Abstract
Innate immunity represents the first line of defence against invading pathogens. It consists of an initial inflammatory response that recruits white blood cells to the site of infection in an effort to destroy and eliminate the pathogen. Some pathogens replicate within host cells, and cell death by apoptosis is an important effector mechanism to remove the replication niche for such microbes. However, some microbes have evolved evasive strategies to block apoptosis, and in these cases host cells may employ further countermeasures, including an inflammatory form of cell death know as necroptosis. This review aims to highlight the importance of the RIP kinase family in controlling these various defence strategies. RIP1 is initially discussed as a key component of death receptor signalling and in the context of dictating whether a cell triggers a pathway of pro-inflammatory gene expression or cell death by apoptosis. The molecular and functional interplay of RIP1 and RIP3 is described, especially with respect to mediating necroptosis and as key mediators of inflammation. The function of RIP2, with particular emphasis on its role in NOD signalling, is also explored. Special attention is given to emphasizing the physiological and pathophysiological contexts for these various functions of RIP kinases.
Similar content being viewed by others
Facts
-
RIP1 mediates the signalling switch between inflammatory gene expression and apoptosis.
-
RIP1 and RIP3 form amyloid filaments to trigger necroptosis.
-
RIP1/RIP3-mediated necroptosis is a defence mechanism but can cause inflammatory disease.
-
RIP1 and RIP3 are important mediators of pattern-recognition receptor (PRR) signalling.
-
RIP2 is a critical mediator of NOD signalling and mucosal immunity.
Open Questions
-
How are the kinase activities of RIP1 and RIP3 regulated to control formation of the necrosome complex?
-
How is RIP3 activated in those pathways that use RIP3 but not RIP1 to induce necroptosis?
-
Apart from virally encoded caspase inhibitors, how is caspase 8 inhibited to promote RIP3-mediated necroptosis and inflammation?
-
How does RIP3 regulate the NLRP3 inflammasome?
-
Can RIP1/RIP3-mediated necroptosis and RIP2 signalling be targeted to treat inflammatory diseases?
The innate immune system is equipped with PRRs that act as the primary sensing systems for invading pathogens by recognizing molecular structures known as pathogen-associated molecular patterns (PAMPs). PRRs include transmembrane Toll-like receptors (TLRs),1 cytosolic NOD-like receptors (NLRs),2 RIG-I-like receptors3 and DNA sensors.4 When engaged by relevant PAMPs, PRRs trigger signal transduction cascades resulting in activation of transcription factors such as NFκB and induction of a plethora of pro-inflammatory genes. Tumour necrosis factor (TNF) and interleukin-1β (IL-1β) are two of the most critical pro-inflammatory cytokines, and their receptors can also activate NFκB to promote further expression of inflammatory genes. This facilitates infiltration of leukocytes into the infected tissue resulting in removal of the pathogen. Cell death can also be an important part of host defence by destroying the replication niche for some invading microbes. Indeed cell killing can integrate closely with inflammation by acting as a potent driving force behind the inflammatory response.5 Although programmed cell death by apoptosis is generally regarded as silent in an inflammatory sense, regulated forms of necrosis result in membrane rupture and release of endogenous danger signals that can act like foreign PAMPS to amplify the inflammatory response.6 It is vitally important that the pathways underlying these inflammatory responses and different types of cell death are tightly controlled and balanced as an exaggerated inflammatory response forms the basis to many inflammatory diseases, and excessive cell death can lead to depletion of protective immune cells and tissue damage. Given the close interplay of inflammation and cell death, it is not surprising that the signalling pathways controlling both processes are highly interconnected and co-ordinated. These pathways can dictate the magnitude and duration of the inflammatory response while also controlling cell fate and deciding on whether cells survive or die. In the latter case, the form of cell death is a critical decision. Significant progress has been made in delineating the components of signalling pathways that underpin the interplay of inflammation and cell death. This review will focus on the emerging importance of the kinase family of receptor interacting proteins (RIPs) as especially critical players in these signalling networks.
The RIP Kinase Family
The RIP kinase family contains seven members with each containing a homologous kinase domain (KD) that is the signature of the family (Figure 1).7 In addition to its N-terminal KD, RIP1 contains a C-terminal death domain (DD) and a bridging intermediate domain (ID) that also harbours a RIP homotypic interaction motif (RHIM). RIP2 also contains the N-terminal KD, an ID (lacking a RHIM) and a C-terminal caspase activation and recruitment domain (CARD). Although RIP3 contains the N-terminal KD, it lacks the ID and instead has a unique C-terminal sequence that contains a RHIM. RIP4 and RIP5 have the KD and ID with both also sharing C-terminal ankyrin domains. RIP6 and RIP7 are less related in structure to the other members, and although both contain the homologous KD, they contain a number of additional and diverse domain structures, such as leucine-rich repeat regions. The functions of RIP 4–7 are poorly understood and are well reviewed elsewhere.7 Briefly, RIP4 was initially identified as a PKCδ-interacting protein8 and was subsequently shown to activate NFκB.9 It has a key role in keratinocyte differentiation10 and cutaneous inflammation.11 Overexpression of RIP5 drives cell apoptosis,12 but its physiological role remains to be delineated. Similarly, the functions of RIP6 and RIP7 (also known as leucine-rich repeat kinases 1 and 2) are unknown although both have been associated with the pathogenesis of Parkinson’s disease.13, 14 Although our understanding of the biology of RIP4–7 is still in its infancy, intensive research has clarified important molecular and physiological roles of RIP1–3 in inflammation and cell death, the core focus of the remainder of this review.
RIP1 and TNF Signalling: Inflammation Versus Apoptosis
Although RIP1 mediates the activation of NFκB in response to a number of death receptors, including TNF-receptor 1 (TNF-R1),15, 16 TNF-related apoptosis-inducing ligand receptor 1 (TRAIL1)17 and Fas,18 the greatest appreciation of the function of RIP1 has emerged from exploring its role in TNF-mediated inflammation and cell death. The stimulation of TNF-R1 with TNF leads to the interaction of TNF-R1-associated death domain protein (TRADD)19 and RIP120 with the TNF-R1 signalling complex (Figure 2). This is followed by the recruitment of a number of E3 ubiquitin ligases to RIP1, including TNF receptor-associated factor 2 (TRAF2) or TRAF5 and the cellular inhibitor of apoptosis proteins (cIAPs) cIAP1 and cIAP2 resulting in the formation of Complex I.21, 22, 23, 24 TRAF225, 26 and cIAPs27, 28, 29, 30 catalyse the polyubiquitination of RIP1. The ubiquitin-decorated RIP1 is recognized by ubiquitin-binding domain containing proteins in the IκB kinase (IKK)31 and TAK1 kinase complexes32, 33, 34 thus facilitating TAK-1-mediated phosphorylation and activation of IKKs. The latter subsequently phosphorylate the IκB proteins, which normally sequester NFκB in an inactive state in the cytoplasm, resulting in ubiquitination and proteasomal degradation of IκB and allowing for nuclear translocation of the liberated NFκB.35, 36, 37 NFκB then drives the transcription of many pro-inflammatory genes that will mediate the inflammatory response. NFκB can also induce anti-apoptotic genes such as cellular FLICE inhibitory protein (c-FLIP) and cIAPs that prevent cell death.38, 39, 40 RIP1 can also mediate TNF-induced activation of the mitogen-activated protein kinases (MAPKs) ERK, p38 and JNK and, interestingly, although the kinase activity of RIP1 is dispensable for activating NFκB, p38 and JNK, it is required to stimulate ERK activity.41
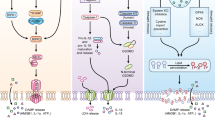
