-Tag (Millipore, Darmstadt, Germany; 05 – 724), α-GFP (Covance, Princeton, NJ, USA; B34), α-p53 DO1 (BD Bioscience, Oxford, UK; #554293), α-p62 (Sigma; p0067), α-BSA (Santa Cruz, Dallas, TX, USA; clone 25G7) and α-COX IV (Abcam; ab16056).
Confocal microscopy
For Immunofluorescence studies, cells were grown on glass coverslips for 2 days, then the coverslips were washed once in PBS and then fixed with 4% p-formaldehyde for 30 min at room temperature. Immunofluorescence staining of cells was carried out as previously described.39 An Olympus FV1000 confocal microscope with a 60 × objective and a Zeiss 710 confocal microscope with × 63 objective were used for taking images.
Antibodies used were as follows: α-Myc-Tag (Millipore; 4A6), α-Myc-Tag (Cell Signaling; #2272), α-LC3B (Cell Signaling; #2775), α-LAMP2 (BD Pharmingen, San Jose, CA, USA; CD107b), α-EEA1 (Abcam, ab2900), α-Calnexin (Cell Signaling; #2679), α-WIPI2 (Bio-rad; MCA578OGA) and α-GM130 (BD Bioscience, #610822).
For live-cell imaging, cells were transfected with GFP-tagged DRAM-3 and RFP-tagged Rab5 or Rab7 at least 2 days before confocal analysis. An Andor spinning disc confocal system with Cairn Optsplit (for simultaneous GFP/RFP imaging) was used for imaging. Videos of endosomes were taken for 1 min, with pictures in 1-s intervals.
Flow cytometry
Flow cytometry for unfixed (cell death assay) and methanol-fixed (cell cycle profile assay) cells was conducted as previously described,37 and a FACS Calibur was used for the analysis. All samples were measured in triplicates. For Annexin V/PI analysis, cells were stained with propidium iodide and Annexin V-Alexa 488 antibody according to the manufacturers instructions (Life Technologies).
Genomic DNA purification and sequencing
Genomic DNA of lentiCRISPR containing cells was isolated and purified using the Qiagen MiniAmp DNA mini Kit. The DRAM-3 sequence targeted by the CRISPR constructs was amplified by PCR, using 20 ng of genomic DNA of each cell line and a HotStar High Fidelity Polymerase kit (Qiagen). PCR was performed for 40 cycles, and the PCR products were run on a 2% agarose gel. After excision, gel purification was performed using the Qiagen QIAquick Gel Extraction Kit and the purified product (~500 bp) was sequenced.
PCR primers were as follows: fw: 5′-CGC TCC TTT CTC AAA CCC AG-3′; rev: 5′-GAG CAA AGG AAT CCA GCC G-3′ Sequencing primers: fw: 5′-AGG AGT CCA TGC CCC AGG-3′; rev: 5′-TAA GCC TAT GGC CAC GGG G-3′.
Mitophagy assay
Parkin-wt or Parkin-YFP containing cells were treated once (CRISPR cells) for 16 h or twice (Saos-2 pWZL cells) for a total of 24 h with 12.5 μM of the mitochondrial depolarizer CCCP (Sigma). Mitochondrial content was measured via COX IV Western blotting.
BCA uptake
To measure general endocytic uptake, cells were starved in serum-free media for 3.5 h and then refed with 1 mg/ml BSA (BP1605-100, Thermo Fisher Scientific, Waltham, MA, USA) and collected at different time points post-refeeding. BSA uptake was measured via Western blot detection of BSA. Data from 3 experiments were used to produce uptake kinetics.
Abbreviations
- DRAM-1:
-
damage-regulated autophagy modulator 1
- DRAM-2:
-
damage-regulated autophagy modulator 2
- DRAM-3:
-
DRAM-related/associated member 3
- Atg:
-
autophagy-related
- LC3:
-
microtubule-associated protein 1A/1B-light chain 3
- p62/SQSTM1:
-
sequestosome 1
- NBR1:
-
neighbor of BRCA1 gene 1
- OPTN:
-
optineurin
- LAMP2:
-
lysosome-associated membrane protein 2
- EEA1:
-
early endosome antigen 1
- TNFα:
-
tumor necrosis factor
- IFN-γ:
-
interferon-gamma
- COX IV:
-
cyctochrome c oxidase subunit IV
- XIAP:
-
X-linked inhibitor of apoptosis
- CCCP:
-
carbonyl cyanide m-chlorophenyl hydrazone
- Crispr:
-
clustered regularly interspaced short palindromic repeats
- Cas9:
-
Crispr-associated protein 9
- PI:
-
propidium iodide
- GFP:
-
green fluorescent protein
- YFP:
-
yellow fluorescent protein
References
Mizushima N, Levine B, Cuervo AM, Klionsky DJ . Autophagy fights disease through cellular self-digestion. Nature 2008; 451: 1069–1075.
Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182: 685–701.
Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010; 141: 656–667.
Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC . Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 2010; 12: 747–757.
