Abstract
Serine/arginine-rich splicing factor 3 (SRSF3), a member of the serine/arginine (SR)-rich family of proteins, regulates both alternative splicing of pre-mRNA and export of mature mRNA from the nucleus. Although its role in nuclear mRNA processing is well understood, the mechanism by which it alters the fate of cytoplasmic mRNA molecules remains elusive. Here, we provide evidence that SRSF3 not only regulates the alternative splicing pattern of programmed cell death 4 (PDCD4) mRNA, but also modulates its translational efficiency in the cytoplasm by lowering translation levels. We observed a marked increase in PDCD4 mRNA in translating polysome fractions upon silencing of SRSF3, and, conversely, ectopic overexpression of SRSF3 shifted PDCD4 mRNA into non-translating ribosomal fractions. In live cells, SRSF3 colocalized with PDCD4 mRNA in P-bodies (PBs), where translationally silenced mRNAs are deposited, and this localization was abrogated upon SRSF3 silencing. Furthermore, using two different reporter systems, we showed that SRSF3 interacts directly with PDCD4 mRNA and mediates translational repression by binding to the 5′-untranslated region (5′-UTR). In summary, our data suggest that the oncogenic potential of SRSF3 might be realized, in part, through the translational repression of PDCD4 mRNA.
Similar content being viewed by others
Main
Serine/arginine(SR)-rich proteins are members of a family of RNA-binding proteins that function in the constitutive and alternative splicing of pre-mRNAs.1, 2, 3 These proteins harbor one or two RNA-recognition motifs (RRMs) at the N-terminus and the characteristic serine-arginine dipeptide repeats (the RS domain) at the C-terminus. Research on SR proteins has focused primarily on the role of these proteins in nuclear processes, including transcription, co-transcriptional mRNA processing, genome stability control,4 and others.5, 6 Intriguingly, recent studies point to critical roles for SR proteins in post-transcriptional regulatory processes such as nuclear export,7 surveillance,5 stability,8 and translational control.9, 10 SR proteins may act as coordinators and key regulators of the extensive network of post-transcriptional processes, but how this coordination is carried out has not been fully elucidated.
SRSF3 (SRp20), the smallest SR protein, has been shown to modulate alternative splicing events for a variety of genes including itself, and its potential binding sequences has been identified using iCLIP (in vivo UV crosslinking and immunoprecipitation). In addition, cytoplasmic functions of SRSF3 have been reported, as it continuously shuttles between the nucleus and the cytoplasm,11 and involves mRNA export through TAP-dependent manner.12 More recently, SRSF3 displayed a positive role for viral IRES-mediated translation.13 However, a specific role for SRSF3 in these cytoplasmic events has remained undefined, and direct binding to particular cytoplasmic mRNA molecules has not been demonstrated.
PDCD4 (programmed cell death 4) is a neoplastic transformation inhibitor protein. Various apoptotic stimuli,14 with the exception of UV exposure and topoisomerase inhibitor treatment, activate PCDC4 gene expression.15 The role of PDCD4 as a tumor suppressor has been of particular interest because of its antiproliferative and tumor-suppressive effects in many different cell types, although its role in cancer cells is debatable.16 Apoptotic cell death caused by an overexpression of PDCD4 is seemingly cell-type specific.17 Furthermore, there is no clear correlation between PDCD4 mRNA and protein levels among different cancer cell types,18 suggesting that transcriptional or post-transcriptional regulation of PDCD4 varies. This variability between mRNA and protein levels is likely due to differing regulatory mechanisms employed between cell types.
In the present study, we identified PDCD4 mRNA as a target for SRSF3 binding by silencing and gene expression profiling experiments. Further analyses revealed that SRSF3 regulates not only the alternative splicing but also the translation of PDCD4 transcript. Moreover, we demonstrated that the 5′-untranslated region (5′-UTR) of PDCD4 mRNA is necessary for the interaction between SRSF3 and PDCD4 mRNA. We also observed that the depletion of SRSF3 led to potent apoptotic cell death mediated by the elevation of PDCD4 protein levels. In summary, we propose that SRSF3 has an anti-apoptotic role, through the translational repression of tumor suppressor such as PDCD4.
Results
SRSF3 regulates apoptosis in cancer cells
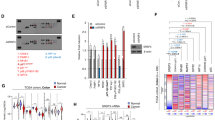
A role for SRSF3 in malignant cancer cell proliferation has been described.18 To further define this role, we tested the effect of SRSF3 silencing on apoptosis using two different siRNAs (siSRSF3-1 and siSRSF3-2) and the cancer cell lines SW480 (human colon adenocarcinoma) and U2OS (human osteosarcoma). As shown in Figure 1a, caspase-3 cleavage was significantly higher in both cancer cell lines when SRSF3 was silenced, but not when control siRNA (siCONT) was used.
Depletion of SRSF3 induces apoptotic cell death by modulating regulatory genes involved in the apoptosis process. (a) Control siRNA (siCONT) and siRNAs specific for SRSF3 (siSRSF3-1, siSRSF3-2) were transfected into either SW480 or U2OS cells for 72 h and analyzed by western blot analysis to detect protein levels. Actin was used as a loading control. (b) HEK-293 cells were transfected with siCONT or siSRSF3-1 and subjected to nuclei staining using Hoechst 33258. Yellow arrows indicate cells with fragmented nuclei. (c) The integrity of chromosomal DNA from siCONT or siSRSF3-1-transfected SW480 cells was assessed through agarose gel electrophoresis. (d) Hierarchical clustering of normalized microarray expression data from U2OS cells transfected with siCONT or siSRSF3 siRNAs. Genes with more than twofold differential expression (a total of 655 genes) between SRSF3 and control knockdown cells are shown. Silencing of SRSF3 was confirmed by western blot analysis (right panel). (e) Gene ontology analysis with microarray expression profiles using David 6.739 was carried out, and twenty functional annotations with the lowest P-value are shown. (f) U2OS cells were transfected with control (siCONT) or siRNAs for SRSF3 (siSRSF3-1, siSRSF3-S) for 72 h, and western blot analysis was performed to measure protein levels
We observed condensed and fragmented nuclei in siSRSF3-treated cells stained with Hoechst33258 (Figure 1b and Supplementary Figure 1a) and direct evidence of DNA fragmentation using agarose gel analysis (Figure 1c). Furthermore, cell proliferation was significantly inhibited, as measured by crystal violet staining (Supplementary Figure 1c). Together, these results demonstrate that decreased level of SRSF3 induces apoptosis and reduces cell proliferation.
