Abstract
Tumor Protein p53-Induced Nuclear Protein 1 (TP53INP1) is a tumor suppressor that modulates the p53 response to stress. TP53INP1 is one of the key mediators of p53 antioxidant function by promoting the p53 transcriptional activity on its target genes. TP53INP1 expression is deregulated in many types of cancers including pancreatic ductal adenocarcinoma in which its decrease occurs early during the preneoplastic development. In this work, we report that redox-dependent induction of p53 transcriptional activity is enhanced by the oxidative stress-induced SUMOylation of TP53INP1 at lysine 113. This SUMOylation is mediated by PIAS3 and CBX4, two SUMO ligases especially related to the p53 activation upon DNA damage. Importantly, this modification is reversed by three SUMO1-specific proteases SENP1, 2 and 6. Moreover, TP53INP1 SUMOylation induces its binding to p53 in the nucleus under oxidative stress conditions. TP53INP1 mutation at lysine 113 prevents the pro-apoptotic, antiproliferative and antioxidant effects of TP53INP1 by impairing the p53 response on its target genes p21, Bax and PUMA. We conclude that TP53INP1 SUMOylation is essential for the regulation of p53 activity induced by oxidative stress.
Similar content being viewed by others

Main
Maintaining homeostasis in response to the broad range of intrinsic and extrinsic aggressions is a challenge for cells. Cellular outcomes are varied and involve cell-cycle arrest, apoptosis, DNA repair, autophagy, senescence, antioxidant activity, cell migration, differentiation, embryo implantation, metabolism and angiogenesis.1, 2, 3, 4, 5, 6, 7, 8, 9 An inadequate cell response can lead to cellular transformation and cancer initialization. Tumor Protein p53-Induced Nuclear Protein 1 (TP53INP1) is a p53 cofactor. The gene TP53INP1 encodes for two protein isoforms generated by alternative splicing, named TP53INP1α and TP53INP1β. TP53INP1 is a p53 target gene and its expression is induced in response to several physical and chemical stresses.10, 11, 12 TP53INP1 directly interacts with p53 and also binds kinases, such as HIPK2 and PKCδ, which modulate p53 transcriptional activity by phosphorylation, thereby creating a positive feedback loop between p53 and TP53INP1.13, 14 Our laboratory demonstrated the role of TP53INP1 as a tumor suppressor as TP53INP1-deficient mice present an increased susceptibility to tumor development. Moreover, TP53INP1 expression is lost at very early steps of pancreatic carcinogenesis due to miR155 activity and, when its expression is restored in pancreatic cancer cells, it suppresses growth of xenograft tumors by favoring apoptotic cell death.15, 16 Interestingly, the role of TP53INP1 as a tumor suppressor is associated with its ability to enhance the p53 antioxidant function.17 In accordance, TP53INP1 transcriptional induction upon oxidative stress is strictly dependent on p53, and TP53INP1-deficient cells accumulate more intracellular reactive oxygen species (ROS) than wild-type cells. This ROS accumulation is associated with a decreased expression of several p53 targets, including p21, Sesn2, PUMA and Bax. In summary, TP53INP1 is a major regulator of p53 response to oxidative stress.17
Post-translational modifications of proteins by ubiquitin and ubiquitin-like proteins have emerged since the last years as major regulators of proteins activity, localization and interactions.18, 19, 20 Ubiquitin-like conjugations are highly dynamic modifications, and multiple isopeptidases are able to remove the modifier from its substrate. Among all members of ubiquitin-like family, SUMO1 (Small Ubiquitin-Related Modifier) is a 11-kDa protein, well known for playing an essential role in the regulation of a number of cellular processes, including transcription, nuclear-cytoplasmic transport, apoptosis, response to stress and progression through the cell cycle.21, 22, 23
Bioinformatic analysis of the primary structure of TP53INP1 reveals the presence of a putative SUMOylation site on lysine 113. In this paper, we show that SUMOylation of TP53INP1 is tightly regulated by oxidative stress and is important for the p53 activity on cell-cycle arrest and cell death induction. Altogether, those results strongly suggest that modification of TP53INP1 by SUMO1 is crucial for its tumor suppressive abilities, as well as for the p53 response to oxidative stress.
Results
SUMO1 is conjugated to TP53INP1 on lysine 113
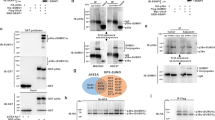
A bioinformatics analysis revealed the presence of a SUMOylation consensus sequence (ΨKXE)24, 25 at positions 112–115 of both isoforms of TP53INP1 (Figure 1a). To experimentally address this prediction, we have used U2OS cells, a human osteosarcoma cell line expressing wild-type p53. These cells were transfected with vectors expressing GFP-TP53INP1α or GFP-TP53INP1β fusion proteins along with a vector expressing a hexahistidine (6His) and Flag-tagged version of SUMO1 (6HF-SUMO1). 6HF-SUMO1 as well as 6HF-SUMO1-modified proteins was isolated using Ni2+-NTA resin, under strong denaturing conditions to prevent the action of specific proteases. An input of the sample was simultaneously saved to detect non-modified TP53INP1, as described under Materials and methods. As illustrated in Figure 1b, both TP53INP1 isoforms migrate under the 100-kDa marker (input panel). Following Ni2+ pull down, we detect a major band at higher molecular weight (over 100 kDa) for both isoforms of TP53INP1, corresponding probably to one SUMO1 molecule conjugated to TP53INP1α and β (Figure 1b). An identical profile was obtained when using an anti-TP53INP1 monoclonal antibody generated in the laboratory (Supplementary Figure 1A). To confirm that high molecular weight isoforms of TP53INP1α and β correspond to SUMO1-conjugated TP53INP1, we used site-directed mutagenesis to change the putative SUMOylated lysine 113 to arginine. The resulting mutant (K113R), as well as the wild-type (WT) form of TP53INP1, was expressed in U2OS cells along with the 6HF-SUMO1 construct, and TP53INP1 SUMOylation was analyzed as described above. As expected, mutation of lysine 113 to arginine completely prevented the SUMOylation of TP53INP1α and β (Figure 1c, Ni2+ pull down; Supplementary Figures 1B and 2), establishing TP53INP1 as a SUMO1 substrate via its lysine 113. Finally, we wondered whether the K113 could also be modified by other post-translational modifiers such as ubiquitin or the ubiquitin-like protein Nedd8. To answer this question, WT or K113R TP53INP1 was transfected in U2OS cells along with vectors expressing 6HF-SUMO1, or a 6His and Flag-tagged version of ubiquitin and Nedd8 (6HF-Ub and 6HF-Nedd8, respectively). Isolation of 6HF-ubiquitin-like modified proteins was performed as previously, and TP53INP1 was revealed using an anti-TP53INP1 antibody (Figure 1d; Supplementary Figure 1C). This experiment showed ladders of poly-ubiquitinated and poly-Neddylated forms of TP53INP1 when co-expressed with 6HF-Ubiquitin or 6HF-Nedd8 (Figure 1d). However, and as shown previously, whereas the K113R mutation completely prevented TP53INP1 SUMOylation, no effect was observed regarding TP53INP1 ubiquitinylation or Neddylation profiles (Figure 1d). Altogether, these results demonstrate that TP53INP1 is specifically conjugated to SUMO1 on its lysine 113 in cellulo.