Tooze SA, Yoshimori T . The origin of the autophagosomal membrane. Nat Cell Biol 2010; 12: 831–835.
Xie Z, Klionsky DJ . Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9: 1102–1109.
Ohsumi Y, Mizushima N . Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol 2004; 15: 231–236.
Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19: 5720–5728.
Stolz A, Ernst A, Dikic I . Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 2014; 16: 495–501.
Eskelinen EL, Saftig P . Autophagy: A lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta 2009; 1793: 664–673.
Mizushima N . Autophagy: process and function. Genes Dev 2007; 21: 2861–2873.
Levine B, Kroemer G . Autophagy in the pathogenesis of disease. Cell 2008; 132: 27–42.
Wilkinson S, Ryan KM . Autophagy: an adaptable modifier of tumourigenesis. Curr Opin Genet Dev 2010; 20: 57–64.
Kissova I, Deffieu M, Manon S, Camougrand N . Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem 2004; 279: 39068–39074.
Lemasters JJ . Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res 2005; 8: 3–5.
Lum JJ, DeBerardinis RJ, Thompson CB . Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol 2005; 6: 439–448.
Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 2005; 25: 1025–1040.
Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T et al. The role of autophagy during the early neonatal starvation period. Nature 2004; 432: 1032–1036.
Kamada Y, Sekito T, Ohsumi Y . Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr Top Microbiol Immunol 2004; 279: 73–84.
Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006; 126: 121–134.
Crighton D, Wilkinson S, Ryan KM . DRAM links autophagy to p53 and programmed cell death. Autophagy 2007; 3: 72–74.
O'Prey J, Skommer J, Wilkinson S, Ryan KM . Analysis of DRAM-related proteins reveals evolutionarily conserved and divergent roles in the control of autophagy. Cell Cycle 2009; 8: 2260–2265.
Corpet F . Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 1988; 16: 10881–10890.
Ryan KM, Ernst MK, Rice NR, Vousden KH . Role of NF-kappaB in p53-mediated programmed cell death. Nature 2000; 404: 892–897.
Mah LY, O'Prey J, Baudot AD, Hoekstra A, Ryan KM . DRAM-1 encodes multiple isoforms that regulate autophagy. Autophagy 2012; 8: 18–28.
Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544.
Baudot AD, Haller M, Mrschtik M, Tait SW, Ryan KM . Using enhanced-mitophagy to measure autophagic flux. Methods 2014; 75: 105–111.
Harlin H, Reffey SB, Duckett CS, Lindsten T, Thompson CB . Characterization of XIAP-deficient mice. Mol Cell Biol 2001; 21: 3604–3608.
Levine B, Mizushima N, Virgin HW . Autophagy in immunity and inflammation. Nature 2011; 469: 323–335.
Choi AM, Ryter SW, Levine B . Autophagy in human health and disease. N Engl J Med 2013; 368: 1845–1846.
Crighton D, O'Prey J, Bell HS, Ryan KM . p73 regulates DRAM-independent autophagy that does not contribute to programmed cell death. Cell Death Differ 2007; 14: 1071–1079.
Hehlgans S, Haase M, Cordes N . Signalling via integrins: implications for cell survival and anticancer strategies. Biochim Biophys Acta 2007; 1775: 163–180.
Bell LA, O'Prey J, Ryan KM . DNA-binding independent cell death from a minimal proapoptotic region of E2F-1. Oncogene 2006; 25: 5656–5663.
Ryan KM, O'Prey J, Vousden KH . Loss of nuclear factor-kappaB is tumor promoting but does not substitute for loss of p53. Cancer Res 2004; 64: 4415–4418.
Tait SW, Oberst A, Quarato G, Milasta S, Haller M, Wang R et al. Widespread mitochondrial depletion via mitophagy does not compromise necroptosis. Cell Rep 2013; 5: 878–885.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823.
Rosenfeldt MT, Bell LA, Long JS, O'Prey J, Nixon C, Roberts F et al. E2F1 drives chemotherapeutic drug resistance via ABCG2. Oncogene 2013; 33: 4164–4172.
Long JS, Crighton D, O'Prey J, Mackay G, Zheng L, Palmer TM et al. Extracellular adenosine sensing-a metabolic cell death priming mechanism downstream of p53. Mol Cell 2013; 50: 394–406.
Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature 2013; 504: 296–300.
Acknowledgements
We thank the members of the Tumour Cell Death Laboratory for advice and for critical reading of the manuscript. Work in the Tumour Cell Death Laboratory is supported by Worldwide Cancer Research, Astellas Pharma Inc. and Cancer Research UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Kroemer
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Mrschtik, M., O'Prey, J., Lao, L. et al. DRAM-3 modulates autophagy and promotes cell survival in the absence of glucose. Cell Death Differ 22, 1714–1726 (2015). https://doi.org/10.1038/cdd.2015.26
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2015.26
This article is cited by
-
TMEM150B is dispensable for oocyte maturation and female fertility in mouse
Scientific Reports (2020)
-
Mannose impairs tumour growth and enhances chemotherapy
Nature (2018)