Given the marked increase in apoptotic cell death in response to an SRSF3 knockdown, we predicted that the regulation of gene expression of particular splicing targets was responsible. To test this hypothesis, gene expression profiling (using Affymetrix Human Genome U133A 2.0 arrays, Santa Clara, CA, USA) was performed using RNA samples extracted from U2OS cells transfected with siSRSF3-1, siSRSF3-2, siSRSF3-S (SMARTpool, a mixture of siRNAs targeting SRSF3), or siCONT. As shown in Figure 1d, the heatmap revealed that 381 genes (449 probes) were upregulated and 274 genes (328 probes) were downregulated at least twofold in response to a silencing of SRSF3 as compared with controls (see Supplementary Table 2). Consistent with the observation that apoptosis was induced in SRSF3 knockdown cells, genes involved in cell death processes were upregulated, further suggesting a critical involvement of SRSF3 in this process (Figures 1d and e).
PDCD4 expression is markedly increased when SRSF3 is silenced
Microarray analyses indicated that the expression of a number of genes involved in apoptotic or anti-apoptotic processes were highly affected in the SRSF3-depleted cells (Figure 1e). Intriguingly, of the transcripts that were upregulated, PDCD4 was one of the most highly affected, with an increase of 5.1-, 3.6-, and 2.2-fold (as a very conservative estimation) in response to siSRSF3-1, siSRSF3-2, and siSRSF3-S, respectively. As PDCD4 was reported to be involved in cellular processes such as antiproliferation, apoptosis, and antimetastasis in various cancer cells through an unknown mechanism,16, 19 we hypothesized that the observed effects in response to SRSF3 depletion might be, in part, due to the upregulation of PDCD4. To investigate this possibility, we used western blot analysis to measure PDCD4 protein levels in U2OS cells transfected with siCONT, siSRSF3-1, or siSRSF3-S. As expected, in both of the SRSF3 knockdowns, there was a remarkable increase in PDCD4 levels as compared with the control (Figure 1f). To test if the increase in PDCD4 protein expression from U2OS cell line can also be reflected to other cell lines including SW480 and HEK-293 utilized in apoptosis assay and one additional HeLa, we performed SRSF3 silencing followed by western analysis. As shown in Supplementary Figure 2, the elevated expression of PDCD4 protein at least twofold in response to silencing of SRSF3 was evident in U2OS, SW480, and HEK-293 but not in HeLa.
The human PDCD4 gene generates two different mRNA isoforms through alternatively splicing events (Figure 2a). As the four probes used in the microarray analysis cannot distinguish PDCD4 isoforms 1 and 2, and given the increased levels of PDCD4 protein in SRSF3 knockdown cells, we asked whether or not splicing of the PDCD4 mRNA is affected by SRSF3. By employing real-time quantitative PCR analysis (RT-qPCR) using total RNA prepared from SW480 cells transfected with either siCONT or siSRSF3, we observed that the expression of PDCD4 isoform 2 was increased approximately twofold in response to the knockdown. In contrast, PDCD4 isoform 1 was present at a level similar to that of the control cells (Figure 2b). Western blot analysis of these same strains demonstrated that, at the protein level, both PDCD4 isoforms are elevated in response to silencing SRSF3. This observation was striking because there was a significant increase (two- to five-fold) in protein levels for isoform 1 without any corresponding change in the mRNA levels, suggesting a potential role for SRSF3 in the management of either the stability or the translation of cytosolic PDCD4 mRNA (Figure 2c). We observed similar results in U2OS cells (Figure 1f and Supplementary Figure 2). We will focus on potential roles of SRSF3 on the regulation of PDCD4 isoform 1 mRNA at the post-transcriptional level for the rest of this study.
Silencing of SRSF3 changes the alternative splicing pattern of PDCD4 transcripts. (a) A schematic representation of the alternative splicing of PDCD4 transcripts. (b) The expression levels of total PDCD4, isoform 1, or isoform 2 were assessed by RT-qPCR using SW480 cells transfected with siSRSF3-1 siRNA. The levels of endogenous PDCD4 mRNAs were normalized to GAPDH mRNA. Three independent experiments were performed, and results are reported as means ±S.D. (c) Protein expression levels of PDCD4 isoform 1 and isoform 2 were measured for the same samples using western blot analysis. β-Actin was used as a loading control
To examine a possible role for SRSF3 in controlling the stability of PDCD4 mRNA, we measured the half-life of PDCD4 mRNA from SW480 cells that were transfected with either siCONT or siSRSF3-1. Total RNA was collected at various time points after actinomycin D (ActD) treatment and analyzed by RT-qPCR using primers specific for both isoforms. The stability of PDCD4 mRNA from siSRSF3-1-transfected cells (T1/2=11.3±0.05) was not significantly different from that of siCONT-transfected cells (T1/2=9.3±0.02) (Supplementary Figure 3a).