TP53INP1 is conjugated to SUMO1 on lysine 113. (a) Schematic representation of the two isoforms of TP53INP1. Positions of lysines in the sequence are indicated by red bars. (b) U2OS cells were transfected with GFP-TP53INP1α, GFP-TP53INP1β and 6HF-SUMO1 expressing constructs. Twenty-four hours post transfection, 80% of cells were lysed in Guanidine-HCl containing buffer and 6HF-SUMO1 conjugates were isolated on Ni2+-NTA agarose beads. The remaining 20% were lysed in non-denaturing buffer to detect non-modified TP53INP1, as described under Materials and methods. SUMOylated proteins and input sample were resolved by SDS-PAGE, and TP53INP1 was revealed by western blotting using anti-GFP antibodies. (c) U2OS cells were transfected with the wild type (WT) or the K113R mutant TP53INP1α together with the 6HF-SUMO1 construct. 6HF-SUMO1 conjugates were isolated under strong denaturing conditions as in (b), and TP53INP1 was detected using the monoclonal anti-TP53INP1 F8 antibody. (d) U2OS cells were transfected with 6HF-SUMO1, 6HF-Ubiquitin and 6HF-Nedd8 constructs, together with the WT or K113R mutant TP53INP1α. 6HF-modified proteins were isolated as in (b), and TP53INP1 was revealed using the F8 antibody
TP53INP1 SUMOylation is regulated by oxidative stress and enable its antiproliferative effect
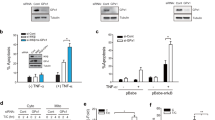
Both TP53INP1 expression and SUMO1 modification of proteins are tightly regulated by oxidative stress,26 which led us to investigate whether oxidative stress could be involved in the regulation of TP53INP1 modification by SUMO1. To answer this question, U2OS cells overexpressing TP53INP1 were transfected with the 6HF-SUMO1 vector. Cells were then treated by supplementation with the antioxidant N-acetylcysteine (NAC) in their culture medium and TP53INP1 SUMOylation was analyzed. As previously shown, TP53INP1α is constitutively SUMOylated in U2OS cells under standard culture conditions (Figure 2a), but NAC treatment strongly decreased this conjugation (Figure 2a). To confirm the observation that SUMOylation of TP53INP1 depends on oxidative stress, we used a U2OS cellular model in which TP53INP1 is induced by ponasterone A, leading to endogenous-like expression levels lower than stably transfected cells (see Supplementary Figure 3). Hence, TP53INP1 expression was induced, the 6HF-SUMO1 vector was expressed and cells were treated with increasing amounts of H2O2. As previously observed, TP53INP1α is constitutively SUMOylated in cells under standard culture conditions (Figure 2b). When 0.1 mM of H2O2 was added to the culture medium, we failed to observe a significant effect on TP53INP1 SUMOylation, probably due to the fact that 10% fetal calf serum present in the medium could interfere with the oxidative capacity of H2O2, but addition of 1 mM was sufficient to overcome this problem and induced the SUMOylation of TP53INP1 (Figure 2b). These experiments indicate that TP53INP1 SUMOylation is dependent on the redox status of the cell, an interesting feature given the fact that TP53INP1 is a stress protein whose expression is induced by several stresses, including oxidative stress.12 Since its discovery, TP53INP1 was described to induce cell-cycle arrest and apoptosis in stressed but also in unstressed cells.10, 12, 13, 14 To investigate whether constitutive SUMO1 conjugation to TP53INP1 could be involved in such cellular processes, we performed a clonogenicity assay with U2OS cells transfected with WT or K113R TP53INP1, or with an empty vector as a control. As shown in Figure 2c, expression of WT TP53INP1 strongly decreased the number of colonies compared with cells transfected with an empty vector, but this effect was prevented by the K113R mutation. This result suggested that SUMOylation on lysine 113 is required for TP53INP1 effect on cell death and/or cell-cycle arrest. To confirm this observation, U2OS cells were transfected with WT or K113R TP53INP1α, or with an empty vector as a control, cells were treated with H2O2, and caspase-3/7 activity was assessed. As expected, H2O2 treatment triggered caspase-3/7 activity in U2OS cells transfected with an empty vector (Figure 2d). However, induction of caspase-3/7 activity was almost two-fold increased when WT TP53INP1 was expressed, while the K113R mutation completely abrogated this effect (Figure 2d). To confirm that TP53INP1 SUMOylation is crucial for its effects on cell death during oxidative stress, we performed TUNEL assays on U2OS cells stably expressing the WT or K113R mutant TP53INP1. These cells were left untreated or exposed to H2O2 for 1 h, and 24 h later, the number of TUNEL-positive nuclei was counted. Under standard culture conditions, the number of apoptotic cells was low and similar in cells expressing either WT or K113R mutant TP53INP1 (Figure 2e). As expected, H2O2 treatment triggered apoptosis in both cell lines. However, the number of TUNEL-positive nuclei was almost twice as important in cells expressing WT TP53INP1 compared with the K113R mutant (Figure 2e). These results demonstrate that SUMOylation of TP53INP1 is regulated by oxidative stress, and also that the SUMO1 conjugation is crucial for the pro-apoptotic activity of TP53INP1 under such conditions.