Regulatory roles of RIP1 and RIP3 in TNF signalling. Stimulation of cells with TNF leads to recruitment of TRADD and RIP1 to TNF-R1. RIP1 is ubiquitinated in a complex I containing TRAF2, TRAF5, cIAP1 and cIAP2 leading to TAK1/IKK-mediated activation of NFκB. The latter induces inflammation (by pro-inflammatory gene expression (e.g., IL-1β, TNF)) and anti-apoptotic proteins such as cFLIP. De-ubiquitination of RIP1 results in the formation of Complex II (or ‘ripoptosome’ in the presence of IAP antagonists) with FADD and procaspase 8. Auto-processing of caspase 8 triggers a downstream caspase cascade and cell death by apoptosis. Pellino3 targets RIP1 to block formation of Complex II and apoptosis. Under the conditions of caspase 8 inhibition, RIP interacts with RIP3 (via their RHIM motifs, indicated in yellow) followed by RIP1/RIP3 phosphorylation (P) and formation of an amyloid filamentous structure known as the necrosome. RIP3 then interacts with MLKL, PYGEL, GLUL and GLUD1 resulting in mitochondrial ROS production. PGAM5 can also be stimulated to interact with the mitochondrial fission factor Drp1 leading to mitochondrial fragmentation and necroptosis, but this may be cell and species dependent. MLKL can also form oligomers that bind to membrane phospholipids resulting in membrane rupture. A FADD/cFLIP/caspase 8 complex can cleave RIP1 and RIP3 to prevent RIP1/RIP3-mediated necroptosis
Although ubiquitinated RIP1 serves to promote downstream activation of NFκB and gene expression that drives inflammation and protects cells from death by apoptosis,42 NFκB can also induce the deubiquitinating enzymes CYLD and A20 that can remove the ubiquitin chains from RIP1 and terminate its ability to activate NFκB.43, 44, 45, 46 In this unmodified form, RIP can leave the TNF-R1 complex to associate with Fas-associated death domain (FADD) and procaspase 8 and form death-inducing signalling complex also known as Complex II.27, 43, 47, 48, 49 The use of IAP antagonists or loss of cIAP proteins generates a similar ripoptosome complex consisting of RIP1, FADD and caspase 8, with components of this complex being subject to ubiquitination and inactivation by cIAPs.49, 50, 51 Complex II and the ripoptosome can promote processing of procaspase 8 to its active form resulting in triggering of the caspase cascade that culminates in cell death by apoptosis.45 Recently, we have demonstrated that a member of the Pellino E3 ubiquitin ligase family, Pellino3, targets RIP1 and impairs Complex II formation in the TNF signalling pathway to suppress cell apoptosis.52 The kinase activity of RIP1 is required for ripoptosome assembly and its downstream triggering of apoptosis.49, 51 Thus the ubiquitination and kinase activity status of RIP1 dictates whether TNF signalling goes down the road of inflammatory gene expression or the terminal path to cell death. Intriguingly, the two pathways counter-regulate each other with NFκB driving anti-apoptotic gene expression, whereas caspase 8 can cleave RIP1 to suppress its ability to activate NFκB,53, 54 with one of the processed forms of RIP1 enhancing the interaction of TRADD and FADD to sensitize cells to the pro-apoptotic effects of TNF.54
RIP1 and RIP3 as Drivers of Necroptosis
Although the induction of apoptosis in virally infected cells represents an important defense system to curtail viral replication and dissemination, cytomegaloviruses are examples of microbes that have evolved evasive strategies to this defense system by encoding inhibitors of caspase 8-mediated apoptosis.55 However, the host has developed further countermeasures to this escape mechanism, including a form of cell death known as necroptosis that is triggered by death receptors under conditions of caspase inhibition. This form of cell death was initially observed when caspase 8 inhibition increased the sensitivity of cells to TNF-induced necrosis.56, 57 Furthermore, targeted deletion of the murine caspase 8 gene resulted in prenatal lethality due to impaired heart muscle development.58 Other death receptor ligands, such as TRAIL and Fas ligand, were also shown to induce caspase-independent cell death.59 This necrotic form of cell death that is induced by death receptors is mediated by RIP1 and is dependent on its kinase activity.59 RIP3 was subsequently shown to be also required for RIP1-induced necrosis,60, 61, 62 with the kinase activity of RIP3 being essential for mediating cell necrosis.61 Interestingly, RIP3 and its catalytic activity facilitate a switch between TNF-induced apoptosis and necrosis,60 with embryonic fibroblasts from RIP3-deficient mice being resistant to TNF-induced necrosis61 and RIP3 kinase dead knock-in mice displaying developmental lethality due to RIP1- and caspase 8-driven apoptosis.63 RIP3 deficiency also rescues the prenatal lethality of caspase 8 knockout mice with double knockouts lacking both caspase 8 and RIP3 surviving and reaching maturity,64, 65 indicating that RIP3 mediates lethality in the absence of caspase 8. This is consistent with the ability of caspase 8 to cleave RIP3 resulting in loss of the kinase domain of RIP3 and abrogation of its ability to trigger caspase-independent cell death.66 Caspase 8 has also been shown to repress necrosis by processing CYLD.67 Interestingly, caspase 8 appears to act in a proteolytically active complex with FADD and cFLIP to block RIP1- and RIP3-mediated necrosis,65, 68 with c-FLIP-69 and FADD-70, 71 deficient cells being highly sensitive to death by necrosis. This is consistent with the developmental lethality, due to cardiac failure, in FADD-deficient embryos,72 with RIP1 deficiency rescuing the embryonic lethality associated with FADD deficiency.71 These studies support a model in which the FADD–caspase 8–c-FLIP complex negatively regulates RIP-kinase-mediated necrosis. This raises the apparent paradox of c-FLIP interacting with caspase 8 to facilitate caspase-mediated processing of RIP kinases while c-FLIP also serves to inhibit caspase 8 in the apoptotic pathway. However, this may relate to auto-processing of caspase 8 being required to trigger apoptosis but not to repress necrosis.73, 74
Many studies have probed the complex functional interplay between RIP1 and RIP3 in regulating cell necrosis. Under resting conditions, RIP1 is proposed to bind to RIP3 to prevent oligomerization of the latter and so prevent spontaneous RIP3 activation and necrosis.75 This may, at least partly, underlie the perinatal lethality associated with RIP1 deficiency but would require that any such protective effects of RIP1 are independent of kinase activity as RIP1 kinase dead knockin mice survive to adulthood.63, 76, 77 In addition, during development the physiological role of RIP1 in regulating RIP3-driven necroptosis appears to be highly dependent on the stage of development with RIP1 being required for TNF-induced necroptosis at E10.578 but inhibiting necroptosis and associated inflammation at later stages of development.78, 79 Although RIP3-driven necroptosis contributes to the perinatal defects associated with RIP1 deficiency, it is not the sole underlying mechanism.63 This is supported by recent studies demonstrating an important role for RIP1 in protecting against TNF- and caspase 8-driven apoptosis.76, 79
Under conditions of TNF stimulation, or during virus infection, that trigger RIP1-dependent necrosis, RIP3 promotes necrosis-specific phosphorylation of RIP1, thus forming a pro-necrotic necrosome complex.62 Phosphorylation-induced activation of the necrosome is dependent on prior de-ubiquitination of RIP1 by CYLD, a step that is proposed to take place in the necrosome itself and not in Complex I.80 De-ubiquitination of RIP1 is a prerequisite for TNF-induced necrosis as NEMO, a regulatory subunit in the IKK complex, can bind to ubiquitinated RIP1 and prevent its engagement with the necrosome.81 Structural studies have shown the RHIMs of RIP1 and RIP3 to mediate their interaction82 and facilitate assembly of heterodimeric filamentous structures, typical of beta-amyloids, and it is these amyloid structures that form the active necrosome complex.83 The authors of the latter study propose that the RHIM sequences may be hidden in resting cells but that these cryptic motifs are revealed in response to RIP1-induced phosphorylation of RIP3 thus relaxing the auto-inhibited state and allowing for the formation of the RHIM-mediated amyloid filaments. In this process, the initial formation of a RIP1-RIP3 heterodimer is insufficient to trigger necroptosis and instead the RIP1-RIP3 amyloid structure must recruit more free RIP3 to the amyloid scaffold resulting in auto-phosphorylation of RIP3 and