As PDCD4 protein can be rapidly phosphorylated by protein kinase S6K1 and subsequently degraded via β-TRCP ubiquitin ligase in response to mitogen signals,20 we investigated a possible role for SRSF3 in this process by measuring the protein stability of PDCD4 in the presence or absence of SRSF3. SW480 cells transfected with siCONT or siSRSF3-1 siRNAs for 48 h were treated with cycloheximide (CHX), after which total protein was extracted at various time points and subjected to western blot analysis. As shown in Supplementary Figures 3b and 4a, PDCD4 protein stability (siCONT, T1/2=5.0±0.1; siSRSF3, T1/2=4.8±0.1) and the phosphorylation status of S6 small ribosomal subunit, another target of S6K1, from both groups were comparable. Taken together, these results suggest that the accumulation of PDCD4 protein that we observed in SRSF3-depleted cells was not due to modifications in either RNA stability or protein stability.
SRSF3 represses the translation of PDCD4 mRNA
As the level of PDCD4 isoform 1 protein was increased in SRSF3 knockdown cells with no corresponding change in the mRNA level, we predicted that regulation by SRSF3 may occur at the translational level. To test this prediction, we used sucrose gradient polysome fractionation to measure the relative association of PDCD4 mRNA with different ribosomal fractions from SW480 cells transfected with either siCONT or siSRSF3-1. A total of 16 fractions were collected, and the PDCD4 mRNA level in each fraction was measured by semiquantitative RT-PCR (Figure 3a). Interestingly, the population of PDCD4 isoform 1 mRNA associated with polysome fractions appeared to be significantly increased in SRSF3 knockdown samples as compared with the control. To monitor detailed movements of tested transcripts, we quantified and plotted the relative abundance of each transcript throughout all the fractions and observed obvious polysomal shift of PDCD4 isoform 1 transcript upon SRSF3 depletion, whereas GAPDH did not show any significant changes in its distribution pattern (Figures 3c and e). Next, we tested if overexpression of SRSF3 would display an opposite effect on a separate polysome analysis. As shown in Supplementary Figure 4, overexpression of Flag-tagged SRSF3 (F-SRSF3) led to lower mRNA levels within polysome fractions.
SRSF3 interacts with PDCD4 isoform 1 mRNA and regulates its translation. (a) SW480 cells were transfected with siCONT and siSRSF3-1 siRNAs and separated by polysome fractionation. The corresponding rRNAs were detected using agarose gel electrophoresis. RNA samples from each fraction were subjected to RT-PCR analyses to detect PDCD4 isoform 1 and isoform 2. GAPDH was used as control. (b) Western blot analysis was performed with total cell extracts from (a). Quantification of RT-PCR product from (a) was performed to observe relative abundance of PDCD4 isoform 1 (c), isoform 2 (d), and GAPDH (e) in each fraction. (f) Levels of PDCD4 mRNA in cell lysates from HEK-293 cells transfected with either vector or F-SRSF3 plasmid were measured by carrying out RNP immunoprecipitation using an anti-Flag tag antibody, followed by RT-PCR using primers specific for either PDCD4 or the control U6 RNA (top panels). Western blot analysis was performed in parallel using a Flag antibody to confirm the presence of F-SRSF3 protein. Figures are representative of at least two independent experiments. (g) RNP immunoprecipitation analysis was performed using a mouse IgG (control) or an anti-SRSF3 antibody on cell extracts from either HEK-293 or SW480 cell lines as described for the previous panel. (h) U2OS cells transfected with either siCONT (top panels) or siSRSF3-1 (bottom panels) were subjected to FISH and indirect immunofluorescence microscopy (see Materials and Methods). PBs were visualized using an anti-Rck/p54 antibody, and PDCD4 mRNAs were visualized using terminal Cy3-labeled PDCD4 mRNA-specific probes. (i) U2OS cells were transfected with either siCONT (top panels) or siSRSF3 (bottom panels) and subjected to FISH and indirect immunofluorescence microscopy as described for the previous panels
To convincingly show the knockdown or overexpression effects of SRSF3 on the regulation of PDCD4 expression, we performed a rescue experiment using combinations of knockdown and overexpression of SRSF3. To do this, we designed a siSRSF3 siRNA targeting 3′-UTR of SRSF3 mRNA (siSRSF3-3′-UTR, see Supplementary Table 1 for sequence information) that cannot target SRSF3 mRNA transcribed from F-SRSF3 overexpression plasmid lacking 3′-UTR of SRSF3. As shown in Supplementary Figure 4g, PDCD4 protein level was significantly elevated in SW480 cells transfected with siSRSF3-3′-UTR together with empty vector. Whereas, co-transfection of F-SRSF3 together with siSRSF3-3′-UTR effectively dampened upregulation of PDCD4 expression. From these data, we concluded that SRSF3 regulates translation of PDCD4 mRNA by restricting PDCD4 mRNA to the non-translating ribosomal fractions.
SRSF3 binds and recruits PDCD4 mRNA to P-bodies
To understand the mechanism by which SRSF3 represses the translation of PDCD4, we first asked whether or not SRSF3 interacts directly with PDCD4 mRNA. Either F-SRSF3 or a vector control was transfected into SW480 cells, and ribonucleoprotein-immunoprecipitation (RNP-IP) was performed using an anti-Flag antibody under conditions that preserve RNP integrity. Subsequent RT-PCR analysis showed a strong signal for PDCD4 in cells containing F-SRSF3, but none in the vector control (Figure 3f). Specificity of the bound PDCD4 mRNA was confirmed by the absence of U6 snRNA amplification from the RNP pellet. To more specifically demonstrate the association of endogenous SRSF3 with PDCD4 mRNA, RNP-IP was performed with either an anti-SRSF3 antibody or mouse IgG as a control. As shown in Figure 3g, PDCD4 mRNA was enriched in the SRSF3 immunoprecipitate but not in IgG controls for both HEK-293 and SW480 cells.