TP53INP1 SUMOylation depends on oxidative stress and participates in the proapoptotic activity of TP53INP1. (a) U2OS cells stably overexpressing GFP-TP53INP1α were transfected with the 6HF-SUMO1 construct. Eight hours later, cells were treated with 10 mM of NAC during 16 h and 6HF-SUMO1 modified proteins were isolated as described previously. TP53INP1 was revealed using the F8 antibody. (b) TP53INP1α was induced by ponasterone A (PonA) in U2OS cells, which were transfected with the 6HF-SUMO1 construct. Twenty-four hours later, cells were treated with the indicated amounts of H2O2 during 15 min. 6HF-SUMO1 modified proteins were isolated and TP53INP1 was revealed using the F8 antibody. (c) U2OS cells were transfected with the WT or K113R mutant GFP-TP53INP1α, or with an empty vector as a control. Transfection efficiency was normalized by GFP epifluorescence, and clonogenicity assays were performed for 2 weeks. The graph represents the number of colonies observed after crystal violet staining at the end of the experiment. Results are presented as the percent of the number of colonies observed with the empty vector ±S.D. Graphs represent data from three independent experiments. *P<0.05. (d) U2OS cells were transfected with the WT or K113R mutant TP53INP1α, or with an empty vector as a control. Sixteen hours later, cells were exposed to 1 mM of H2O2 for 1 h, and incubated for another 24-h period. Caspase 3/7 activity was measured using a pro-fluorescent caspase-3/7 consensus substrate. Results are expressed in fold change compared with the control ±S.D. Graphs represent data from three independent experiments.*P<0.05. (e) U2OS cells stably expressing WT or K113R mutant TP53INP1 were seeded on coverslips, and exposed to 1 mM of H2O2 for 1 h the day after. Cells were fixed 24 h post treatment, and apoptotic cells were stained using a colorimetric TUNEL assay. The × 20 magnification was used to count the number of TUNEL-positive cells. Data presented are the mean of five fields counting (*P⩽0.05) from two independent experiments (n=2)
TP53INP1 SUMOylation is necessary for full activation of p53 transcriptional activity under oxidative stress
TP53INP1 induces cell death and cell-cycle arrest mainly through its interaction with p53, and the subsequent induction of p53 transcriptional activity on its target genes.10, 11, 13 Accordingly, we hypothesized that SUMOylation of TP53INP1 could be important for p53 transcriptional activity, especially under oxidative stress. To test this hypothesis, we performed luciferase reporter assays in which the reporter gene was under the control of p53-binding site (p53-TA-luc) to monitor the activity of p53 in cellulo. Cells were treated or not with H2O2 and luciferase activity was measured. When p53 was expressed alone, oxydative stress led to a moderate increase in the transcriptional activity of p53 (Figure 3a). Co-expression of WT TP53INP1 significantly increased p53 activity whereas this effect was not observed with the K113R mutant, indicating that SUMOylation of TP53INP1 was necessary for the complete activation of p53 transcriptional activity under oxidative stress. To confirm this finding, we decided to monitor the activity of p53 on several pro-apoptotic genes in cellulo. U2OS cells were transfected with p53, as well as with the WT or K113R forms of TP53INP1α. Cells were then treated or not with H2O2, and the mRNA levels of p53 target genes were monitored by quantitative RT-PCR. Under standard culture conditions, we failed to observe any significant difference in the mRNA levels of p21, Bax and PUMA (Figure 3b). However, during oxidative stress, a slight increase in the mRNA levels of p21, Bax and PUMA was observed when p53 was transfected alone. Co-expression of p53 with WT TP53INP1 led to a significant increase in the mRNA levels of p21, Bax and PUMA, which was abolished by the K113R mutation (Figure 3b). Altogether, these data demonstrate that under oxidative stress, SUMO1 conjugation to TP53INP1 is able to enhance p53 activity.
SUMOylation is necessary for TP53INP1-mediated activation of p53 transcriptionnal activity. (a) U2OS cells were co-transfected with p53 and a luciferase reporter gene construct p53-TA-luc. Where indicated, the WT or K113R mutant TP53INP1α was co-transfected with the previous constructs. Sixteen hours later, cells were exposed to H2O2 1 mM for 1 h, and incubated for an additional 24-h period, luciferase activity was measured in cell lysates. The graph represents luciferase activity normalized by a transfection efficiency control as described in Materials and Methods; results are expressed as fold changes compared with the control ±S.D. The assay was performed three times in triplicate with comparable results (*P<0.05). (b) U2OS cells were transfected with the WT or K113R mutant TP53INP1, or with an empty vector as a control and 16 h later, cells were exposed to 1 mM of H2O2 for 1 h. After incubation for another 12-h period total RNA was extracted using TRIzol (Invitrogen) and mRNA levels of p21, Bax and PUMA were analyzed by RT-qPCR. Results are expressed as fold changes compared with the control ±S.D. The assay was performed three times in triplicate with comparable results (*P<0.05)
SUMOylation is crucial for TP53INP1 interaction with p53 under oxidative stress
Our previous results showed that oxidative stress-induced SUMOylation of TP53INP1 is necessary to increase p53 transcriptional activity. We speculated that this SUMOylation could modulate the interaction between TP53INP1 and p53.13 To support this, proximity ligation assays (PLAs) were carried out in U2OS cells stably expressing TP53INP1α WT or K113R mutant, and transiently transfected with p53. It is important to note that PLA immunostaining does not show the intracellular distribution of the two proteins, but reveals whether and where they interact in the cell. A positive signal was detected in the nucleus of cells expressing WT but not K113R mutant TP53INP1, suggesting that the K113 residue is essential for the TP53INP1/p53 interaction (Figures 4a and b). Moreover, we observed that p53 co-immunoprecipitated only with wild-type TP53INP1 but not with the K113R mutant (Figure 4c).
SUMOylation is crucial for TP53INP1 interaction with p53 in cellulo under oxidative stress. (a) U2OS cells stably expressing WT TP53INP1 were transfected with p53 expression construct, seeded on coverslips 24 h post transfection and treated with H2O2 the day after. Cells were then fixed and stained for PLA immunofluorescence experiments. A positive signal in PLA experiments is observed as red dots and requires the spatial proximity between two proteins (<40 nm). Nuclei are stained by blue-fluorescent DAPI. (b) Quantification of TP53INP1-p53 interaction. Quantification of PLA dots was performed in a cell-by-cell manner using the NIS element AR software. A minimum of 15 cells for each condition has been used. PLAs were performed at least three times with comparable results. Graphs represent data from three independent experiments ±S.D. (*P<0.05). (c) U2OS cells stably expressing wild type or K113R mutant GFP-TP53INP1α were transfected with a p53 expressing construct. TP53INP1 was precipitated with anti-GFP antibodies, immunoprecipitated proteins were fractioned on SDS-PAGE, and p53 was detected by western blotting. Amount of precipitated TP53INP1 and expression levels in input samples were controlled
SUMOylation does not affect TP53INP1 function in autophagy
We and others have characterized the importance of TP53INP1 in macroautophagy.27, 28 We previously showed that TP53INP1 colocalizes with LC3 (microtubule-associated protein light chain 3) in autophagosomes and also increases the number of autophagosomes in the cytoplasm of cells leading to an accumulation of the autophagic markers LC3-II and p62. We investigated whether this process was also dependent on SUMO1 conjugation. As shown in Figure 5a and Supplementary Figure 4, TP53INP1 colocalized with endogenous LC3 into autophagosomes, and the K113R mutation did not impair its localization into these organelles (Figure 5a). This suggests that SUMOylation is not necessary for TP53INP1 localization into autophagosomes. To confirm this observation, we analyzed the impact of the K113R mutation on the number of autophagosomes in cells. Immunostaining on endogenous LC3 was performed, and the number of autophagosomes was followed by counting LC3-positive puncta. As expected, EBBS+rapamycin or Bafilomycin A1 treatments led to an increase in the number of autophagosomes per cell when an empty vector was transfected (Figure 5b). When autophagy was induced or blocked by the same treatments, the expression of TP53INP1 increased significantly the number of autophagosomes per cell, and the K113R mutation did not impair this effect (Figure 5b, compare WT or K113R TP53INP1). To further demonstrate that SUMOylation is not necessary for TP53INP1 autophagic activity, we monitored the amount of autophagy-related proteins such as the lipidated form of LC3 (LC3-II) and p62 by western blotting. We have recently shown that expression of TP53INP1 leads to the accumulation of both LC3-II and p62 in cellulo. As shown in Figure 5c, the K113R mutation did not affect the accumulation of the lipidated form of LC3 (LC3-II) and of p62 triggered by TP53INP1 expression. Thus, these data indicate that SUMOylation is not required forTP53INP1-mediated autophagy.