recruitment of mixed lineage kinase domain-like protein (MLKL) to trigger downstream necroptosis.84 The recruitment of MLKL to the necrosome leads to RIP3-mediated phosphorylation of MLKL with the RIP3 inhibitor, necrosulfonamide, blocking necrosis downstream of RIP3 activation85 and MLKL-deficient mice being resistant to necroptosis.86, 87 Various downstream effector mechanisms have been proposed to mediate necroptosis. Phosphorylation of MLKL promotes its oligomerization and translocation to the plasma membrane where it interacts with phospholipids and compromises membrane integrity ultimately resulting in cell rupture.88, 89, 90, 91 MLKL also promotes the generation of reactive oxygen species (ROS) and late phase activation of JNK.92 The increased production of ROS, especially by the mitochondria, has been strongly linked with mediating TNF-induced necrosis.93, 94, 95 Indeed cIAP1 and TAK1 has been shown to block TNF-induced necrosis by inhibiting RIP1/RIP3-mediated production of ROS.96 RIP3 also interacts with and activates a number of metabolic enzymes, including glycogen phosphorylase (PYGEL), glutamate–ammonia ligase (GLUL) and glutamate dehydrogenase 1 (GLUD1) that partly contribute to TNF-induced production of ROS and necroptosis60, 95 (Figure 2). In addition, the RIP1-RIP3 necrosome can interact with the mitochondrial protein phosphatase PGAM5 to drive downstream necrosis.97 This study demonstrated that PGAM5 recruits the mitochondrial fission factor Drp1 to promote its GTPase activity by dephosphorylating serine residue 637 of Drp1. The activation of Drp1 triggers mitochondrial fragmentation, an essential driver of necrosis execution. However, a more recent study has questioned the role of PGAM5 in mediating TNF-induced necrosis, at least in murine fibroblasts.87 Knockdown of PGAM5 expression in these cells failed to affect susceptibility to TNF-driven necroptosis, suggesting alternative or additional mediatory pathways. Such discrepancies may reflect varying effector mechanisms in different cells and species.98
RIP Kinases and TLRs
Although RIP1 and RIP3 have key roles in controlling the outcome of death receptor signalling pathways, they also have important roles in PRR pathways especially TLR3 and TLR4 (Figure 3). This is due to these pathways employing an adaptor protein termed TRIF that contains a RHIM that allows it to interact with and deploy RIP1 and RIP3. All TLRs, except TLR3, uses the MyD88 adaptor protein to promote downstream activation of NFκB.99 TLR4 can also use TRIF to activate NFκB by a MyD88-independent pathway.100, 101 TLR3 is unique in the TLR family in that that it does not use Myd88 but instead exclusively employs TRIF to activate NFκB.102 In the case of both TLR3 and TLR4 signalling, the RHIM motif of TRIF recruits RIP1 to mediate downstream activation of NFκB.102, 103 RIP3 is not required for activation of NFκB in TLR signalling pathways.104 Similar to TNF-R1 signalling, RIP1 needs to be ubiquitinated in order to drive TRIF-induced activation of NFκB, and Pellino1 is a key E3 ligase that ubiquitinates RIP1 in the TLR3 and TLR4 pathways.105
RIP1 and RIP3 in pattern recognition receptor signalling. LPS stimulates TLR4 to allow its Toll/IL-1 receptor (TIR) domain to interact with the TIR adaptor MyD88. This leads to ubiquitination of TRAF6, TAK1/IKK-mediated activation of NFκB and induction of pro-inflammatory genes such as IL-1β and TNF. The TIR domains of TLR3 and TLR4 can recruit another TIR adaptor TRIF, that contains a RHIM motif (in yellow), allowing TRIF to interact with RIP1. This is followed by Pellino1-mediated polyubiquitination of RIP1 allowing for TAK/IKK-induced activation of NFκB. TRIF (in a RIP1-independent manner) can also activate the TBK1/IKKi kinases to phosphorylate IRF3 and induce type I interferons (IFNs). Under conditions of caspase inhibition, the RHIM domain of TRIF can interact with the RHIM of RIP3 to trigger MLKL-mediated necroptosis. RIP1 can also facilitate the direct recruitment of caspase 8 to TLR3 leading to apoptosis. Murine cytomegalovirus (MCMV) can stimulate the DNA sensor DAI (containing two RHIMs) to interact with RIP1 and RIP3 to promote TAK/IKK-mediated activation of NFκB. DAI can also interact with RIP3 to promote MLKL-mediated necroptosis. The MCMV-encoded protein M45 contains a RHIM that allows it to target RIP3 and inhibit DAI- and RIP3-mediated necroptosis. The RNA helicase RIG-I is recruited to the mitochondria by MAVS followed by association with RIP1 and downstream activation of NFκB by TAK/IKK. RIG-1 also triggers TBK1/IKKi-mediated activation of IRF3 and induction of type I IFNs. RIP1 recruits caspase 8 to the RIG-1 complex resulting in RIP1 cleavage and termination of RIG-I signalling
In addition to activation of NFκB, TRIF can also stimulate the TBK1 and IKKi/IKKɛ kinases to activate interferon regulatory factor (IRF) transcription factors that drive expression of anti-viral type I interferons (IFNs)99, 106 (Figure 3). Interestingly, RIP1 is not used by TRIF in its activation of IRFs.102, 103 However, under circumstances of FADD depletion or its phosphorylation on serine residue 191 (during cell cycle arrest) or when caspases are inactivated (as occurs in virus-infected cells), IFNs can feed back on virally infected cells to activate the RNA-responsive protein kinase PKR, which then interacts with RIP1 and triggers RIP1/RIP3-mediated necroptosis.107 When caspases are inhibited, TLR3 and TLR4 can also directly induce necroptosis by virtue of the RHIM motif of TRIF engaging RIP3 and MLKL to trigger downstream necrosis108, 109 (Figure 3). Interestingly, whereas RIP1 is required to mediate TNF-induced RIP3-dependent necroptosis,78 a recent report has indicated that RIP1 blocks TLR3-, TRIF- and IFN-driven necroptosis before birth.79 TLRs that do not employ TRIF can also induce necroptosis in an indirect manner by inducing TNF to trigger necrosis via TNFR-1 as described above.109 In addition, some of these pathways have been associated with cell apoptosis. Overexpression of TRIF results in interaction with RIP1 and RIP3 and induction of apoptosis,110 and in the context of TLR3 signalling in lung cancer cells, the TLR3 ligand dsRNA can induce apoptosis by recruiting caspase 8 to TLR3 in a RIP1-dependent manner.111 The ability of RIP kinases to orchestrate both apoptosis and necroptosis in response to triggering of viral-sensing TLR3 provides a major survival advantage to the host. Although TLR3-induced apoptosis can serve as the initial effort to eliminate virus-infected cells, some viruses encode caspase inhibitors to neutralize this defense system. However, in the absence of caspase activity, necroptosis will be strongly triggered as a contingency measure to deny the virus its home of replication.
RIP Kinases and Nucleic Acid Sensing
DNA-dependent activator of IRFs (DAI, also known as ZBP1 or DLM-1) is a cytosolic DNA sensor that can respond to immunostimulatory DNA to activate NFκB and IRFs and induce pro-inflammatory cytokines and IFNs.112 DAI contains two RHIM motifs that allows it to interact with RIP1 and RIP3 and trigger downstream activation of NFκB113, 114 (Figure 3). The interaction of DAI with RIP3 also sensitizes cells to murine cytomegalovirus (MCMV)-induced necrosis with DAI- and RIP3-deficient cells being resistant to this form of death.115 Intriguingly, MCMV encodes a M45 protein, that also contains a RHIM and targets the DAI–RIP3 interaction to suppress premature killing of endothelial cells during MCMV infection.116, 117 Such findings highlight the importance of RIP-mediated necroptosis to anti-viral immunity.6, 118
The RNA helicase RIG-I also serves as a cytoplasmic viral sensor by recognizing viral RNA.119 Engagement of RIG-I by RNA results in its recruitment by the MAVS adaptor protein to the outer membrane of the mitochondria.3 The assembly of this complex triggers downstream activation of NFκB and IRF3 to induce pro-inflammatory cytokines and IFNs (Figure 3). A recent study has shown RIP1 to be recruited to the RIG-1 mitochondrial complex with ubiquitination of RIP1 serving to provide docking sites for key signalling molecules such as the IKK complex that activates NFκB.120 However, RIP1 can also facilitate recruitment of caspase 8 to the complex, resulting in the cleavage of RIP1 and the generation of an inhibitory RIP1 fragment that represses RIG-I-induced activation of IRF3. Thus RIP1 is a key regulator of the temporal expression of virus-responsive genes.