Given that SRSF3 repressed translation and associated with PDCD4 mRNA, we then asked whether PDCD4 mRNA was targeted to P-bodies (PB), which are cytosolic RNA granules where translationally silenced or short-lived mRNAs aggregate. We employed indirect immunocytochemical analysis using a PB-specific marker (Rck/p54) in conjunction with FISH (fluorescence in situ hybridization) using a PDCD4 mRNA-specific probe to localize intracellular PDCD4 mRNA. As seen in Figure 3h, colocalization of Rck and PDCD4 mRNA was evident in cells transfected with siCONT, whereas this punctate staining pattern was not observed when SRSF3 was depleted. We then performed a similar experiment to look for colocalization of SRSF3 itself with PDCD4 mRNA in PBs. As shown in Figure 3i, colocalization of SRSF3 and PDCD4 mRNA was clear, and the observed cellular location suggested that they associate in PBs. Notably, the punctate pattern of PDCD4 mRNA was less visible in siSRSF3-treated cells, suggesting that SRSF3 was responsible for directing PDCD4 mRNA to PBs. Together, these data support an association between SRSF3 and PDCD4 mRNA in cytoplasmic subfractions including PBs.
SRSF3 preferentially binds to the 5′-UTR of PDCD4 mRNA
In order to define which region of PDCD4 mRNA is bound and recruited to PB by SRSF3, we used RNP-IP in combination with a MS2 tethering system,21 which uses the bacteriophage MS2 coat protein to tether proteins to RNA. We prepared three different chimeric MS2 reporters in which 12 tandem MS2 RNA hairpins are located immediately upstream of the 5′-UTR, the coding region (CR), or the 3′-UTR of PDCD4 mRNA (Figure 4a). To test which region(s) of PDCD4 mRNA could be associated with SRSF3, we performed RNP-IP using a control IgG or the SRSF3 antibody and cell extracts from HEK-293 cells transfected with each of the MS2 reporter constructs and then analyzed the samples using RT-PCR with primers specific for each region of the PDCD4 mRNA. As shown in Figure 4b, the presence of the 5′-UTR led to the strongest signal. There was slightly more binding of SRSF3 to the 3′-UTR as compared with the vector control, but the signal was less prominent than for the 5′-UTR. Neither GAPDH nor U6 RNA precipitated with either the control IgG or the anti-SRSF3 antibody.
SRSF3 interacts with the 5′-UTR of PDCD4 mRNA and represses PDCD4 translation. (a) A schematic representation of the MS2 tethering system from the pMS2 reporter plasmid is shown. Cloned downstream of the 12 MS2 hairpin structures are the 5′-UTR (5′-U), the coding region (CR), or the 3′-UTR (3′-U) of PDCD4 mRNA. A MS2-GFP fusion protein with a nuclear localization signal (NLS) can recognize and bind to the MS2 hairpin structures. (b) HEK-293 cells were transfected with a pMS2 vector (control), pMS2-PDCD4-5′-U, pMS2-PDCD4-CR, or pMS2-PDCD4-3′-U plasmids and subjected to RNP-IP analyses using a mouse IgG (control) or an anti-SRSF3 antibody. RT-PCR of each immunoprecipitate was carried out to measure enrichment of each region of PDCD4 mRNA. GAPDH and U6 were used as non-specific controls. (c) Using immunofluorescence microscopy, U2OS cells expressing MS2, MS2-5′-U, MS2-CR, and MS2-3′-U mRNAs were visualized using a coexpressed MS2-GFP protein (green) to detect the MS constructs and an anti-Rck antibody (red) to detect PBs. Cells were incubated in the presence or absence of sodium arsenite (SA) (0.5 mM, 1 h). (d) A schematic of firefly luciferase reporter (pGL3) constructs in which the 5′-UTR (pGL3-PDCD4-5’-UTR), 3′-UTR (pGL3-PDCD4-3′-UTR), or both regions (pGL3-PDCD4-UTRs) of PDCD4 mRNA were cloned upstream or downstream of the luciferase coding region (Luc). (e) HEK-293 cells were transfected with pGL3 vector, pGL3-PDCD4-5′-UTR, pGL3-PDCD4-3′-UTR, or pGL3-PDCD4-UTRs plasmids and subjected to RNP-IP analyses as decribed for panel (b). Primers used to detect each region are designated to the right of each panel. GAPDH and β-microglobulin (B2M) were used as non-specific controls. (f) Luciferase assays were performed after transfecting each pGL3 construct into SW480 cells transfected with either siCONT or siSRSF3. Renilla luciferase vector was cotransfected for normalization. (g) Caspase-3 activation was assessed in SW480 cells transfected with different combinations of siRNAs (siSRSF3-1 and siPDCD4), and indicated proteins were detected by Western blot analysis. (h) Chromosomal fragmentation analysis of SW480 cells transfected with siCONT, siSRSF3, siPDCD4, or siSRSF3/siPDCD4. DNA size marker (M) is shown in the leftmost lane. (i) A model of SRSF3-mediated post-transcriptional regulation of PDCD4 mRNA
Given that PDCD4 mRNA localized to PBs (Figures 3h and i), we next sought to identify which region(s) of this molecule are necessary for this localization by cotransfecting the MS2 reporter constructs and pMS2-GFP (a vector expressing MS2-GFP fusion protein) into U2OS cells. The association of MS2-GFP with MS2 hairpin structures in reporter mRNAs allows for tracking of their subcellular localization. MS2-GFP was exclusively localized to the nucleus due to the presence of nuclear localization signals in this protein (NLS; Figures 4a and c). In contrast, some of the PDCD4 5′-UTR was retained in the cytoplasm in a punctate pattern, colocalizing with the PB marker Rck. Colocalization of either CR or 3′-UTR was minimal as the signal for each in PBs was almost at background levels. Similar results were obtained when cells were treated with sodium arsenite, which leads to strong oxidative conditions under which PBs are robustly induced (Figure 4c, lower panels).