SUMOylation is not involved in TP53INP1-mediated autophagy. (a) U2OS stably expressing the WT or K113R mutant TP53INP1 were seeded on coverslips, and treated with Bafilomycin A1 100 nM for 6 h to block the autophagic flux. Cells were then fixed and immunostaining was performed using anti-GFP and anti-LC3 antibodies. Green staining represents TP53INP1 and red staining endogenous LC3. Merged images are shown in yellow. (b) U2OS cells were seeded on coverslips and transfected with the WT or K113R mutant TP53INP1, or with an empty vector as a control. Twenty-four hours later, cells were incubated either with rapamycin 10 μM for 6 h followed by EBSS treatment for 30 min, or with Bafilomycin A1 100 nM for 6 h. Cells were then fixed and immunostained using an anti-LC3 antibody. Autophagy was measured by quantification of LC3-positive dots per cell (n=50), data represent LC3 puncta by cell ±S.D. (*P<0.05). (c) U2OS cells were transfected with WT or K113R mutant TP53INP1, or with an empty vector as a control. Cell lysates were prepared and proteins were resolved by SDS-PAGE. Total levels of p62, LC3-I and II, TP53INP1 and β-Tubulin were monitored by western blotting. Right panel shows a densitometric quantification of bands. All assays were performed at least three times with comparable results (*P<0.05). NS, non significant differences
TP53INP1 is SUMOylated by Cbx4 and PIAS3, and deSUMOylated by SENP1, 2 and 6
To identify the enzyme implicated in TP53INP1 SUMOylation, U2OS cells stably expressing TP53INP1α were first transfected with siRNA targeting most of the known SUMO ligases,20 then transfected with the 6HF-SUMO1 construct, and 48 h later SUMOylation of TP53INP1 was analyzed by Ni2+ pull down and western blotting. As shown in Supplementary Figure 5A, we detected a significant decrease in TP53INP1 SUMOylation when PIAS3 and Cbx4 were knocked down (see Supplementary Figure 5B for inhibition efficiency) indicating that both enzymes are involved, and that they can compensate for each other. To confirm this hypothesis, U2OS cells were transfected with siRNA targeting Cbx4 and PIAS3 alone or in combination. An siRNA targeting the SUMO E2 Ubc9 was also used as a control (Figure 6a, compare Scramble and Ubc9 conditions). When Cbx4 and PIAS3 are simultaneously silenced, the amount of SUMO1-conjugated TP53INP1 was similar to the amount observed after Ubc9 silencing. Since Ubc9 is the SUMO E2 acting upstream of SUMO E3 ligases, this suggested that both Cbx4 and PIAS3 are responsible for TP53INP1 SUMOylation.
SUMO1 conjugation to TP53INP1 is mediated by Cbx4 and PIAS3, and deconjugation by SENP1, SENP2 and SENP6. (a) U2OS cells expressing TP53INP1α were transfected with 20 nM of siRNA targeting Cbx4 and PIAS3 alone or together with an siRNA targeting the Ubc9. Eight hours later, cells were transfected with the 6HF-SUMO1 construct, and 6HF-SUMO1 modified proteins were isolated by Ni2+ pull down 48 h later as indicated in the legend of Figure 1. TP53INP1 was revealed by western blotting using the F8 antibody. (b) U2OS cells stably expressing TP53INP1α wild type or the K113R mutant were transfected with the 6HF-SUMO1 construct, alone or in combination with a 3 × Flag-PIAS3 plasmid. 6HF-SUMO1 modified proteins were isolated by Ni2+ pull down, and TP53INP1 was revealed by western blotting using anti-GFP antibody. (c) U2OS cells were transfected with WT TP53INP1α GFP tagged, or with an empty vector as a control. TP53INP1 was precipitated using anti-GFP antibodies, immunoprecipitated proteins were fractioned on SDS-PAGE, and endogenous Cbx4 and PIAS3 was detected by western blotting. Amount of precipitated TP53INP1 and expression levels in input samples were controlled. (d) U2OS cells expressing TP5INP1α were transfected with siRNA targeting SENP1, SENP2 or SENP6 and 8 h later, cells were transfected with the 6HF-SUMO1 construct. The day after, cells were treated with NAC for an additional 24-h period, and 6HF-SUMO1 conjugates were isolated as described above. TP53INP1 was revealed using the anti-GFP antibody. Bottom panel shows a densitometric quantification of bands from the Ni2+ pull down assay
Nevertheless, we observed that siRNA-mediated inhibition of PIAS3 led to a slight decrease in the level of total TP53INP1α (Figure 6a, input panel). To unambiguously show that PIAS3 is implicated in TP53INP1 SUMOylation, U2OS cells stably expressing wild type or K113R mutant TP53INP1α were transfected with the 6HF-SUMO1 construct in combination with a vector expressing PIAS3. As shown in Figure 6b, co-expression of PIAS3 strongly increased TP53INP1α SUMOylation (Ni2+ pull down panel) and even led to the appearance of a heavier form of TP53INP1α with an apparent molecular weight of ∼150 kDa, which was not detected with the K113R mutant. Thus, these results strongly suggest that TP53INP1 is conjugated to SUMO1 on lysine 113 in a Cbx4 and PIAS3-dependent manner. Moreover, we showed that endogenous Cbx4 and PIAS3 co-immunoprecipitate with TP53INP1 (Figure 6c). Altogether, these data demonstrate that TP53INP1 SUMOylation is mediated by Cbx4 and PIAS3 in cellulo.
We next wanted to identify the enzymes that could remove SUMO1 from TP53INP1. In human, six sentrin-specific proteases (SENP) have been described (SENP1–3 and SENP5–7).29 Interestingly, these proteases differ with respect to SUMO isoform.30, 31 We previously showed that treating cells with NAC, a strong antioxidant, leads to a decrease in TP53INP1 SUMOylation (Figure 2a). We therefore hypothesized that under such conditions, SUMO1 is removed from TP53INP1 by endogenous SUMO1-specific proteases, namely SENP1, SENP2 or SENP6. To assess this hypothesis, siRNAs targeting these enzymes were transfected into U2OS cells stably expressing TP53INP1α, followed by the transfection of the 6HF-SUMO1 construct. The day after, cells were treated or not with NAC to induce SUMO1 deconjugation from TP53INP1 and the impact of each enzyme was monitored by Ni2+ pull down. As observed previously in Figure 2a, when a control siRNA was transfected, NAC treatment led to a decrease in TP53INP1 SUMOylation (Figure 6d). However, inhibition of each SENP partly rescued NAC-induced deSUMOylation of TP53INP1 (Figure 6d, see western blot and the corresponding quantification). In particular, silencing of SENP6 was the most potent in blocking NAC-induced deSUMOylation (Figure 6d, quantification panel). This experiment suggests that SENP6 is the main enzyme implicated in TP53INP1 deSUMOylation during NAC treatment. Altogether, these data demonstrate that TP53INP1 is SUMOylated by the two SUMO ligases Cbx4 and PIAS3, and that the three SUMO1-specific proteases SENP1, SENP2 and SENP6 deconjugate SUMO1 from TP53INP1.
