Abstract
Rhabdomyosarcoma (RMS) is a paediatric soft-tissue sarcoma arising from skeletal muscle precursors coexpressing markers of proliferation and differentiation. Inducers of myogenic differentiation suppress RMS tumourigenic phenotype. The Notch target gene HES1 is upregulated in RMS and prevents tumour cell differentiation in a Notch-dependent manner. However, Notch receptors regulating this phenomenon are unknown. In agreement with data in RMS primary tumours, we show here that the Notch3 receptor is overexpressed in RMS cell lines versus normal myoblasts. Notch3-targeted downregulation in RMS cells induces hyper-phosphorylation of p38 and Akt essential for myogenesis, resulting in the differentiation of tumour cells into multinucleated myotubes expressing Myosin Heavy Chain. These phenomena are associated to a marked decrease in HES1 expression, an increase in p21Cip1 level and the accumulation of RMS cells in the G1 phase. HES1-forced overexpression in RMS cells reverses, at least in part, the pro-differentiative effects of Notch3 downregulation. Notch3 depletion also reduces the tumourigenic potential of RMS cells both in vitro and in vivo. These results indicate that downregulation of Notch3 is sufficient to force RMS cells into completing a correct full myogenic program providing evidence that it contributes, partially through HES1 sustained expression, to their malignant phenotype. Moreover, they suggest Notch3 as a novel potential target in human RMS.
Similar content being viewed by others
Main
Rhabdomyosarcoma (RMS) is a skeletal muscle-derived soft-tissue sarcoma affecting children and adolescents. It accounts for approximately 50% of all paediatric soft-tissue sarcomas, and for 7–8% of all childhood malignancies.1 Paediatric RMS includes two major histological subtypes, embryonal and alveolar. Approximately 70% of alveolar RMS harbour chromosomal translocations such as t(2;13) or t(1;13), resulting in PAX3-FKHR or PAX7-FKHR fusion products, and were considered the most aggressive forms. By contrast, recent studies demonstrated that fusion-negative alveolar tumours were molecularly and clinically indistinguishable from embryonal ones.2 Despite aggressive treatment, the management of paediatric RMS, especially the fusion-positive alveolar form, often metastatic at diagnosis and resistant to conventional therapy, remains challenging.
Similar to myogenic progenitors, RMS cells express muscle-differentiating factors but have lost the ability to terminally differentiate thus proliferating indefinitely.3 Promoting redifferentiation of RMS to its original skeletal muscle tissue type is a recognized strategy to suppress transformed phenotype.4, 5 In this view, gaining of knowledge on pathways involved in physiological myogenesis and that are found deregulated in RMS is pivotal for the identification of selected factors sustaining the pro-tumoural phenotype.
Notch proteins have fundamental roles in balancing proliferation versus differentiation.6 Mammals harbour four Notch genes, each encoding a type I transmembrane Notch receptor paralog (Notch1–4). Notch activation begins with the binding of the extracellular domain of transmembrane ligands of the Delta and Jagged family to Notch receptors on neighbouring cells. This interaction allows Notch to undergo sequential proteolytic cleavages, the last one being mediated by a γ-secretase complex that releases an active Notch intracellular domain (NotchIC). NotchIC translocates into the nucleus, where it activates the expression of target genes together with the DNA-binding RBP-Jk protein (also known as CSL, for CBF1/Su(H)/Lag1).7 Among Notch target genes are those encoding the enhancer of split group of transcriptional repressors, which are termed hairy and enhancer of split (HES) 1–7 and HES-related repressor (HEY) 1,2 and L in mammals.8
Notch activation in muscle progenitors impairs the transcription of myogenic factors, preventing cell differentiation and promoting proliferation.9 Consistently, inhibition of γ-secretase activity leads to myoblasts differentiation.10, 11, 12 Recently, Sang et al.13 have shown that HES1 is upregulated in primary RMS and its inhibition, as well as the blockade of γ-secretase activity, induces cell cycle arrest and differentiation of RMS cell lines. Combined with the fact that Notch signalling modulates critical aspects of cancer progression in many types of tumours,14 these data suggested that aberrant Notch signalling could underlie the development of RMS. However, the mechanism responsible for the anti-differentiative function of HES1 in RMS remained to be clarified. Here we identify the Notch3 receptor as a major player in this process. We report that, in agreement with data on RMS primary tumours,15 Notch3 is overexpressed and activated in RMS cell lines compared with their normal counterpart. Moreover, downregulation of Notch3 in RMS cells induces inhibition of G1-to-S cell cycle progression resulting in skeletal muscle-like differentiation and the expression of myogenic markers such as Myosin Heavy Chain (MHC) and troponin. These processes follow a marked increase of p38 MAPK (mitogen-activated protein kinase) phosphorylation and cyclin-dependent kinase (CDK) inhibitor p21Cip1 expression. HES1 is an important downstream effector of these phenomena as (i) HES1 level is noticeably decreased after Notch3 silencing and (ii) HES1-forced overexpression partly overcomes Notch3 depletion-associated phenotype. Finally, Notch3 silencing in RMS cells compromises their tumourigenic ability in vitro and tumour growth in vivo. Taken together, these results provide evidence for a pro-tumourigenic role of Notch3 signalling pathway in RMS.