To further investigate the effect of SRSF3 on the translation of PDCD4 mRNA, we utilized a luciferase reporter system in which the 5′-UTR, the 3′-UTR, or both of PDCD4 were cloned into the appropriate locations adjacent to a luciferase open reading frame (ORF) (Figure 4d). First, we tested the ability of SRSF3 to interact with transcripts from each construct. In a RNP-IP experiment followed by RT-PCR analysis, reporter transcripts harboring the 5′-UTR region were precipitated by an SRSF3 antibody (Figure 4e). Neither of the controls, GAPDH nor β-microglobulin (B2M) mRNA, was detected in any of the precipitated samples.
Next, the luciferase reporter constructs were employed again, and enzyme activity was measured as an indication of the level of protein translation from each. SW480 cells containing each transcript were transfected with either siCONT or siSRSF3 to assess the effect of SRSF3 depletion. As expected, the reporter harboring the 5′-UTR (or both of the UTRs) had a markedly higher level of luciferase activity in an SRSF3 knockdown than in the control (Figure 4f). Cells containing the vector alone or the 3′-UTR reporter construct had the same amount of luciferase activity, regardless of SRSF3 depletion. It is interesting to note that the luciferase activity from the 5′-UTR construct was approximately fourfold higher than the vector control in cells transfected with siCONT, whereas that from the 3′-UTR was more than approximately fivefold lower, suggesting the existence of additional regulatory protein(s) that may associate with these regions of PDCD4 mRNA. Collectively, these results support the hypothesis that SRSF3 interacts and represses PDCD4 mRNA translation by binding to the 5′-UTR.
As PDCD4 is known to be a neoplastic transformation inhibitor, SRSF3 depletion-mediated antiproliferation and apoptotic cell death might be due to abnormally high expression levels of PDCD4, and perhaps PDCD4 alone (Supplementary Figure 5). If this is the case, elevated PDCD4 should cause apoptotic cell death in SRSF3-depleted cells. To test whether or not a knockdown of both SRSF3 and PDCD4 would prevent apoptosis, siPDCD4 and siSRSF3 were cotransfected into SW480 cells and the presence or absence of apoptotic characteristics were assessed. Caspase-3 activation was lower than that in cells transfected with siSRSF3 alone (Figure 4g). Moreover, the amount of fragmented chromosomal DNA observed in siSRSF3-treated cells, seen as a ladder pattern on an agarose gel, was significantly lower in siSRSF3/siPDCD4-cotransfected cells (Figure 4h). Together, these data suggest that abnormally high levels of PDCD4 protein itself may contribute significantly to cell death in SRSF3-depleted cells.
Discussion
Here we describe for the first time that the intracellular role of SRSF3 is not simply in alternative splicing of pre-mRNA but also in regulating the translation of PDCD4 mRNA. Of two alternatively spliced transcripts of PDCD4, isoform 1 alone was affected at the translational level by SRSF3, so we focused our study on this particular isoform. We provide evidence that the translational efficiency of PDCD4 mRNA was directly altered by the presence of SRSF3. Intriguingly, we observe that SRSF3 exerts its effect on PDCD4 mRNA through a strong interaction with the 5′-UTR and recruitment to PBs. The luciferase assay with the reporter constructs harboring the 5′-UTR reinforced these observations.
Our findings on a novel cytoplasmic function for SRSF3 are consistent with an emerging paradigm in which various stages of gene expression are coordinated with one another and that there is a connection between nuclear transcription and cytoplasmic post-transcriptional events.22, 23 Except from the well-known nuclear RNA processing function of SR proteins, what may be the most intriguing new findings on cytoplasmic SR protein functions are that SR proteins are directly involved in the process of mRNA translation. In experiments involving sucrose gradient polysome profiling analysis, both SRSF1 and SRSF3 associated with ribosomes.9 SRSF1 had a stronger affinity for the polysome fractions than SRSF3, suggesting that it has a more active role in translation. Furthermore, the translation of a luciferase reporter gene fused to exonic-splicing enhancer (ESE) was strongly enhanced both in vivo and in vitro by SRSF1.9 It was further demonstrated that SRSF1 recruits mTOR kinase to the target mRNA, leading to the hyper-phosphorylation of eIF4E-binding protein (4E-BP), which then relieves inhibition of translation.10 Although no cytoplasmic mRNAs have been identified as targets for regulation by SRSF3, this protein has been shown to be involved in an internal ribosome entry site (IRES)-mediated translation of viral RNA by interacting with PCBP2, an IRES-binding protein.13
SR proteins such as SRSF1 and SRSF3 shuttle between the nucleus and the cytoplasm and migrate into RNA granules, such as PBs and stress granules (SGs), where translationally silenced mRNAs and regulatory proteins are deposited.24, 25, 26 These RNA granules harbor both mRNAs that have disassembled from translationally active polysomes and proteins including a eukaryotic translation initiation factor 4E (eIF4E), XRN1 (5′-3′ exonuclease), FAST (Fas-activated serine/threonine kinase), TTP (tristetraprolin), and, notably, Argonaute, a key component of RISC (RNA-induced silencing complex), a complex involved in miRNA-mediated translational repression.27, 28 We show that SRSF3 and PDCD4 mRNA are colocalized to PBs. Furthermore, we observed that when SRSF3 was depleted, PBs disappeared and translational repression of PDCD4 mRNA was relieved. These observations suggest that SRSF3 recruits PDCD4 mRNA to PBs as a mechanism for fine-tuning PDCD4 expression in actively growing cells under specific conditions (e.g., in response to apoptotic stimuli).