Discussion
Mammalian cells developed a complex network of oxidative stress response to ensure their homeostasis and avoid DNA, proteins and lipids oxidative damages. These mechanisms enable cells exposed to an unusual amount of ROS, either to come back to a normal redox state or to undergo cell-cycle arrest and/or cell death, depending on the severity of the stress-associated damages. The transcription factor p53 and its cofactors have a critical and context-dependent role in the regulation of the cellular response to oxidative stress.32 TP53INP1 has previously been reported to be a major regulator of the p53 response to oxidative stress,17 but the mechanism responsible for this modulation remains elusive. In this work, we show evidence indicating that the redox-dependent regulation of p53 by TP53INP1 depends on a redox-sensitive SUMOylation of TP53INP1 at position K113.
At low levels of ROS, p53 exhibits antioxidant activity to eliminate free radicals ensuring cell survival, however, p53 can induce cell death in response to high oxidative stress.32 This switch in p53 functions is allowed by a tight regulation of its transcriptional activity mediated by post-translational modifications, subcellular re-localization and interaction or not with a broad range of regulator proteins.33, 34 TP53INP1 seems to have a key role in helping p53 response against oxidative stress. We showed in a previous work that cells exposed to an oxidative challenge trigger TP53INP1 gene expression, and that the newly synthesized TP53INP1 protein interacts with p53 thereby enhancing its transcriptional activity.17 Two of the p53 activated target genes are sestrin1 and 2 (Sesn1 and Sesn2), which protect cells from H2O2-induced damages through the generation of peroxiredoxins.35 In agreement with this in cellulo data, TP53INP1-deficient mice undergo chronic oxidative stress.16 Moreover, TP53INP1 is also able to enhance the p53 transcriptional activity on cell cycle-related genes such as p21 and pro-apoptotic genes like PUMA and Bax. Data presented in this work suggest that TP53INP1 SUMOylation is one of the molecular mechanisms driving p53 response to oxidative stress. However, the in vivo biological relevance of the stress-induced TP53INP1-mediated augmentation of transcriptional activities of p53 needs to be further studied in other models. The stress conditions used in this work are very stringent and many of the effects observed were obtained under overexpression conditions.
In addition to its function on p53 regulation, we recently showed that TP53INP1 possesses a cytoplasmic role associated with the autophagic process.27 When TP53INP1 expression is strongly induced by high oxidative stress, it interacts with LC3 in the autophagosomes, displacing p62, and inducing autophagic-dependent cell death. However, under low oxidative stress conditions, the intracellular concentration of TP53INP1 is very low (negligible in comparison with p62), and TP53INP1 does not displace p62 from the autophagosomes and therefore autophagy works as a cell survival mechanism.36 Interestingly, we show in this paper that TP53INP1 SUMOylation is not involved in the autophagic process. TP53INP1 K113R mutant, although not SUMOylated, colocalizes with LC3 into the autophagosomes, allowing the increase in the number of autophasomes in the cytoplasm of cells and triggering an accumulation of the autophagic marker LC3-II. We observed p62 accumulation with the wild type and the mutant TP53INP1, suggesting that both proteins interact with LC3 and displace p62 from the autophagosomes. This observation indicates that post-translational modification by SUMO does not affect TP53INP1 role in autophagy and is only associated with its nuclear function when cooperating with p53 in the antioxidant response.
In the model shown in Figure 7 we describe our hypothesis in which TP53INP1 acts as a dampening mechanism for the p53-mediated activation of apoptosis through a redox-sensitive post-translational modification. We are now studying whether the SUMOylation of TP53INP1 and the subsequent effects on p53 functions are exclusively redox dependents or could also be induced by other stresses that usually induce the transcriptional activity of p53.
Oxidative stress-induced p53 activity by a redox-sensitive TP53INP1 SUMOylation. During oxidative stress, TP53INP1 expression is induced by p53. If stress induces autophagy, then TP53INP1 interacts with LC3 in the cytoplasm, goes to the autophagososmes to displace p62 triggering autophagic-associated cell death. If stress induces oxidation, then TP53INP1 is SUMOylated, interacting with p53 in the nucleus and favoring p53 transcriptional activity on pro-apoptotic genes
Post-translational modifications by SUMO are well known to be tightly regulated by the intracellular redox status. The steady-state SUMOylation of cellular substrates results from an equilibrium between SUMO conjugation and deconjugation. Both conjugation machinery, including E1 (Aos1/Uba2) and E2 (Ubc9) enzymes, and deconjugation enzymes, such as sentrine protease (SENP) family members, are redox-sensitive proteins.26 Moreover, the effect of oxidative stress on SUMO conjugation is dose dependent. Indeed, low levels of oxidative stress could prevent the SUMOylation of cellular substrates while higher concentrations of ROS could impair the deconjugation and could lead to an accumulation of SUMO conjugates. Following this principle, it is important to note that the accumulation of SUMOylated TP53INP1 we observed during oxidative stress could be due to either activation of the ligases or inactivation of proteases. For the moment, our work cannot distinguish between these two possibilities. However, we show that SUMOylation of TP53INP1 is indeed related to the intracellular oxidative level, and SUMOylation process, which is well controlled by oxidative thresholds, could be a more general mechanism functioning as an oxidative stress sensor for p53. In this work, we also report a post-translational modification of TP53INP1 by ubiquitine and Nedd8, but their implication regarding the molecular role of TP53INP1 remains unknown. TP53INP1 seems to be a key intracellular sensor of ROS, which is regulated at multiple levels. It is first regulated at the transcriptional level by p53, p73 and E2F1,10, 12, 37 which promote its expression upon cellular stress conditions. Second, it is regulated at post-transcriptional levels by a broad range of micro-RNA, including miR-17, miR-93, miR-125b, miR-130b and miR-155.15, 38, 39, 40 In this work, we add a third layer on the complex TP53INP1 regulation, through its redox-dependent post-translational modifications. This reinforces the hypothesis that p53 regulators such as TP53INP1 are very important in cell fate decision, and shed light to their tumor suppressor mechanism of action.