Results
Notch signalling components are transcriptionally deregulated in human RMS cell lines and primary tumours
Two PAX3-FKHR fusion-positive alveolar (RH30 and RH41) and three fusion-negative embryonal (RD, RH18 and A204) human RMS cell lines were characterized for the expression and activation of Notch components and compared with healthy primary human skeletal myoblasts (SkMC, control cells).
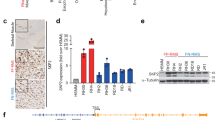
As reported in Figure 1a, all tumour cell lines had increased levels of both Notch1 and Notch3 activated forms , Notch1IC and Notch3IC, whereas Notch2IC levels were lower than those of SkMC cells. The activation of Notch signalling was confirmed by the detection of NotchIC forms of all three receptors in nuclear-enriched fractions (Figure 1b). A specific anti-Notch1IC antibody was used. Moreover, Supplementary Figure 1 identifies Notch3IC and Notch2IC bands as decreased in nuclear-enriched extracts from cell treated with an inhibitor of γ-secretase (i.e., DAPT). In RD cells, both Notch3IC and Notch2IC were detected in cytoplasmic fractions as well, suggesting cell type-specific regulation of Notch signalling/trafficking.16
Upregulation of Notch signaling components in rhabdomyosarcoma (RMS) cell lines. (a, left) Western blotting of Notch1-3 intracellular cleaved domains (IC), Jagged1, HES1 and β-actin (loading control) in whole-cell lysates from embryonal (ERMS) and alveolar (ARMS) RMS cell lines and normal human myoblasts SkMCs as control. Arrows indicate the IC domains of Notch1-3. Notch1IC was detected using the antibody that recognizes only the activated form (cleaved at Val1744). (a, right) Histograms report densitometric analysis of Notch1IC (Val1744), Notch2IC, Notch3IC, HES1, and Jagged1 bands normalized to β-actin of three independent experiments. (b) Western blot analysis of nuclear (N) and cytoplasmic (C) -enriched cell fractions of embryonal (ERMS) and alveolar (ARMS) RMS cell lines. Notch1IC (Val1744), Notch2IC and Notch3IC forms were detected in all cell lines. β-actin and topoisomerase IIβ were used as loading controls to discriminate the different cell fractions. (c) mRNA levels of Notch1-3, HES1 and Jagged1 (real time RT-PCR) were normalized to β-actin levels and expressed as fold increase over control SkMC (black column; 1 arbitrary unit). Columns, means; Bars, S.D. Results from three independent experiments are shown
Consistently, Notch1 and Notch3 were transcriptionally upregulated in all tumour cell lines whereas Notch2 mRNA level was only modestly increased in three tumour cell lines, that is, embryonal RH18 and A204 and alveolar RH41 (Figure 1c). In agreement with Sang et al.,13 the Notch target gene HES1 was significantly upregulated in all RMS cell lines. Notch ligand Jagged1 was overexpressed with different extents (Figures 1a and b) whereas Delta1 and Notch4IC were under the threshold of detection in both control and RMS cells (data not shown). These findings suggest that Notch1 and Notch3 signalling could be deregulated in RMS cell lines, irrespective of their subtype.