RNA regulatory proteins are known to affect the expression of specific target mRNAs either positively or negatively to allow for fine-tuning of expression levels.22 Understanding the conditions that affect the mRNA targets of SRSF3 would provide insight into the role of this protein. An oncogenic role for SRSF3 in cancer has been proposed by several research groups.29, 30, 31 The first observation in support of this role was that a decrease in endogenous SRSF3 levels through RNA interference (RNAi) led to antiproliferation effects, cellular senescence, and eventually apoptotic cell death.32, 33 Furthermore, recent studies have suggested that altered expression of several SR family proteins affects the expression of many apoptotic genes, oncogenes, and tumor suppressors in renal carcinoma.34 Consistent with these observations, our microarray analysis identified pro-apoptotic and anti-apoptotic genes as those most affected by the presence or absence of SRSF3. It was also interesting to note that SRSF3 is a target of the Wnt/β-catenin signaling pathway in colorectal cancer.35 Increase in β-catenin/TCF transcriptional activity was highly correlated with the enhancement of SRSF3 expression at both the mRNA and protein levels, suggesting an important role in colon cancer physiology. More mechanistic details must be elucidated to understand the complex interplay between oncogenic signaling and SR proteins during tumorigenesis.
Materials and Methods
Cell culture and transfection
The human colorectal adnocarcinoma (SW480), human cervical adenocarcinoma (HeLa), human osteosarcoma (U2OS), and human embryonic kidney (HEK-293T) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 500 U/ml penicillin-streptomycin at 37 °C in a humidified atmosphere of 5% CO2 in air. SW480, HeLa, and U2OS cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) or Fugene 6 (Roche, Indianapolis, IN, USA). After 36 h, the cells were lysed by using RIPA buffer (150 mM sodium cloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris-HCl pH 8.0).
Reagents and antibodies
Actinomycin D and cycloheximide were from Sigma (St Louis, MO, USA), and crystal violet was from Samchun Chemical (Pyeongtaek, South Korea). The SRSF3 (7B4) was purchased from Santa Cruz (Dallas, TX, USA). The Caspase-3, p-S6 (Ser240/244), and eIF6 antibodies were purchased from Cell Signaling (Danvers, MA, USA). The PDCD4, β-actin (AC15), eIF4A1, and anti-HA antibodies were purchased from Abcam (Cambridge, MA, USA). PTEN (6H2.1) was purchased from Millipore (Billerica, MA, USA). Rck/DDX6 was purchased from Bethyl (Montgomery, TX, USA). The FLAG-M2 antibody was purchased from Sigma.
Preparation of cell lysate and western blot analysis
The SW480, U2OS, HeLa, and HEK-293T cells were washed with PBS twice and lysed in RIPA buffer supplemented with protease and phosphatase inhibitor cocktails. After incubation on ice for 30 min, the supernatants were collected by centrifugation at 13 000 r.p.m. for 15 min. Protein concentration was determined using a Bradford protein assay (BioRad, Hercules, CA, USA). Aliquots containing 20∼35 μg protein were separated on a 12% SDS-polyacrylamide gel and transferred to a PVDF membrane. The protein-bound membrane was incubated with an antibody, followed by HRP-conjugated secondary antibodies, and visualized by chemiluminescence (ECL).
Immunofluorescence microscopic analysis
Cells were grown on glass coverslips, transfected, and incubated as indicated above, then washed twice in PBS-A (1 × PBS with 0.02% sodium azide), immediately fixed with 4% (v/v) formaldehyde in PBS for 15 min at 4 °C, and permeabilized with 0.5% (v/v) Triton X-100 in PBS for 15 min at RT. Protein was detected using a primary antibody for overnight at 4 °C, then washed twice in PBS-A, briefly stained with Cy2- or Cy3-conjugated immunoglobulin (Jackson ImmunoResearch, West Grove, PA, USA) and Hoechst 33258 (Sigma) for 1 h at RT in dark condition, washed twice in PBS-A, and the coverslips were mounted on slide glasses. Fluorescence microscopic images were taken using a Zeiss Axioplan2 fluorescence microscope (63 × ) and compiled using Adobe Photoshop software (San Jose, CA, USA).
RT-PCR and quantitative RT-PCR
Total RNA was extracted from cell lysates with the TriZol (RBC), and 2 μg of total RNA was reverse transcribed to cDNA using an oligo dT primer (Fermantas, Pittsburgh, PA, USA) and M-MuLV Reverse Transcriptase (Fermantas). Oligomers used for amplifying PCR products and primer sequences are listed in Supplementary Table 1.
Microarrays
Affymetrix Human Genome U133A 2.0 Arrays were used for gene expression analysis. Sample preparation, array hybridization, incubation, washing, and scanning were performed as described in manufacturer’s instructions at the Microarray Core Facility at the Dana Farber Cancer Institute. The CEL files were imported into dChIP software36 for data normalization and extraction of expression values. The normalized expression data were clustered by Cluster 3.0 and displayed by Java Treeview. dChip, Cluster 3.0, and Treeview are public software downloaded from http://www.hsph.harvard.edu/cli/complab/dchip/ and http://www.eisenlab.org/ websites.
Polysome profiling analysis
Cell lysates of SW480 for sucrose gradient centrifugation were prepared as polysome profiling buffer (20 mM HEPES (pH 7.6), 125 mM KCl, 5 mM MgCl2, 2 mM DTT, and DEPC water). Extracts were incubated on ice for 10 min, and insoluble material was pelleted by centrifugation at 13 000 r.p.m. for 10 min. The resulting supernatant extracts were then loaded onto a 17.5∼50% sucrose gradient prepared with polysome profiling buffer and ultra-centrifuged for 2.5 h at 35 000 r.p.m. in a SW41-Ti rotor (Beckman, Brea, CA, USA). Following centrifugation, the gradients were fractionated using a fraction collector (Brandel, Gaithersburg, MD, USA), and their quality was monitored at 254 nm using a UA-6 Absorbance Detector (Isco, Lincoln, NE, USA). Total RNA in each fraction was extracted by using TriZol.
RNA interference
U2OS or SW480 cells were transfected with siRNAs (40 nM) using Lipofectamine 2000 (Invitrogen). After 40 h, cells were trypsinized, reseeded, and transfected again for another 40 h. For single transfection, cells were treated for 48∼72 h and processed for the next step. Knockdown efficiencies were verified by western blot analysis. siRNAs were designed using published recommendation and target mRNA coding region. SiRNA sequences used in this study are listed in the Supplementary Table 1.