RIP Kinases and the Inflammasome
IL-1β is one of the key pro-inflammatory cytokines that drives inflammation.121 The secretion of mature IL-1β requires two signals. First, innate receptors, like TLR4, promote increased transcription of the gene encoding IL-1β, resulting in expression of an inactive pro-IL-1β precursor. A second signal requires the generation of a signalling platform termed the inflammasome consisting of a NLR protein such as NLRP3 that recruits the adaptor protein ASC and caspase 1 into a complex. Caspase 1 in this inflammasome complex will process pro-IL-1β precursor into the mature secreted form of IL-1β and will also effect an inflammatory form of cell death termed pyroptosis. A recent report has suggested that the inflammasome can be regulated by RIP1 and RIP3. Caspase 8 deficiency in dendritic cells enhanced TLR-4 induced formation and activation of the NLRP3 inflammasome by a mechanism that was dependent on RIP1, RIP3, MLKL and PGAM5.122 This resulted in augmented LPS-induced expression of mature IL-1β and exacerbation of LPS-induced septic shock in mice with dendritic cell-specific deletion of the caspase 8 gene. Interestingly, these effects were proposed to be independent of necroptosis. Such findings suggest that caspase 8 has dualist roles in targeting RIP kinases to control inflammation. Caspase 8 acts to suppress RIP1/RIP3-driven necroptosis and the ensuing inflammatory fall-out from cell necrosis while also controlling RIP1/RIP3-mediated activation of the NLRP3 inflammasome and production of IL-1β. However, the role of caspase 8 is complex and context dependent as the causative agent of plague Yersinia pestis and its outer protein YopJ employs caspase 8, RIP1 and RIP3 to trigger cell death and caspase 1 activation.123, 124
Interestingly, another study demonstrated that pharmacological or genetic depletion of the cIAP proteins in macrophages, in conjunction with TLR stimulation, resulted in augmented processing of pro-IL-1β into its mature form.125 The processing of IL-1β was driven by two independent pathways involving NLRP3/caspase 1 and caspase 8. Both pathways were dependent on RIP3 and ROS. Thus under conditions of ripoptosome formation, as occurs with cIAP depletion, RIP3 can strongly drive IL-1β production further extending the pro-inflammatory potential of RIP3 beyond its ability to drive inflammatory cell death by necroptosis. However, the physiological circumstances under which cIAP proteins are depleted or inhibited in the presence of TLR stimuli remain to be characterized. These findings suggest that IAP proteins serve important regulatory roles in tempering the pro-inflammatory potential of RIP3. This is further supported by recent reports demonstrating that XIAP limits RIP3-dependent cell death and IL-1β expression in response to TNF126 while cIAPs and XIAP control RIP1 and RIP3-dependent pro-inflammatory cytokine production in myeloid cells.127
RIP1 and RIP3 in a Pathophysiological Context
Given that necroptosis results in plasma membrane rupture and the release of endogenous danger signals that can activate PRRs, this form of cell death is regarded as being strongly pro-inflammatory in nature. Consequently, many studies have explored the potential contribution of RIP1/RIP3-mediated necroptosis to inflammatory diseases. To this end, necrostatins, inhibitors of the kinase activity of RIP1 and of necroptosis, have been evaluated in various disease models.128 Necrostatins ameliorate pathology in a number of inflammatory disease models, including brain ischaemia,129 mycocardial infarction130 and head trauma.131 Inhibition of RIP1 or RIP3 deficiency reduces mortality during TNF-induced systemic inflammatory response syndrome and pathology in the caecal ligation and puncture model of polymicrobial sepsis highlighting the potential value of targeting RIP1/RIP3 in sepsis.132 RIP3-deficient mice are also free of inflammation in an acute pancreatitis model61 and show reduced macrophage necroptosis to ameliorate atherosclerosis development.133 Keratinocyte-specific deletion of FADD results in serious inflammatory skin lesions via RIP3-mediated necroptosis.134 Furthermore, conditional deletion of caspase 8 in the intestinal epithelium resulted in great levels of RIP3 and TNF-driven necroptosis and increased susceptibility to colitis.135 Interestingly, the latter study also demonstrated high levels of RIP3 and necroptosis in the terminal ileum of patients with Crohn’s disease, suggesting that RIP3-induced necroptosis may be a valuable therapeutic target in human disease. Absence or inhibition of RIP3 also reduces liver damage in response to ethanol136 or acetaminophen.137 Finally, RIP1 and RIP3 drive necrotic cell death in retinal pigment epithelial cells and photoreceptor cells, and these effects likely have key roles in vision problems, such as macular degeneration, retinitis pigmentosa and retinal detachment.138, 139, 140, 141, 142 All of these studies emphasize the potential roles of RIP1 and RIP3 in driving diverse inflammatory diseases, and future research is faced with the challenge of exploiting these kinases as therapeutic targets.
RIP2 and NOD Signalling
RIP2 was initially identified as a RIP-like kinase that, when overexpressed, could activate NFκB and MAP kinases and augment caspase 8-mediated apoptosis.143, 144, 145 RIP2 contains an N-terminal kinase domain and C-terminal CARD domain (Figure 1). The kinase activity of RIP2 is dispensable to manifest its activation of NFκB but is required to mediate the activation of ERK MAPK145, 146 and to stabilize RIP2 itself.147, 148 Early studies demonstrated that RIP2-deficient mice are viable but show impaired activation of NFκB in response to TLR signalling and are more resistant to LPS-induced lethal sepsis.149, 150 However, a more recent report, using synthetic and highly purified forms of TLR ligands, contend that TLR signalling is intact in cells from RIP2 null mice, but loss of RIP2 leads to abrogation of signalling in response to stimulation of nucleotide-binding oligomerization domain-containing protein 1 (NOD1) and NOD2 by their specific ligands or the intracellular pathogen Listeria monocytogenes.151 These findings indicate that RIP2 mediates NOD1 and NOD2 signalling but not TLR signal transduction.
NOD1 and NOD2 are cytosolic receptors for bacterial peptidoglycan derivatives such as muramyl dipeptide (MDP) and are expressed highly in mucosal epithelium.152, 153, 154 Loss-of-function mutations in NOD2 are associated with greatly increased susceptibility to Crohn’s disease,155, 156, 157 whereas gain-of-function mutations are linked to early onset sarcoidosis and Blau syndrome.158, 159 NOD2 contains an N-terminal CARD domain, a central NACHT region and C-terminal LRRs.160 Upon binding of MDP to the LRRs of NOD2, the NACHT regions are exposed, allowing for self-oligomerization of NOD2 molecules, followed by homotypic interactions between the CARD domains of NOD2 and RIP2161, 162 (Figure 4). This results in ubiquitination of RIP2 followed by recruitment of the TAK1 and IKK complexes and downstream activation of NFκB and MAP kinase pathways by an analogous mechanism to that described above for RIP1 signalling in the TNFR-1 pathway.148, 163, 164, 165 This results in the expression of a range of inflammatory proteins, anti-bacterial proteins, activation of autophagy and antigen presentation.166, 167, 168 Notably, RIP2 is required to mediate all of the in vivo host responses to MDP.169
RIP2 and NOD signalling. Bacterial invasion of epithelial cells results in the stimulation of NOD proteins by peptidoglycan-derived peptides such as MDP. MDP binds to the leucine-rich repeat regions of NOD2 allowing for NACHT domains to mediate NOD oligomerization. The CARD domains of oligomerized NOD proteins interact with the CARD domains of RIP2 kinase molecules followed by binding of TRIP protein to RIP2 and XIAP and Pellino3-mediated polyubiquitination of RIP2. This facilitates recruitment of TAK1 and IKK complexes and downstream activation of NFκB, MAPKs and AP-1. The transcription factors drive expression of cytokines, chemokines and anti-bacterial peptides. NOD signalling can also result in cell autophagy. RIP2 is targeted by various negative regulatory proteins: ITCH catalyzes ubiquitination of RIP2 to inhibit NFκB activation; SHIP-1 disrupts the interaction between XIAP and RIP2; Free ubiquitin competes with RIP2 for the binding of NOD1; the autophagy protein ATG16L1 interferes with the polyubiquitination of RIP2 and the recruitment of RIP2 into NOD-signalling complexes; MEKK4 inhibits the basal interaction of RIP2 with NOD2; and Caspase 12 targets RIP2 and inhibits downstream signalling
The ubiquitination of RIP2 is a critical step in mediating activation of these NOD2 pathways, especially activation of NFκB,164, 165 and a number of E3 ubiquitin ligases, including TRAF6,164 cIAP and XIAP proteins170, 171, 172 and ITCH173 have been proposed to catalyse ubiquitination of RIP2. However, other studies have questioned the importance of many of these E3 ligases in the context of ubiquitinating RIP2 and activating NFκB. Thus the ubiquitination of RIP2 is intact in TRAF6-deficient cells,165 pharmacological depletion of cIAP1 and cIAP2 has no effect on RIP2 ubiquitination171 and ITCH-mediated ubiquitination of RIP2 is associated with negative regulation of RIP2-mediated NFκB signalling.173 We have recently described a key role for the E3 ubiquitin ligase Pellino3 in directly ubiquitinating RIP2 and mediating NOD2 downstream signalling, including its activation of NFκB and protective effects in colitis174, 175 (Figure 4). We also showed that Pellino3 protein expression is greatly reduced in the colons of Crohn’s disease subjects consistent with a protective role in human disease.174 We have proposed functional cooperation between Pellino3 and XIAP in that the former promotes the formation of polyubiquitin chains on RIP2 in which the isopeptide linkages between adjacent ubiquitin molecules are linked via lysine 63 of ubiquitin and XIAP facilitates linear ubiquitination of components of the RIP2 complex in which individual ubiquitin proteins are joined head to tail. This shows remarkable similarity to the RIP1-containing complex I in the TNFR-1 signalling pathway in which components of the complex are initially modified by lysine 63-linked chains followed by LUBAC-mediated linear ubiquitination that serves to stabilize the complex and further enhance downstream signalling pathways, such as NFκB and inflammatory gene expression.176
The ubiquitination pathway in NOD–RIP2 signalling is subject to various forms of regulation. Thus the inositol phosphatase SHIP-1 disrupts the interaction between XIAP and RIP2 to inhibit NOD2-induced NFκB activation.177 In addition, free ubiquitin can compete with RIP2 for the binding of NOD1.178 Furthermore, the autophagy protein ATG16L1, which has also been linked to Crohn’s disease, interferes with the polyubiquitination of RIP2 and the recruitment of RIP2 into NOD-signalling complexes, resulting in impaired downstream signalling.179 The allelic form of ATG16L1, which is associated with Crohn’s disease, fails to regulate NOD-mediated inflammatory signalling, suggesting that the targeting of RIP2 is important in controlling intestinal pathogenesis. The LIM domain-containing protein TRIP can interact with RIP2 to positively regulate NOD1 signalling.180 Finally, the MAP3K, MEKK4, interacts with RIP2 to preclude basal interaction of the latter with NOD2 while stimulation of cells with the NOD2 ligand MDP promotes dissociation of RIP2 from MEKK4 allowing for interaction of RIP2 with NOD2.181
The NOD–RIP2 pathway is also targeted by caspases, and this is especially interesting given that NOD proteins belong to the large NLR family, many members of which are components of caspase-containing inflammasomes. Caspase 12 has been shown to target RIP2 and inhibit downstream signalling in response to NOD2 stimulation.182 This results in an impaired mucosal antimicrobial response to enteric pathogens due to reduced production of antimicrobial peptides, cytokine and chemokines. Intriguingly, some patients with variants of the caspase 1 gene, which encode for forms of procaspase 1 with greatly reduced or absent enzymatic activity, frequently exhibit fever even though levels of IL-1β are low.183 The latter study demonstrated that the CARD domain of these procaspase 1 variants binds to the CARD of RIP2 to trigger activation of NFκB and presumably downstream inflammatory responses that underpin the regular febrile episodes.