Notch3 silencing induces myogenic differentiation of RMS cells
Murine and human myoblasts have been shown to lose the ability to undergo skeletal muscle differentiation when Notch3 is activated; conversely Notch3 downregulation preserves this differentiation potential.17 Therefore, we focus on Notch3 to evaluate whether it could be involved in the inability of RMS cells to terminally differentiate. To this end, Notch3 was silenced and in vitro RMS cell differentiation was monitored. As shown in Figures 2a, b and c, numerous multinucleated fibres were detected in Notch3 siRNA-transfected RD and RH30 cell lines along with de novo expression of MHC and troponin, consistent with myotube fusion. This effect was associated with an increase of the muscle marker Myogenin in both cell lines (Figure 2d, right panel). Moreover, Notch3 silencing resulted in enhanced phosphorylation of p38 MAPK and serine-threonine kinase Akt, both essential for terminal muscle differentiation18, 19, 20 (Figure 2d, right panel). Consistently, also the Akt downstream target serine-threonine kinase mTOR (mammalian target of Rapamycin) was hyper-phosphorylated.20 Notch3 siRNA efficiency was attested by drastic decrease of mRNA levels (data not shown) and the reduced Notch3IC and HES1 protein levels in both cell lines (Figure 2d). Silencing Jagged1 by siRNA lowered Notch3IC and HES1 protein levels, especially in RD cells, suggesting that Notch3 activation is due, at least in part, to the binding with this ligand (Figure 2e).
Notch3 downregulation promotes RMS cell differentiation. (a) RD and RH30 cells cultured in complete medium (i.e., supplemented with 10% of fetal calf serum) were analyzed after 6 days of Notch3 or control (CTR) siRNA treatment. Representative immunofluorescence showing de novo expression of endogenous myosin heavy chain (MHC, green) in multinucleated fibers of Notch3 siRNA-transfected RD and RH30 cells (white arrows). Representative of three assays. (b) Western blotting showing de novo expression of Troponin I and β-actin (loading control) in Notch3 siRNA RD and RH30 cells treated as in (a). (c) Representative light microscopy pictures of RD and RH30 cells showing elongated multinucleated structures in Notch3 siRNA-treated cells treated as in (a). (d) Western blotting showing levels of Notch3IC and HES1 (left) and Myogenin along with the phosphorylation of p38, Akt and mTOR (right) in RD and RH30 cells 24 and 48 h after CTR or Notch3 siRNA transfection. β-actin was the loading control. Representative of three independent experiments. (e) Western blotting showing levels of Notch3IC and HES1 in RD and RH30 cells 24 and 48 h after CTR or Jagged1 siRNA transfection. β-actin was the loading control. Representative of three independent experiments
Despite the fact that Notch1 has not been reported significantly overexpressed in RMS primary tumours with respect to muscle tissue,15 it appeared activated in RMS cell lines (Figures 1a and b). To clarify its role in RMS cells differentiation, we knocked down Notch1 expression and monitored the induction of myogenesis in RD and RH30 cells. Forty-eight hours post-Notch1 siRNA, we detected an increase in Myogenin expression in RD cells (Supplementary Figure 2a, upper panel). This resulted in the formation of muscle-like multinucleated myotubes expressing MHC, albeit more modestly if compared with that obtained after Notch3 downregulation (Supplementary Figures 2b and c). Differently, neither increase in Myogenin levels nor myotube-like formation could be evidenced in Notch1 siRNA-transfected RH30 cells. Interestingly, a marked downregulation of Myogenin levels was detected up to 48 h after Notch2 silencing, ruling out the possibility of Notch2 involvement in the inhibition of myogenic program in RMS (Supplementary Figure 2a, lower panel). This finding is consistent with our observation that Notch2 is not markedly more activated/expressed in RMS tumour cells as compared with normal skeletal myoblasts (Figures 1a and c). Notably, protein and transcript levels of HES1 did not show any decrease after Notch1 siRNA transfection (Supplementary Figure 2a, upper panel and data not shown). In Notch2 siRNA cells, HES1 protein levels were slightly increased with no significant changes in mRNA in both cell lines as compared with control siRNA-treated cells (Supplementary Figure 2a, lower panel and data not shown). Therefore, a differential regulation of HES1 by different Notch paralogs is present in RMS.
Altogether, these results suggest that Notch3 expression/activation is able to prevent the differentiation of both alveolar and embryonal RMS cells.
Downregulation of Notch3 reduces RMS cell proliferation and soft-agar colony formation
To evaluate the impact of Notch3 siRNA-induced differentiation on cell growth rate, cell proliferation and cell cycle were analyzed. Notch3 knockdown markedly decreased cell growth in both RD and RH30 cells (by almost 50% as early as 3 days after transfection when compared with control siRNA) (Figure 3a). Similar results were obtained in embryonal A204 and alveolar RH41 cell lines (Supplementary Figure 3). We also observed that the ability of Notch3 siRNA-treated cells to form colonies in soft-agar was significantly impaired (Figure 3b). Interestingly, forcing Notch3 activation by ectopic expression of a Notch3IC in both RD and RH30 cell lines led to opposite molecular and functional results (data not shown). These findings strongly indicate that Notch3 expression promotes proliferation and supports tumourigenic features of RMS cells in vitro.