Ribonucleoprotein-immunoprecipitation (RNP-IP)
The method has described previously.37 Cells were washed in cold PBS and then resuspended in RNP-IP lysis buffer (50 mM Tris-Cl (pH7.5), 1% NP-40, 0.5% Sodium deoxycholate, 0.05% SDS, 1 mM EDTA(pH8.0),150 mM NaCl, 1 × Protease Inhibitor Cocktail (Sigma-Aldrich), 40 U/ml RiboLock RNase Inhibitor (Fermentas) in DEPC water). After incubating for 20 min, samples were spun at 13 000 r.p.m. for 15 min. The extracts were immunoprecipitated with normal IgG or anti-FLAG or anti-SRSF3 at 4 °C overnight. Beads were washed with RNP-IP lysis buffer for five times, and then beads extracted in Trizol. RNA samples were treated with DNase (TURBO DNA-free) and used in RT-PCR reaction.
Immunoprecipitation
Collected cells were washed with ice-cold PBS and lysed in IP-150 lysis buffer (20 mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM EDTA (pH8.0), and 0.5% NP-40) supplemented with protease and phosphase inhibitor cocktails. After incubating for 20 min, samples were spun at 13 000 r.p.m. for 15 min. The extracts were immunoprecipitated with an antibody and Protein G-Sepharose 4 Fast Flow (GE Healthcare, Fairfield, CT, USA) at 4 °C for overnight. Beads were washed with IP-150 lysis buffer for five times. The immunoprecipitates were resolved on a 12% SDS-PAGE and analyzed by western blot analysis for coimmunoprecipitation or proteins of interest.
Flow cytometry
Cells were washed twice in cold PBS and fixed with ice-cold 70% ethanol. Cells were stored at −20 °C in fixative for at least 2 h. Cells were centrifugated for 2 min at 5000 r.p.m., and ethanol was aspirated. The cell pellets were washed with PBS, resuspended in propidium iodide (PI)-staining solution (3 μg/ml PI, 0.5 mg/ml RNase A, and 0.25% Triton X-100 in 1 × PBS), and incubated at RT for 30 min before flow cytometric analysis system (Guava, Millipore).
Nuclear–cytoplasmic fraction
Nuclear–cytoplasm fraction was prepared as described.38 In brief, cells were washed in cold PBS and then resuspended in nuclear extract buffer A (20 mM HEPES, 10 mM KCl, 1 mM EDTA, 0.2% NP-40, 10% glycerol, 1 mM DTT, and protease inhibitor). After incubating for 5 min on ice, samples were spun at 2400 × g for 5 min. The supernatant was considered the cytoplasmic fraction. The pellet was resuspended in nuclear extract buffer B (20 mM HEPES, 10 mM KCl, 1 mM EDTA, 350 mM NaCl, 20% glycerol, 1 mM DTT, and protease inhibitor) and incubated for 10 min on ice. Sample was spun at 13 000 r.p.m. for 15 min, and the supernatant represents the soluble nuclear fraction.
Luciferase reporter assay
Cells seeded in 12-well plates were transiently cotransfected with the firefly luciferase reporter (50 ng), renilla luciferase reporter (5 ng), and siRNA (40 nM). Luciferase activity was determined with a dual-lucifersae assay system (Promega, Madison, WI, USA). Activity was determined using a Glomax 20/20 luminometer (Promega).
RNA fluorescence in situ hybridization
All RNA-FISH reactions were performed in RNase-free condition. Cells on cover slips were transfected with siRNA (40 nM) for 72 h, fixed with 4% paraformaldehyde in PBS at RT for 15 min, and permeabilized with 0.5% Triton X-100 in PBS at RT for 5 min. Pre-hybridizations were done with hybridization buffer (3% BSA, 100 μg/ml salmon sperm DNA (Stratagene) in 4 × SSC) at 42 °C for 1 h. Hybridizations were performed with Cy3-conjugated 5′-UTR and 3′-UTR FISH-probe mixture (200 nM) in hybridization buffer at 42 °C for overnight. Slides were washed with SSC at RT. For signal detection, primary antibody was treated for overnight at 4 °C, washed twice in PBS-A, briefly stained with Cy3-conjugated IgG (Jackson) and Hoechst 33258 (Sigma) for 1 h at RT in dark condition. Samples were washed twice in PBS supplemented with 0.02% sodium azide, and coverslips were mounted. Probe sequences are PDCD4 5′-UTR, 5′-Cy3-CCCTTCTCGCTCTGTTTGTTTT-3′; PDCD4 3′-UTR, 5′-Cy3-AGGAGTGGCAGTAGTCAGACATCAA-3′.
Abbreviations
- PDCD4:
-
programmed cell death 4
- SR:
-
serine/arginine(SR)-rich
- SRSF3:
-
serine/arginine-rich splicing factor 3
- PB:
-
P-body
- SG:
-
stress granule
- ActD:
-
actinomycin D
- CHX:
-
cycloheximide
- UTR:
-
untranslated region
- RNP-IP:
-
ribonucleoprotein-immunoprecipitation
- GAPDH:
-
glyceraldehyde 3-phosphate dehydrogenase
- B2M:
-
β-microglobulin
- IgG:
-
immunoglobulin G
References
Shepard PJ, Hertel KJ . The SR protein family. Genome Biol 2009; 10: 242.
Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD . SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 2009; 35: 1–10.
Manley JL, Krainer AR . A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev 2010; 24: 1073–1074.
Li X, Manley JL . Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 2005; 122: 365–378.
Zhang Z, Krainer AR . Involvement of SR proteins in mRNA surveillance. Mol Cell 2004; 16: 597–607.
Huang Y, Steitz JA . SRprises along a messenger's journey. Mol Cell 2005; 17: 613–615.
Huang Y, Gattoni R, Stevenin J, Steitz JA . SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell 2003; 11: 837–843.