RIP2 in Inflammation and Disease
Although the NOD–RIP2 signalling pathways are of particular relevance to the control of intestinal inflammation, RIP2 also has important roles in mucosal immunity in the respiratory system. Thus RIP2-deficient mice show impaired bacterial clearance in an E. Coli pneumonia infection model184 and Chlamydophila pneumoniae-induced pneumonia.185 In both models, the absence of RIP2 resulted in impaired expression of various pro-inflammatory mediators, reduced neutrophil infiltration and increased bacterial burden. However, under certain circumstances, the role of RIP2 in mediating an anti-bacterial response can be damaging to the host. This applies in the case of secondary bacterial infection following an initial viral infection.186 In this case, viral challenge leads to production of type I IFNs that strongly upregulate NOD1, NOD2 and RIP2 resulting in an exaggerated inflammatory response to secondary infection with E. Coli. Thus the NOD–RIP2 pathway likely has key roles in the increased lethality and morbidity that is clinically observed in secondary bacterial infections.
RIP2 has been associated with other inflammatory disease states and models. Levels of RIP2 are elevated in the non-T-cell fraction of blood from multiple sclerosis (MS) subjects,187 and pathogenesis in murine models of MS have been shown to be dependent on NOD1, NOD2 and RIP2 with the latter having an especially important role in activating CNS dendritic cells.188 Intriguingly, peptidoglycan has been detected within antigen-presenting cells, including dendritic cells, in the brain of MS patients189 suggesting that the peptidoglycan–NOD–RIP2 axis in CNS may contribute to MS pathogenesis. RIP2 has also been implicated as a driver in experimental allergic airway inflammation by activating NFκB and inflammatory gene expression.190 Furthermore, RIP2-deficient macrophages, although showing weaker inflammatory signalling, display increased lipid accumulation that contributes to more severe atherosclerosis in recipient mice,191 implicating a potential role for RIP2 in cardiovascular disease.
A complex picture thus emerges of the role of RIP2 in inflammation and immunity. Given its critical role in NOD pathways, RIP2 clearly has a protective role in mucosal immunity and homeostasis as deficiency in NOD signalling is linked to Crohn’s disease and loss of RIP2 leads to increased bacterial burden in pulmonary infection models. However, high levels of RIP2 can lead to damaging inflammatory responses as indicated by pathogenesis in models of MS and secondary bacterial infections. Such opposing roles of RIP2 highlight the need for efficient regulatory mechanisms to avoid the potential damaging consequences of RIP2 action.
Conclusion and Perspective
Although much research has focused on the role of RIP proteins in cell death, it is clear that these kinases have many physiological and pathophysiological functions that are derived from their important roles in inflammation and innate immunity. Furthermore, many of the regulatory roles of RIP kinases in inflammation are mediated by their effects on cell death. Thus, while RIP1 is a key determinant in deciding whether a cell produces pro-inflammatory mediators or dies by apoptosis, RIP3 can direct a cell to die by the more inflammatory process of necroptosis. Although the latter provides a safeguard death mechanism against intracellular pathogens that encode for factors that interfere with apoptosis, necroptosis is also emerging as a key player in a number of inflammatory diseases. Thus, like RIP2, RIP1and RIP3 must be tightly controlled, with loss of this control leading to hyper-inflammation and pathology. RIP kinase thus emerge as lead therapeutic targets in a number of diseases. The first inhibitors of RIP kinases have emerged over the past number of years.128 The challenge and opportunity sit side by side to translate our increased understanding of RIP kinase biology into RIP-targeted therapeutics to treat inflammatory diseases.
Abbreviations
- PAMP:
-
pathogen-associated molecular pattern
- PRR:
-
pattern-recognition receptor
- TLR:
-
Toll-like receptor
- NOD:
-
nucleotide-binding oligomerization domain-containing protein
- NLR:
-
NOD-like receptor
- TNF:
-
tumour necrosis factor
- IL-1β:
-
interleukin-1β
- RIP:
-
receptor interacting protein
- DD:
-
death domain
- ID:
-
intermediate domain
- RHIM:
-
RIP homotypic interaction motif
- CARD:
-
C-terminal caspase activation and recruitment domain
- TNF-R1:
-
TNF-receptor 1
- TRAIL:
-
TNF-related apoptosis-inducing ligand receptor
- TRADD:
-
TNF-R1-associated death domain protein
- cIAP:
-
cellular inhibitor of apoptosis protein
- MLKL:
-
mixed lineage kinase domain-like protein
- MDP:
-
muramyl dipeptide
References
O'Neill LA, Golenbock D, Bowie AG . The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol 2013; 13: 453–460.
Chen G, Shaw MH, Kim YG, Nunez G . NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol 2009; 4: 365–398.
Loo YM, Gale M Jr . Immune signaling by RIG-I-like receptors. Immunity 2011; 34: 680–692.
Bhat N, Fitzgerald KA . Recognition of cytosolic DNA by cGAS and other STING-dependent sensors. Eur J Immunol 2014; 44: 634–640.
Lamkanfi M, Dixit VM . Manipulation of host cell death pathways during microbial infections. Cell Host Microbe 2010; 8: 44–54.
Mocarski ES, Kaiser WJ, Livingston-Rosanoff D, Upton JW, Daley-Bauer LP . True grit: programmed necrosis in antiviral host defense, inflammation, and immunogenicity. J Immunol 2014; 192: 2019–2026.
Zhang D, Lin J, Han J . Receptor-interacting protein (RIP) kinase family. Cell Mol Immunol 2010; 7: 243–249.
Bhr C, Rohwer A, Stempka L, Rincke G, Marks F, Gschwendt M . DIK, a novel protein kinase that interacts with protein kinase Cdelta. Cloning, characterization, and gene analysis. J Biol Chem 2000; 275: 36350–36357.
Meylan E, Martinon F, Thome M, Gschwendt M, Tschopp J . RIP4 (DIK/PKK), a novel member of the RIP kinase family, activates NF-kappa B and is processed during apoptosis. EMBO Rep 2002; 3: 1201–1208.
Holland P, Willis C, Kanaly S, Glaccum M, Warren A, Charrier K et al. RIP4 is an ankyrin repeat-containing kinase essential for keratinocyte differentiation. Curr Biol 2002; 12: 1424–1428.
Rountree RB, Willis CR, Dinh H, Blumberg H, Bailey K, Dean C Jr et al. RIP4 regulates epidermal differentiation and cutaneous inflammation. J Invest Dermatol 2010; 130: 102–112.
Zha J, Zhou Q, Xu LG, Chen D, Li L, Zhai Z et al. RIP5 is a RIP-homologous inducer of cell death. Biochem Biophys Res Commun 2004; 319: 298–303.
Seol W . Biochemical and molecular features of LRRK2 and its pathophysiological roles in Parkinson's disease. BMB Rep 2010; 43: 233–244.
Gandhi PN, Chen SG, Wilson-Delfosse AL . Leucine-rich repeat kinase 2 (LRRK2): a key player in the pathogenesis of Parkinson's disease. J Neurosci Res 2009; 87: 1283–1295.
Ting AT, Pimentel-Muinos FX, Seed B . RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J 1996; 15: 6189–6196.
Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P . The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity 1998; 8: 297–303.
Lin Y, Devin A, Cook A, Keane MM, Kelliher M, Lipkowitz S et al. The death domain kinase RIP is essential for TRAIL (Apo2L)-induced activation of IkappaB kinase and c-Jun N-terminal kinase. Mol Cell Biol 2000; 20: 6638–6645.