Materials and Methods
Cell culture and reagents
Cells were maintained in DMEM Glutamax medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS), in a humidified atmosphere with 5% CO2 at 37°C. HEK293T (human embryonic kidney cells), MCF7 (breast cancer cells) and U2OS (human osteosarcoma containing wild-type p53) cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). TP53INP1α-inducible U2OS cells were obtained as previously indicated15 and were cultured in the presence of zeocin (0.05 mg/ml) and G418 (0.2 mg/ml). U2OS stably expressing TP53INP1αWT and TP53INP1αK113R was obtained by transfection of vectors expressing the desired proteins and selection in the presence of G418 (1 mg/ml), and were cultured with G418 (0.2 mg/ml). In TP53INP1α-inducible U2OS cells, TP53INP1α-GFP expression was induced using 10 μM ponasterone A (Invitrogen) for 24 h. Cells were treated as indicated with NAC (Sigma-Aldrich, St. Louis, MO, USA), H2O2 (Sigma-Aldrich), MG132 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), bafilomycin A1 (LC Laboratories, Woburn, MA, USA), rapamycin (LC Laboratories) and EBSS (Invitrogen).
Transfections and siRNA
DNA and siRNA transfections were performed respectively using FuGENE HD (Promega, Madison, WI, USA) and INTERFERin (Polyplus Transfection, Illkirch, France) following the manufacturer’s instructions. siRNA was synthetized by Eurofins MWG Operon (Ebersberg, Germany). Sequences of the siRNA used are indicated in Supplementary Table 1.
DNA constructs
TP53INP1α and β cDNAs were subcloned into the pEGFP-N1 vector (Clontech, Mountain View, CA, USA). TP53INP1K113R mutants were obtained by PCR site-directed mutagenesis. SUMO1, Ubiquitin and Nedd8 cDNAs were ligated into the pCCL-WPS-PGK vector modified to express 6-histidines and a Flag tag at the N-terminus of the expressed protein. pcDNA-p53 was a kind gift from A Sparks (University of Dundee, UK). pHRLuc-C2 (Perkin Elmer Life and Analytical Sciences, Boston, MA, USA) was used as a transfection control. pCMV-3 × Flag-PIAS3 was a kind gift from Laura Corbo (Centre de Recherche en Cancerologie de Lyon, Lyon, France).
Isolation of 6Histidine-ubiquitin-like conjugates
Purification of 6His-ubiquitin-like conjugates was performed as described by Rodriguez et al.24 U2OS cells were seeded in 10 cm dishes and transfected using FuGENE HD as indicated. Twenty-four hours post transfection, cells were washed twice with PBS and scrapped in 1 ml of PBS. Twenty percent of cell suspension was lysed in non-denaturing buffer and used to control expression of transfected proteins (see below). The remainder was lysed in 6 ml of 6 M Guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 8.0, 15 mM imidazole and 10 mM β-mercaptoethanol (βME) (Buffer 1). After sonication of lysates to reduce viscosity, 50 μl of Ni2+-NTA resin (Qiagen, Chatsworth, CA, USA) pre-washed with buffer 1 was added and lysates were rotated at room temperature (RT) for 4 h. The beads were successively washed for 5 min in each step at RT with 750 μl of each of the following buffers: buffer 1; buffer 2 (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 8.0, 10 mM βME); buffer 3 (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 6.3, 10 mM βME) plus 0.2% Triton X-100; buffer 3 and then buffer 3 plus 0.1% Triton X-100. After the last wash, 6His-ubiquitin-like conjugates were eluted by incubating the beads in 50 μl of buffer 4 (200 mM imidazole, 0.15 M Tris/HCl pH 6.7, 30% glycerol, 0.72 M βME, 5% SDS) for 20 min at RT. Eluates were analyzed by western blotting.
Non-denaturing cell lysis
The cell pellet obtained from the 20% cell suspension described above was lysed in cold phosphate lysis buffer (50 mM NaH2PO4, 150 mM NaCl, 1% Tween-20, 5% Glycerol, pH 8.0) supplemented with protease cocktail inhibitor (Roche Applied Science, Meylan, France; 1:200), 10 mM N-ethylmaleimide (NEM) and 2 mM phenylmethylsulfonyl fluoride (PMSF). After 5 min of incubation on ice, lysates were centrifuged 10 min at 13 000 r.p.m. at 4°C and pellets were discarded. Protein concentration in the supernatant was determined by Bradford assay (Bio-Rad), and equal amounts of total proteins were used for western blot analysis.
Co-immunoprecipitations
U2OS cells were transfected using FuGENE HD (Promega) following the manufacturer’s instructions. Briefly, 70% confluent cells seeded in 60 mm dishes were transfected with a total of 4 μg of the indicated plasmids. Cells were lysed 24 h post transfection in lysis buffer (50 mM Hepes, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100, 25 mM NaF, 10 μM ZnCl2, 1 mM Na3VO4, protease inhibitors cocktail (Roche Applied Science), 10 mM NEM and 2 mM PMSF) and centrifuged at 13 000 r.p.m. for 10 min at 4°C. Pellet was discarded and protein concentration in the supernatant was adjusted using Bradford assay (Bio-Rad). In all, 1 mg of proteins was used for immunoprecipitations. Lysates were cleared with 60 μl of protein G-sepharose beads for 45 min, and cleared lysates were incubated with primary antibodies for 2 h at 4°C, followed by 45 min incubation with 20 μl of protein G-sepharose beads. After washing the beads five times in cold lysis buffer, the complexes were dissolved in Laemmli sample buffer and boiled for 5 min. Eluates were analyzed by western blotting as described.
Western blots
Proteins were resolved by SDS-PAGE, transferred onto nitrocellulose filters, blocked 1 h at RT in Tris-buffered saline/5% non-fat dry milk/0.1% Tween-20, and blotted overnight with primary antibodies at 1 : 1000. After extensive washings in TBS/0.1% Tween-20, filters were incubated 1 h at RT with HRP-conjugated secondary antibodies at 1 : 3000 before being revealed with ECL. Acquisition was performed with a Fusion FX7 imager (Vilber-Lourmat, France). For Flag (6HF constructs) and β-Tubulin immunoblots, SNAP i.d. protein detection system (Millipore, Molsheim, France) was used following the manufacturer’s instructions. Antibodies used for immunoblotting were anti-GFP (11814460001; Roche Applied Science), anti-LC3 (2775; Cell Signaling Technology, Danvers, MA, USA), anti-p62 (610833; BD Biosciences, San Diego, CA, USA), anti-Flag M2 (F3165; Sigma-Aldrich), anti-p21 (sc-397; Santa Cruz Biotechnology), anti-bax (5023; Cell Signaling Technology), anti-PUMA (4976, Cell Signaling Technology), anti-p53 (2527; Cell Signaling Technology), anti-PIAS3 (C-12, sc46682; Santa Cruz Biotechnology), anti-Cbx4 (HPA008228; Sigma-Aldrich), anti-p53 (7F5; Cell Signaling Technology) and anti-TP53INP1 (rat monoclonal antibody generated in our laboratory, clone F8). Densitometry quantification of bands was done using the ImageJ software (http://rsb.info.nih.gov/ij). Experiments were performed at least three times with comparable results.