Lemaire R, Prasad J, Kashima T, Gustafson J, Manley JL, Lafyatis R . Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev 2002; 16: 594–607.
Sanford JR, Gray NK, Beckmann K, Caceres JF . A novel role for shuttling SR proteins in mRNA translation. Genes Dev 2004; 18: 755–768.
Michlewski G, Sanford JR, Caceres JF . The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell 2008; 30: 179–189.
Caceres JF, Screaton GR, Krainer AR . A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev 1998; 12: 55–66.
Huang Y, Steitz JA . Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell 2001; 7: 899–905.
Bedard KM, Daijogo S, Semler BL . A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J 2007; 26: 459–467.
Shibahara K, Asano M, Ishida Y, Aoki T, Koike T, Honjo T . Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene 1995; 166: 297–301.
Onishi Y, Hashimoto S, Kizaki H . Cloning of the TIS gene suppressed by topoisomerase inhibitors. Gene 1998; 215: 453–459.
Lankat-Buttgereit B, Goke R . The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol Cell 2009; 101: 309–317.
Vikhreva PN, Shepelev MV, Korobko EV, Korobko IV . Pdcd4 tumor suppressor: properties, functions, and their application to oncology. Mol Gen Mikrobiol Virusol 2010; 2: 3–11.
Jia R, Li C, McCoy JP, Deng CX, Zheng ZM . SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci 2010; 6: 806–826.
Cmarik JL, Min H, Hegamyer G, Zhan S, Kulesz-Martin M, Yoshinaga H et al. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc Natl Acad Sci USA 1999; 96: 14037–14042.
Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M . S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006; 314: 467–471.
Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol 2003; 13: 161–167.
Keene JD . RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 2007; 8: 533–543.
Maniatis T, Reed R . An extensive network of coupling among gene expression machines. Nature 2002; 416: 499–506.
Delestienne N, Wauquier C, Soin R, Dierick JF, Gueydan C, Kruys V . The splicing factor ASF/SF2 is associated with TIA-1-related/TIA-1-containing ribonucleoproteic complexes and contributes to post-transcriptional repression of gene expression. FEBS J 2010; 277: 2496–2514.
Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P . A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 2008; 10: 1224–1231.
Yoon SP, Kim HH, Kim J, Park RY, Ohn T . Regulation of cellular RNA nano-particle assembly by splicing factor SRp20. J Nanosci Nanotechnol 2013; 13: 184–187.
Anderson P, Kedersha N . Stress granules: the Tao of RNA triage. Trends Biochem Sci 2008; 33: 141–150.
Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 2005; 169: 871–884.
Fischer DC, Noack K, Runnebaum IB, Watermann DO, Kieback DG, Stamm S et al. Expression of splicing factors in human ovarian cancer. Oncol Rep 2004; 11: 1085–1090.
Stickeler E, Kittrell F, Medina D, Berget SM . Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene 1999; 18: 3574–3582.
He X, Ee PL, Coon JS, Beck WT . Alternative splicing of the multidrug resistance protein 1/ATP binding cassette transporter subfamily gene in ovarian cancer creates functional splice variants and is associated with increased expression of the splicing factors PTB and SRp20. Clin Cancer Res 2004; 10: 4652–4660.
He X, Arslan AD, Pool MD, Ho TT, Darcy KM, Coon JS et al. Knockdown of splicing factor SRp20 causes apoptosis in ovarian cancer cells and its expression is associated with malignancy of epithelial ovarian cancer. Oncogene 2011; 30: 356–365.
Tang Y, Horikawa I, Ajiro M, Robles AI, Fujita K, Mondal AM et al. Downregulation of splicing factor SRSF3 induces p53beta, an alternatively spliced isoform of p53 that promotes cellular senescence. Oncogene 2012; 32: 2792–2798.
Piekielko-Witkowska A, Wiszomirska H, Wojcicka A, Poplawski P, Boguslawska J, Tanski Z et al. Disturbed expression of splicing factors in renal cancer affects alternative splicing of apoptosis regulators, oncogenes, and tumor suppressors. PLoS One 2010; 5: e13690.
Goncalves V, Matos P, Jordan P . The beta-catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression. RNA 2008; 14: 2538–2549.
Schadt EE, Li C, Ellis B, Wong WH . Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem Suppl 2001; 37: 120–125.
Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA . Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo . Methods 2002; 26: 182–190.
Kim J, Cantor AB, Orkin SH, Wang J . Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat Protoc 2009; 4: 506–517.
Huang da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Acknowledgements
We thank Dr. Xiang-Dong Fu for critical comments on the manuscript. This research was supported by grants (NRF-2011-0013484 and NRF-2011-0029629) from the National Research Foundation of Korea (NRF) to TB and NRF grants (2013-003436 and 2013-003868) and National R&D Program for Cancer Control grant from Ministry for Health and Welfare, Republic of Korea (1320270) to SJ.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Willis
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, J., Park, R., Chen, JK. et al. Splicing factor SRSF3 represses the translation of programmed cell death 4 mRNA by associating with the 5′-UTR region. Cell Death Differ 21, 481–490 (2014). https://doi.org/10.1038/cdd.2013.171
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2013.171
Keywords
This article is cited by
-
Circular RNA hsa_circ_0050386 suppresses non-small cell lung cancer progression via regulating the SRSF3/FN1 axis
Journal of Translational Medicine (2024)
-
A dual role of RBM42 in modulating splicing and translation of CDKN1A/p21 during DNA damage response
Nature Communications (2023)
-
Splicing factor SRSF3 represses translation of p21cip1/waf1 mRNA
Cell Death & Disease (2022)
-
Analysis of Pelagia noctiluca proteome Reveals a Red Fluorescent Protein, a Zinc Metalloproteinase and a Peroxiredoxin
The Protein Journal (2017)
-
NEDDylation promotes stress granule assembly
Nature Communications (2016)