Kreuz S, Siegmund D, Rumpf JJ, Samel D, Leverkus M, Janssen O et al. NFkappaB activation by Fas is mediated through FADD, caspase-8, and RIP and is inhibited by FLIP. J Cell Biol 2004; 166: 369–380.
Hsu H, Xiong J, Goeddel DV . The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell 1995; 81: 495–504.
Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV . TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 1996; 4: 387–396.
Chen G, Goeddel DV . TNF-R1 signaling: a beautiful pathway. Science 2002; 296: 1634–1635.
Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, Liu Z . The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity 2000; 12: 419–429.
Karin M . Nuclear factor-kappaB in cancer development and progression. Nature 2006; 441: 431–436.
Muppidi JR, Tschopp J, Siegel RM . Life and death decisions: secondary complexes and lipid rafts in TNF receptor family signal transduction. Immunity 2004; 21: 461–465.
Lee TH, Shank J, Cusson N, Kelliher MA . The kinase activity of Rip1 is not required for tumor necrosis factor-alpha-induced IkappaB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J Biol Chem 2004; 279: 33185–33191.
Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 2010; 465: 1084–1088.
Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell 2008; 30: 689–700.
Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci USA 2008; 105: 11778–11783.
Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K et al. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem 2008; 283: 24295–24299.
Park SM, Yoon JB, Lee TH . Receptor interacting protein is ubiquitinated by cellular inhibitor of apoptosis proteins (c-IAP1 and c-IAP2) in vitro. FEBS Lett 2004; 566: 151–156.
Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ . Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 2006; 22: 245–257.
Chen ZJ . Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol 2005; 7: 758–765.
Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K . Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J 2003; 22: 6277–6288.
Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ . TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 2001; 412: 346–351.
DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M . A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature 1997; 388: 548–554.
Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J et al. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 1997; 278: 860–866.
Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M . Identification and characterization of an IkappaB kinase. Cell 1997; 90: 373–383.
Chu ZL, McKinsey TA, Liu L, Gentry JJ, Malim MH, Ballard DW . Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-kappaB control. Proc Natl Acad Sci USA 1997; 94: 10057–10062.
Kreuz S, Siegmund D, Scheurich P, Wajant H . NF-kappaB inducers upregulate cFLIP, a cycloheximide-sensitive inhibitor of death receptor signaling. Mol Cell Biol 2001; 21: 3964–3973.
Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS Jr . NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 1998; 281: 1680–1683.
Devin A, Lin Y, Liu ZG . The role of the death-domain kinase RIP in tumour-necrosis-factor-induced activation of mitogen-activated protein kinases. EMBO Rep 2003; 4: 623–627.
Li H, Kobayashi M, Blonska M, You Y, Lin X . Ubiquitination of RIP is required for tumor necrosis factor alpha-induced NF-kappaB activation. J Biol Chem 2006; 281: 13636–13643.
Declercq W, Vanden Berghe T, Vandenabeele P . RIP kinases at the crossroads of cell death and survival. Cell 2009; 138: 229–232.
Zhang SQ, Kovalenko A, Cantarella G, Wallach D . Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKgamma) upon receptor stimulation. Immunity 2000; 12: 301–311.
Wilson NS, Dixit V, Ashkenazi A . Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol 2009; 10: 348–355.
Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 2004; 430: 694–699.
Micheau O, Tschopp J . Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 2003; 114: 181–190.
O'Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT . Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr Biol 2007; 17: 418–424.
Wang L, Du F, Wang X . TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008; 133: 693–703.
Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 2011; 43: 449–463.
Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 2011; 43: 432–448.
Yang S, Wang B, Tang LS, Siednienko J, Callanan JJ, Moynagh PN . Pellino3 targets RIP1 and regulates the pro-apoptotic effects of TNF-alpha. Nat Commun 2013; 4: 2583.
Lin Y, Devin A, Rodriguez Y, Liu ZG . Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev 1999; 13: 2514–2526.
Martinon F, Holler N, Richard C, Tschopp J . Activation of a pro-apoptotic amplification loop through inhibition of NF-kappaB-dependent survival signals by caspase-mediated inactivation of RIP. FEBS Lett 2000; 468: 134–136.
Fliss PM, Brune W . Prevention of cellular suicide by cytomegaloviruses. Viruses 2012; 4: 1928–1949.
Vercammen D, Beyaert R, Denecker G, Goossens V, Van Loo G, Declercq W et al. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med 1998; 187: 1477–1485.
Luschen S, Ussat S, Scherer G, Kabelitz D, Adam-Klages S . Sensitization to death receptor cytotoxicity by inhibition of fas-associated death domain protein (FADD)/caspase signaling. Requirement of cell cycle progression. J Biol Chem 2000; 275: 24670–24678.
Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 1998; 9: 267–276.
Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol 2000; 1: 489–495.
Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009; 325: 332–336.
He S, Wang L, Miao L, Wang T, Du F, Zhao L et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009; 137: 1100–1111.
Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123.
Newton K, Dugger DL, Wickliffe KE, Kapoor N, de Almagro MC, Vucic D et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 2014; 343: 1357–1360.
Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 2011; 471: 368–372.
Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 2011; 471: 363–367.
Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N et al. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal 2007; 19: 2056–2067.
O'Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R et al. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol 2011; 13: 1437–1442.
Dillon CP, Oberst A, Weinlich R, Janke LJ, Kang TB, Ben-Moshe T et al. Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell Rep 2012; 1: 401–407.
He MX, He YW . A role for c-FLIP(L) in the regulation of apoptosis, autophagy, and necroptosis in T lymphocytes. Cell Death Differ 2013; 20: 188–197.
Osborn SL, Diehl G, Han SJ, Xue L, Kurd N, Hsieh K et al. Fas-associated death domain (FADD) is a negative regulator of T-cell receptor-mediated necroptosis. Proc Natl Acad Sci USA 2010; 107: 13034–13039.
Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J . Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 2011; 471: 373–376.
Yeh WC, de la Pompa JL, McCurrach ME, Shu HB, Elia AJ, Shahinian A et al. FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 1998; 279 (5358): 1954–1958.
Kang TB, Oh GS, Scandella E, Bolinger B, Ludewig B, Kovalenko A et al. Mutation of a self-processing site in caspase-8 compromises its apoptotic but not its nonapoptotic functions in bacterial artificial chromosome-transgenic mice. J Immunol 2008; 181: 2522–2532.
Khan N, Lawlor KE, Murphy JM, Vince JE . More to life than death: molecular determinants of necroptotic and non-necroptotic RIP3 kinase signaling. Curr Opin Immunol 2014; 26: 76–89.
Orozco S, Yatim N, Werner MR, Tran H, Gunja SY, Tait SWG et al. RIPK1 both positively and negatively regulates RIPK3 oligomerization and necroptosis. Cell Death Differ 2014 e-pub ahead of print 6 June 2014 doi:10.1038/cdd.2014.76.
Kaiser WJ, Daley-Bauer LP, Thapa RJ, Mandal P, Berger SB, Huang C et al. RIP1 suppresses innate immune necrotic as well as apoptotic cell death during mammalian parturition. Proc Natl Acad Sci USA 2014; 111: 7753–7758.
Berger SB, Kasparcova V, Hoffman S, Swift B, Dare L, Schaeffer M et al. Cutting edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J Immunol 2014; 192: 5476–5480.
Rickard JA, O'Donnell JA, Evans JM, Lalaoui N, Poh AR, Rogers T et al. RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 2014; 157: 1175–1188.
Dillon CP, Weinlich R, Rodriguez DA, Cripps JG, Quarato G, Gurung P et al. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell 2014; 157: 1189–1202.
Moquin DM, McQuade T, Chan FK . CYLD deubiquitinates RIP1 in the TNFalpha-induced necrosome to facilitate kinase activation and programmed necrosis. PLoS One 2013; 8: e76841.
O'Donnell MA, Hase H, Legarda D, Ting AT . NEMO inhibits programmed necrosis in an NFkappaB-independent manner by restraining RIP1. PLoS One 2012; 7: e41238.
Sun X, Yin J, Starovasnik MA, Fairbrother WJ, Dixit VM . Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J Biol Chem 2002; 277: 9505–9511.
Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 2012; 150: 339–350.
Wu X-N, Yang Z-H, Wang X-K, Zhang Y, Wan H, Song Y et al. Distinct roles of RIP1–RIP3 hetero- and RIP3–RIP3 homo-interaction in mediating necroptosis. Cell Death Differ 2014 e-pub ahead of print 6 June 2014 doi:10.1038/cdd.2014.77.
Sun L, Wang H, Wang Z, He S, Chen S, Liao D et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012; 148: 213–227.
Wu J, Huang Z, Ren J, Zhang Z, He P, Li Y et al. Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res 2013; 23: 994–1006.
Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 2013; 39: 443–453.
Chen X, Li W, Ren J, Huang D, He WT, Song Y et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res 2014; 24: 105–121.
Cai Z, Jitkaew S, Zhao J, Chiang HC, Choksi S, Liu J et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol 2014; 16: 55–65.
Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell 2014; 54: 133–146.
Dondelinger Y, Declercq W, Montessuit S, Roelandt R, Goncalves A, Bruggeman I et al. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep 2014; 7: 971–981.
Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci USA 2012; 109: 5322–5327.
Lin Y, Choksi S, Shen HM, Yang QF, Hur GM, Kim YS et al. Tumor necrosis factor-induced nonapoptotic cell death requires receptor-interacting protein-mediated cellular reactive oxygen species accumulation. J Biol Chem 2004; 279: 10822–10828.
Schulze-Osthoff K, Bakker AC, Vanhaesebroeck B, Beyaert R, Jacob WA, Fiers W . Cytotoxic activity of tumor necrosis factor is mediated by early damage of mitochondrial functions. Evidence for the involvement of mitochondrial radical generation. J Biol Chem 1992; 267: 5317–5323.
Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G . Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010; 11: 700–714.
Vanlangenakker N, Vanden Berghe T, Bogaert P, Laukens B, Zobel K, Deshayes K et al. cIAP1 and TAK1 protect cells from TNF-induced necrosis by preventing RIP1/RIP3-dependent reactive oxygen species production. Cell Death Differ 2011; 18: 656–665.
Wang Z, Jiang H, Chen S, Du F, Wang X . The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 2012; 148: 228–243.
Remijsen Q, Goossens V, Grootjans S, Van den Haute C, Vanlangenakker N, Dondelinger Y et al. Depletion of RIPK3 or MLKL blocks TNF-driven necroptosis and switches towards a delayed RIPK1 kinase-dependent apoptosis. Cell Death Dis 2014; 5: e1004.
Moynagh PN . TLR signalling and activation of IRFs: revisiting old friends from the NF-kappaB pathway. Trends Immunol 2005; 26: 469–476.
Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K et al. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J Immunol 2002; 169: 6668–6672.
Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K et al. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol 2003; 171: 4304–4310.
Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA . Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-{kappa}B activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem 2005; 280: 36560–36566.
Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M et al. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol 2004; 5: 503–507.
Newton K, Sun X, Dixit VM . Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol Cell Biol 2004; 24: 1464–1469.
Chang M, Jin W, Sun SC . Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol 2009; 10: 1089–1095.
Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 2003; 4: 491–496.
Thapa RJ, Nogusa S, Chen P, Maki JL, Lerro A, Andrake M et al. Interferon-induced RIP1/RIP3-mediated necrosis requires PKR and is licensed by FADD and caspases. Proc Natl Acad Sci USA 2013; 110: E3109–E3118.
He S, Liang Y, Shao F, Wang X . Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc Natl Acad Sci USA 2011; 108: 20054–20059.
Kaiser WJ, Sridharan H, Huang C, Mandal P, Upton JW, Gough PJ et al. Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J Biol Chem 2013; 288: 31268–31279.
Kaiser WJ, Offermann MK . Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J Immunol 2005; 174: 4942–4952.
Estornes Y, Toscano F, Virard F, Jacquemin G, Pierrot A, Vanbervliet B . dsRNA induces apoptosis through an atypical death complex associating TLR3 to caspase-8. Cell Death Differ 2012; 19: 1482–1494.
Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007; 448: 501–505.
Kaiser WJ, Upton JW, Mocarski ES . Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J Immunol 2008; 181: 6427–6434.
Rebsamen M, Heinz LX, Meylan E, Michallet MC, Schroder K, Hofmann K et al. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep 2009; 10: 916–922.
Upton JW, Kaiser WJ, Mocarski ES . DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 2012; 11: 290–297.
Upton JW, Kaiser WJ, Mocarski ES . Cytomegalovirus M45 cell death suppression requires receptor-interacting protein (RIP) homotypic interaction motif (RHIM)-dependent interaction with RIP1. J Biol Chem 2008; 283: 16966–16970.
Upton JW, Kaiser WJ, Mocarski ES . Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 2010; 7: 302–313.
Kaiser WJ, Upton JW, Mocarski ES . Viral modulation of programmed necrosis. Curr Opin Virol 2013; 3: 296–306.
Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006; 441: 101–105.
Rajput A, Kovalenko A, Bogdanov K, Yang SH, Kang TB, Kim JC et al. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by caspase-8-mediated cleavage of the RIP1 protein. Immunity 2011; 34: 340–351.
Lamkanfi M, Dixit VM . Mechanisms and functions of inflammasomes. Cell 2014; 157: 1013–1022.
Kang TB, Yang SH, Toth B, Kovalenko A, Wallach D . Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity 2013; 38: 27–40.
Philip NH, Dillon CP, Snyder AG, Fitzgerald P, Wynosky-Dolfi MA, Zwack EE et al. Caspase-8 mediates caspase-1 processing and innate immune defense in response to bacterial blockade of NF-kappaB and MAPK signaling. Proc Natl Acad Sci USA 2014; 111: 7385–7390.
Weng D, Marty-Roix R, Ganesan S, Proulx MK, Vladimer GI, Kaiser WJ et al. Caspase-8 and RIP kinases regulate bacteria-induced innate immune responses and cell death. Proc Natl Acad Sci USA 2014; 111: 7391–7396.
Vince JE, Wong WW, Gentle I, Lawlor KE, Allam R, O'Reilly L et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity 2012; 36: 215–227.
Yabal M, Muller N, Adler H, Knies N, Gross CJ, Damgaard RB et al. XIAP restricts TNF- and RIP3-dependent cell death and inflammasome activation. Cell Rep 2014; 7: 1796–1808.
Wong WW, Vince JE, Lalaoui N, Lawlor KE, Chau D, Bankovacki A et al. cIAPs and XIAP regulate myelopoiesis through cytokine production in an RIPK1- and RIPK3-dependent manner. Blood 2014; 123: 2562–2572.
Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313–321.
Chavez-Valdez R, Martin LJ, Flock DL, Northington FJ . Necrostatin-1 attenuates mitochondrial dysfunction in neurons and astrocytes following neonatal hypoxia-ischemia. Neuroscience 2012; 219: 192–203.
Lim SY, Davidson SM, Mocanu MM, Yellon DM, Smith CC . The cardioprotective effect of necrostatin requires the cyclophilin-D component of the mitochondrial permeability transition pore. Cardiovasc Drugs Ther 2007; 21: 467–469.
You Z, Savitz SI, Yang J, Degterev A, Yuan J, Cuny GD et al. Necrostatin-1 reduces histopathology and improves functional outcome after controlled cortical impact in mice. J Cereb Blood Flow Metab 2008; 28: 1564–1573.
Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 2011; 35: 908–918.
Lin J, Li H, Yang M, Ren J, Huang Z, Han F et al. A role of RIP3-mediated macrophage necrosis in atherosclerosis development. Cell Rep 2013; 3: 200–210.
Bonnet MC, Preukschat D, Welz PS, van Loo G, Ermolaeva MA, Bloch W et al. The adaptor protein FADD protects epidermal keratinocytes from necroptosis in vivo and prevents skin inflammation. Immunity 2011; 35: 572–582.
Gunther C, Martini E, Wittkopf N, Amann K, Weigmann B, Neumann H et al. Caspase-8 regulates TNF-alpha-induced epithelial necroptosis and terminal ileitis. Nature 2011; 477: 335–339.
Roychowdhury S, McMullen MR, Pisano SG, Liu X, Nagy LE . Absence of receptor interacting protein kinase 3 prevents ethanol-induced liver injury. Hepatology 2013; 57: 1773–1783.
Li JX, Feng JM, Wang Y, Li XH, Chen XX, Su Y et al. The B-Raf(V600E) inhibitor dabrafenib selectively inhibits RIP3 and alleviates acetaminophen-induced liver injury. Cell Death Dis 2014; 5: e1278.
Hanus J, Zhang H, Wang Z, Liu Q, Zhou Q, Wang S . Induction of necrotic cell death by oxidative stress in retinal pigment epithelial cells. Cell Death Dis 2013; 4: e965.
Murakami Y, Matsumoto H, Roh M, Giani A, Kataoka K, Morizane Y et al. Programmed necrosis, not apoptosis, is a key mediator of cell loss and DAMP-mediated inflammation in dsRNA-induced retinal degeneration. Cell Death Differ 2014; 21: 270–277.
Murakami Y, Matsumoto H, Roh M, Suzuki J, Hisatomi T, Ikeda Y et al. Receptor interacting protein kinase mediates necrotic cone but not rod cell death in a mouse model of inherited degeneration. Proc Natl Acad Sci USA 2012; 109: 14598–14603.
Trichonas G, Murakami Y, Thanos A, Morizane Y, Kayama M, Debouck CM et al. Receptor interacting protein kinases mediate retinal detachment-induced photoreceptor necrosis and compensate for inhibition of apoptosis. Proc Natl Acad Sci USA 2010; 107: 21695–21700.