Clonogenicity assay
U2OS cells in 10 cm dishes were transfected with TP53INP1αWT-GFP, TP53INP1αK113R-GFP and pEGFP-N1 as a control. Transfection efficiency was controlled by GFP fluorescence. After 24 h of transfection, cells were harvested, diluted and distributed in new 10 cm dishes (250 000 GFP fluorescent cells per dish), with G418 (1 mg/ml) as selection pressure. Cells were allowed to grow during 2 weeks under selection pressure and then were stained with crystal violet to quantify surviving colonies. These experiments were performed three times with comparable results.
Caspase-3/7 activity assay
U2OS cells in 12-well plates were transfected with TP53INP1αWT-GFP, TP53INP1αK113R-GFP and pEGFP-N1 as a control. Sixteen hours after transfection, cells were exposed 1 h to 1 mM H2O2. Caspase-3/7 activity was measured 40 h after transfection (24 h after treatment) using the APO-One Homogeneous Caspase-3/7 Assay Kit (Promega) according to the manufacturer’s instructions.
TUNEL assay
U2OS cells stably expressing either TP53INP1α WT or the K113R mutant were seeded in 24-well plates on glass coverslips, at 105 cells per well. The day after cells were exposed 1 h to 1 mM of H2O2, and 24 h post treatment cells were fixed in 4% paraformaldehyde (PFA) in PBS. Apoptotic cells were detected using the DeadEnd Colorimetric TUNEL System (Promega, G7130) following the manufacturer’s instructions, and microscopy was performed by using a Nikon Eclipse 90i fluorescence microscope (Nikon Instruments Europe B.V., Champigny-sur-Marne, France). Images were captured in 16 bit TIFF format with a Nikon DS-1QM camera and visualized using the NIS element AR software (Nikon Instruments Europe B.V.). For TUNEL-positive cells counting, five independent fields per condition were examined using the × 20 objective. Results are presented as a mean value of TUNEL-positive nuclei per field.
RNA extraction and quantitative PCR
U2OS cells in 10 cm dishes were transfected with TP53INP1αWT-GFP, TP53INP1αK113R-GFP and pEGFP-N1 as a control. Sixteen hours after transfection, cells were exposed 1 h to 1 mM H2O2 and total RNA was extracted using TRIzol (Invitrogen) 12 h after treatment. For siRNA validation, U2OS cells in 6-well plates were transfected by siRNA and RNA extraction was performed 48 h later. cDNA synthesis was performed using the Improm-II kit (Promega) according to the manufacturer’s protocol, and quantitative RT-PCR was performed in a MX3005P machine (Agilent, Massy, France) using the SYBR Premix Ex Taq and ROX reference dye (Takara Bio, Otsu, Shiga, Japan). Primers were synthetized by Eurofins MWG Operon. Sequences of the primers used for quantitative PCR are indicated in Supplementary Table 2.
Transcriptional activation assay
U2OS cells in 12-well plates were co-transfected by a Firefly luciferase reporter gene construct controlled by p53-binding site multimers (p53-TA-luc) with pcDNA-p53 and TP53INP1αWT-GFP or TP53INP1αK113R-GFP as indicated, together with 0.1 μg of pHRLuc-C2 as a transfection control. All co-transfections were balanced with pcDNA4 empty vector. Sixteen hours after transfection, cells were treated for 1 h with 1 mM H2O2, and 24 h later cells were lysed and Firefly luciferase activity was measured in cell lysate using the Luciferase Assay System Kit (Promega) according to the manufacturer’s instructions. Renilla luciferase activity (from pHRluc-C2) was also measured after adding cœlenterazin-h (Promega) to a final concentration of 5 μM in cell lysates, and was used to normalize Firefly luciferase activity. Luminescence measurements were done using a LB941 Tristar reader (Berthold France SA, Thoiry, France).
Immunofluorescence
Cells on glass coverslips were transfected or induced for TP53INP1 expression. After 24 h, cells were treated as indicated and then fixed with 4% PFA in PBS. Cells were permeabilized (0.2% Triton X-100 in PBS), incubated for 30 min in blocking buffer (5% FBS), and then incubated 1 h with the following antibodies diluted in blocking buffer: anti-GFP (11814460001; Roche Applied Science) or anti-LC3 (PM063; MBL, Woburn, MA, USA). Cells were then washed with PBS and incubated 1 h with secondary antibodies (Alexa fluor 488 anti-mouse and Alexa fluor 568 anti-rabbit; Invitrogen), washed with PBS, stained with DAPI (Invitrogen) for 5 min and mounted in ProLong Gold antifade reagent (Invitrogen) for imaging. Fluorescent images were captured using a Nikon Eclipse 90i microscope equipped with a Nikon DS-1QM camera. Quantification of LC3-positive puncta per cell was done by counting 50 cells by condition in three independent experiments.
Proximity ligation assay
U2OS cells stably expressing the WT or K113R mutant TP53INP1 were seeded in 10 cm dishes and transfected with 3 μg of p53 expression construct using FuGENE HD (Promega). Twenty four hours post transfection, cells were seeded on coverslips and fixed the day after with 4% PFA in PBS. Free aldehydes were quenched by the addition of 50 mM NH4Cl in PBS, and cells were permeabilized in 0.2% Triton X-100. Immunostaining using the DuoLink kit (Olink Bioscience, Uppsala, Sweden) was performed following the manufacturer’s protocol. The preparations were mounted using Prolong Gold with DAPI reagent (Invitrogen) and examined with Nikon microscope Eclipse 90I. Z-stack pictures were obtained by using a Nikon Digital Sight DS-1QM camera controlled by NIS element AR software, using the × 40 objective and same exposure times, and deconvolution algorithm ‘Richardson-Lucy’ from Lab imaging was applied. For each field, z-images with clear PLA signals were fused and quantification of PLA signals was performed in a cell-by-cell manner using the NIS element AR software. A minimum of 15 cells for each condition has been used.
Abbreviations
- βME:
-
β-mercaptoethanol
- FBS:
-
fetal bovine serum
- hnRNP-K:
-
heterogeneous ribonucleoprotein-K
- LC3:
-
microtubule-associated protein light chain 3
- NAC:
-
N-acetylcysteine
- ROS:
-
reactive oxygen species
- SUMO:
-
small ubiquitin-like modifier
- TP53INP1:
-
tumor protein 53-induced nuclear protein1
- PLA:
-
proximity ligation assay
References
Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B . A model for p53-induced apoptosis. Nature 1997; 389: 300–305.
Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006; 126: 121–134.
Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O et al. p53 regulates mitochondrial respiration. Science 2006; 312: 1650–1653.
Roger L, Gadea G, Roux P . Control of cell migration: a tumour suppressor function for p53? Biol Cell 2006; 98: 141–152.
Teodoro JG, Parker AE, Zhu X, Green MR . p53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science 2006; 313: 968–971.
Hu W, Feng Z, Teresky AK, Levine AJ . p53 regulates maternal reproduction through LIF. Nature 2007; 450: 721–724.
Vousden KH, Prives C . Blinded by the light: the growing complexity of p53. Cell 2009; 137: 413–431.
Demidenko ZN, Korotchkina LG, Gudkov AV, Blagosklonny MV . Paradoxical suppression of cellular senescence by p53. Proc Natl Acad Sci USA 2010; 107: 9660–9664.
Kang HN, Oh SC, Kim JS, Yoo YA . Abrogation of Gli3 expression suppresses the growth of colon cancer cells via activation of p53. Exp Cell Res 2012; 318: 539–549.
Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y et al. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell 2001; 8: 85–94.
Tomasini R, Samir AA, Pebusque MJ, Calvo EL, Totaro S, Dagorn JC et al. P53-dependent expression of the stress-induced protein (SIP). Eur J Cell Biol 2002; 81: 294–301.
Tomasini R, Samir AA, Vaccaro MI, Pebusque MJ, Dagorn JC, Iovanna JL et al. Molecular and functional characterization of the stress-induced protein (SIP) gene and its two transcripts generated by alternative splicing. SIP induced by stress and promotes cell death. J Biol Chem 2001; 276: 44185–44192.
Tomasini R, Samir AA, Carrier A, Isnardon D, Cecchinelli B, Soddu S et al. TP53INP1s and homeodomain-interacting protein kinase-2 (HIPK2) are partners in regulating p53 activity. J Biol Chem 2003; 278: 37722–37729.
Yoshida K, Liu H, Miki Y . Protein kinase C delta regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage. J Biol Chem 2006; 281: 5734–5740.
Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA 2007; 104: 16170–16175.
Gommeaux J, Cano C, Garcia S, Gironella M, Pietri S, Culcasi M et al. Colitis and colitis-associated cancer are exacerbated in mice deficient for tumor protein 53-induced nuclear protein 1. Mol Cell Biol 2007; 27: 2215–2228.
Cano CE, Gommeaux J, Pietri S, Culcasi M, Garcia S, Seux M et al. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res 2009; 69: 219–226.
Haglund K, Dikic I . Ubiquitylation and cell signaling. EMBO J 2005; 24: 3353–3359.
Welchman RL, Gordon C, Mayer RJ . Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol 2005; 6: 599–609.
Kerscher O, Felberbaum R, Hochstrasser M . Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 2006; 22: 159–180.
Seeler JS, Dejean A . Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 2003; 4: 690–699.
Verger A, Perdomo J, Crossley M . Modification with SUMO. A role in transcriptional regulation. EMBO Rep 2003; 4: 137–142.
Hay RT . SUMO: a history of modification. Mol Cell 2005; 18: 1–12.
Rodriguez MS, Dargemont C, Hay RT . SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 2001; 276: 12654–12659.
Sampson DA, Wang M, Matunis MJ . The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem 2001; 276: 21664–21669.
Bossis G, Melchior F . Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell 2006; 21: 349–357.
Seillier M, Peuget S, Gayet O, Gauthier C, N’Guessan P, Monte M et al. TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagy-dependent cell death. Cell Death Differ 2012; 19: 1525–1535.
Sancho A, Duran J, Garcia-Espana A, Mauvezin C, Alemu EA, Lamark T et al. DOR/Tp53inp2 and Tp53inp1 constitute a metazoan gene family encoding dual regulators of autophagy and transcription. PLoS One 2012; 7: e34034.
Hickey CM, Wilson NR, Hochstrasser M . Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 2012; 13: 755–766.
Shen LN, Dong C, Liu H, Naismith JH, Hay RT . The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem J 2006; 397: 279–288.
Reverter D, Lima CD . Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol 2006; 13: 1060–1068.
Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM . The antioxidant function of the p53 tumor suppressor. Nat Med 2005; 11: 1306–1313.
Sionov RV, Haupt Y . The cellular response to p53: the decision between life and death. Oncogene 1999; 18: 6145–6157.
Murray-Zmijewski F, Slee EA, Lu X . A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol 2008; 9: 702–712.
Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM . Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science 2004; 304: 596–600.
Maiuri MC, Zalckvar E, Kimchi A, Kroemer G . Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8: 741–752.
Hershko T, Chaussepied M, Oren M, Ginsberg D . Novel link between E2F and p53: proapoptotic cofactors of p53 are transcriptionally upregulated by E2F. Cell Death Differ 2005; 12: 377–383.
Yeung ML, Yasunaga J, Bennasser Y, Dusetti N, Harris D, Ahmad N et al. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res 2008; 68: 8976–8985.
Enomoto Y, Kitaura J, Hatakeyama K, Watanuki J, Akasaka T, Kato N et al. Emu/miR-125b transgenic mice develop lethal B-cell malignancies. Leukemia 2011; 25: 1849–1856.
Bomben R, Gobessi S, Dal BoM, Volinia S, Marconi D, Tissino E et al. The miR-17 approximately 92 family regulates the response to Toll-like receptor 9 triggering of CLL cells with unmutated IGHV genes. Leukemia 2012; 26: 1584–1593.
Acknowledgements
We thank Patricia Spoto and Bruno Olivier for technical help, Laurence Borge and the platform of cellular culture at the Cancer Research Center of Marseille, and Julie Leca for technical assistance with TUNEL experiments. We also thank Dr Laura Corbo (Centre de Recherche en Cancérologie de Lyon) for the gift of PIAS3 expressing construct. Authors acknowledged also the reviewers of this manuscript, some sentences included in the discussion were directly inspired on their comments. This work was supported by La Ligue Contre le Cancer, the INCa, the SIRIC and the INSERM. SP was supported by CFP, ARC and la Ligue Contre le Cancer and TB was supported by la Ligue Contre le Cancer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Oren
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Peuget, S., Bonacci, T., Soubeyran, P. et al. Oxidative stress-induced p53 activity is enhanced by a redox-sensitive TP53INP1 SUMOylation. Cell Death Differ 21, 1107–1118 (2014). https://doi.org/10.1038/cdd.2014.28
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.28
This article is cited by
-
Long non-coding RNA GAS5 promotes cisplatin-chemosensitivity of osteosarcoma cells via microRNA-26b-5p/TP53INP1 axis
Journal of Orthopaedic Surgery and Research (2023)
-
GDF11 slows excitatory neuronal senescence and brain ageing by repressing p21
Nature Communications (2023)
-
Food-seeking behavior is triggered by skin ultraviolet exposure in males
Nature Metabolism (2022)
-
TGF-β and WNT signaling pathways in cardiac fibrosis: non-coding RNAs come into focus
Cell Communication and Signaling (2020)
-
Increased nuclear permeability is a driver for age-related motoneuron loss
GeroScience (2020)