Sato K, Li S, Gordon WC, He J, Liou GI, Hill JM et al. Receptor interacting protein kinase-mediated necrosis contributes to cone and rod photoreceptor degeneration in the retina lacking interphotoreceptor retinoid-binding protein. J Neurosci 2013; 33: 17458–17468.
Inohara N, del Peso L, Koseki T, Chen S, RICK Nunez G . a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J Biol Chem 1998; 273: 12296–12300.
McCarthy JV, Ni J, Dixit VM . RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J Biol Chem 1998; 273: 16968–16975.
Thome M, Hofmann K, Burns K, Martinon F, Bodmer JL, Mattmann C et al. Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr Biol 1998; 8: 885–888.
Navas TA, Baldwin DT, Stewart TA . RIP2 is a Raf1-activated mitogen-activated protein kinase kinase. J Biol Chem 1999; 274: 33684–33690.
Nembrini C, Kisielow J, Shamshiev AT, Tortola L, Coyle AJ, Kopf M et al. The kinase activity of Rip2 determines its stability and consequently Nod1- and Nod2-mediated immune responses. J Biol Chem 2009; 284: 19183–19188.
Windheim M, Lang C, Peggie M, Plater LA, Cohen P . Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem J 2007; 404: 179–190.
Chin AI, Dempsey PW, Bruhn K, Miller JF, Xu Y, Cheng G . Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature 2002; 416: 190–194.
Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 2002; 416: 194–199.
Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol 2007; 178: 2380–2386.
Rubino SJ, Selvanantham T, Girardin SE, Philpott DJ . Nod-like receptors in the control of intestinal inflammation. Curr Opin Immunol 2012; 24: 398–404.
Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem 2003; 278: 5509–5512.
Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 2003; 278: 8869–8872.
Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 2001; 411: 599–603.
Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 2001; 411: 603–606.
Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S et al. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet 2001; 357: 1925–1928.
Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R et al. CARD15 mutations in Blau syndrome. Nat Genet 2001; 29: 19–20.
Kanazawa N, Okafuji I, Kambe N, Nishikomori R, Nakata-Hizume M, Nagai S et al. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-kappaB activation: common genetic etiology with Blau syndrome. Blood 2005; 105: 1195–1197.
Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J 2004; 23: 1587–1597.
Abbott DW, Wilkins A, Asara JM, Cantley LC . The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol 2004; 14: 2217–2227.
Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G . Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem 2001; 276: 4812–4818.
Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC . Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol 2007; 27: 6012–6025.
Yang Y, Yin C, Pandey A, Abbott D, Sassetti C, Kelliher MA . NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J Biol Chem 2007; 282: 36223–36229.
Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J 2008; 27: 373–383.
Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med 2010; 16: 90–97.
Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 2010; 11: 55–62.
Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, Turner LJ et al. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe 2014; 15: 623–635.
Magalhaes JG, Lee J, Geddes K, Rubino S, Philpott DJ, Girardin SE . Essential role of Rip2 in the modulation of innate and adaptive immunity triggered by Nod1 and Nod2 ligands. Eur J Immunol 2011; 41: 1445–1455.
Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M . Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity 2009; 30: 789–801.
Damgaard RB, Nachbur U, Yabal M, Wong WW, Fiil BK, Kastirr M et al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell 2012; 46: 746–758.
Krieg A, Correa RG, Garrison JB, Le Negrate G, Welsh K, Huang Z et al. XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci USA 2009; 106: 14524–14529.
Tao M, Scacheri PC, Marinis JM, Harhaj EW, Matesic LE, Abbott DW . ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr Biol 2009; 19: 1255–1263.
Yang S, Wang B, Humphries F, Jackson R, Healy ME, Bergin R et al. Pellino3 ubiquitinates RIP2 and mediates Nod2-induced signaling and protective effects in colitis. Nat Immunol 2013; 14: 927–936.
Moynagh PN . The roles of Pellino E3 ubiquitin ligases in immunity. Nat Rev Immunol 2014; 14: 122–131.
Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 2009; 36: 831–844.
Conde C, Rambout X, Lebrun M, Lecat A, Di Valentin E, Dequiedt F et al. The inositol phosphatase SHIP-1 inhibits NOD2-induced NF-kappaB activation by disturbing the interaction of XIAP with RIP2. PLoS One 2012; 7: e41005.
Ver Heul AM, Fowler CA, Ramaswamy S, Piper RC . Ubiquitin regulates caspase recruitment domain-mediated signaling by nucleotide-binding oligomerization domain-containing proteins NOD1 and NOD2. J Biol Chem 2013; 288: 6890–6902.
Sorbara MT, Ellison LK, Ramjeet M, Travassos LH, Jones NL, Girardin SE et al. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity 2013; 39: 858–873.
Li L, Bin LH, Li F, Liu Y, Chen D, Zhai Z et al. TRIP6 is a RIP2-associated common signaling component of multiple NF-kappaB activation pathways. J Cell Sci 2005; 118 (Pt 3): 555–563.
Clark NM, Marinis JM, Cobb BA, Abbott DW . MEKK4 sequesters RIP2 to dictate NOD2 signal specificity. Curr Biol 2008; 18: 1402–1408.
LeBlanc PM, Yeretssian G, Rutherford N, Doiron K, Nadiri A, Zhu L et al. Caspase-12 modulates NOD signaling and regulates antimicrobial peptide production and mucosal immunity. Cell Host Microbe 2008; 3: 146–157.
Heymann MC, Winkler S, Luksch H, Flecks S, Franke M, Russ S et al. Human procaspase-1 variants with decreased enzymatic activity are associated with febrile episodes and may contribute to inflammation via RIP2 and NF-kappaB signaling. J Immunol 2014; 192: 4379–4385.
Balamayooran T, Batra S, Balamayooran G, Cai S, Kobayashi KS, Flavell RA et al. Receptor-interacting protein 2 controls pulmonary host defense to Escherichia coli infection via the regulation of interleukin-17A. Infect Immun 2011; 79: 4588–4599.
Shimada K, Chen S, Dempsey PW, Sorrentino R, Alsabeh R, Slepenkin AV et al. The NOD/RIP2 pathway is essential for host defenses against Chlamydophila pneumoniae lung infection. PLoS Pathog 2009; 5: e1000379.
Kim YG, Park JH, Reimer T, Baker DP, Kawai T, Kumar H et al. Viral infection augments Nod1/2 signaling to potentiate lethality associated with secondary bacterial infections. Cell Host Microbe 2011; 9: 496–507.
Satoh J, Nakanishi M, Koike F, Miyake S, Yamamoto T, Kawai M et al. Microarray analysis identifies an aberrant expression of apoptosis and DNA damage-regulatory genes in multiple sclerosis. Neurobiol Dis 2005; 18: 537–550.
Shaw PJ, Barr MJ, Lukens JR, McGargill MA, Chi H, Mak TW et al. Signaling via the RIP2 adaptor protein in central nervous system-infiltrating dendritic cells promotes inflammation and autoimmunity. Immunity 2011; 34: 75–84.
Schrijver IA, van Meurs M, Melief MJ, Wim Ang C, Buljevac D, Ravid R et al. Bacterial peptidoglycan and immune reactivity in the central nervous system in multiple sclerosis. Brain 2001; 124 (Pt 8): 1544–1554.
Goh FY, Cook KL, Upton N, Tao L, Lah LC, Leung BP et al. Receptor-interacting protein 2 gene silencing attenuates allergic airway inflammation. J Immunol 2013; 191: 2691–2699.
Levin MC, Jirholt P, Wramstedt A, Johansson ME, Lundberg AM, Trajkovska MG et al. Rip2 deficiency leads to increased atherosclerosis despite decreased inflammation. Circ Res 2011; 109: 1210–1218.
Acknowledgements
This work is supported by a grant from Science Foundation Ireland (12/IA/1736).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict to interest.
Additional information
Edited by H-U Simon
Rights and permissions
About this article
Cite this article
Humphries, F., Yang, S., Wang, B. et al. RIP kinases: key decision makers in cell death and innate immunity. Cell Death Differ 22, 225–236 (2015). https://doi.org/10.1038/cdd.2014.126
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.126
This article is cited by
-
Effects of fine particulate matter on bone marrow-conserved hematopoietic and mesenchymal stem cells: a systematic review
Experimental & Molecular Medicine (2024)
-
5-Iodotubercidin sensitizes cells to RIPK1-dependent necroptosis by interfering with NFκB signaling
Cell Death Discovery (2023)
-
Delta (B1.617.2) variant of SARS-CoV-2 induces severe neurotropic patterns in K18-hACE2 mice
Scientific Reports (2023)
-
PRMT5-mediated regulatory arginine methylation of RIPK3
Cell Death Discovery (2023)
-
Rubiarbonol B induces RIPK1-dependent necroptosis via NOX1-derived ROS production
Cell Biology and Toxicology (2023)