Notch3 depletion reduces proliferation and impairs RMS cell lines tumourigenic features in vitro. (a) RD and RH30 cells were transfected (t0) with Notch3 siRNA or control (CTR) siRNA, cultured in complete medium (i.e. supplemented with 10% of fetal calf serum) and harvested and counted at the indicated time points. *P<0.05; Bars, SD. (b) 2 × 104 RD living cells (evidenced by trypan blue exclusion) were seeded on soft-agar in 35 mm petri dishes 48 h after transfection with CTR or Notch3 siRNA. Histogram represents the mean number of colonies per field counted under a light inverted microscope 4 weeks after seeding (two independent experiments in triplicate). At least five fields per dish were checked at 100 × magnification. Bars, S.D. **P<0.001
Consistently, cell cycle analysis showed that after Notch3 knockdown, about 80% of RMS cells were in G1 phase compared with only about 45% of control siRNA cells (Figure 4a). Also the percentage of cells in S phase was reduced from about 45% to about 10% in both cell lines. In line with this, Notch3 silencing triggered a marked increase of p21Cip1 as early as 24 h after treatment (Figure 4b). p21Cip1 mRNA level was not significantly changed within 2 days from Notch3 depletion, ruling out early transcriptional effects (data not shown). As shown in Figure 4c, Notch3 silencing also caused an increase of the under-phosphorylated (active) form of the retinoblastoma tumour suppressor protein (pRb) and of the phosphatase and tensin homolog deleted on chromosome ten (PTEN). Consistent with a slowing down of cell proliferation, Notch3 siRNA also decreased extracellular signalling-regulated kinases ERK1/2 phosphorylation in both cell lines compared with control siRNA. Collectively, these results suggest that Notch3 stimulates cell cycle progression by promoting the G1-to-S phase transition, and that it regulates the ERK pathway in RMS cells.
Notch3 downregulation prevents the G1-to-S phase transition in RMS cell lines. (a) RD and RH30 cells were transfected with Notch3 siRNA or control (CTR) siRNA and, 48 h later, stained with 5-ethynyl-2′-deoxyuridine (EdU)-Alexa 488/Cell cycle 633 for analysis by flow cytometry gating on living cells. Left, representative diagrams. Right, the histogram depicts the percentage of control (CTR) or Notch3 siRNA transfected RD and RH30 cells in G1, S and G2 phases. Representative of three independent experiments in duplicate. (b and c) Western blotting showing expression of p21Cip1, PTEN and the Ser380 phophorylated form of PTEN, and pRb along with the phosphorylation of ERK1/2 in whole-cell extracts after CTR or Notch3 siRNA transfection. Arrows (c) point to bands corresponding to hyper- (upper) and hypo-phosphorylated (lower) pRb species. β-actin was the loading control
HES1 has a role in Notch3 signalling in RMS cells
In the attempt to evaluate whether the function of Notch3 in RMS is mediated, at least in part, by HES1, we performed loss- and gain-of-function experiments either silencing or forcing the expression of HES1 in RMS cells. HES1 downregulation by siRNAs in RD cells led to an enhancement of p21Cip1 levels 48 h after siRNA transfection (Figure 5a). This happened although HES1 protein levels were modestly decreased 48 h after siRNAs treatment when transcript reduction reached about 60 and 70% in RD and RH30 cells, respectively, compared with control siRNA cells (data not shown). HES1 downregulation did not induce any increase in p21Cip1 and Myogenin transcript levels up to 48 h after knocking down. In RH30 cells, these molecular modulations were undetectable. However, after 6 days of HES1 knocking down, multinucleated fibres positive for MHC were seen in both cell lines compared with control siRNA cells (Figure 5b). Interestingly, reduction of HES1 levels in Notch3-depleted cells reinforced all these phenomena suggesting that HES1 cooperates with Notch3 signalling to regulate them in RMS.
HES1 downregulation in RMS cells induces cell differentiation mimicking the effect of Notch3 knockdown. RD and RH30 cells were transfected with a Notch3 or control (CTR) siRNA, and 24 h later they were transfected with a HES1 or control (CTR) siRNA* in complete medium (i.e., supplemented with 10% of fetal calf serum). (a) Western blotting showing expression of p21Cip1 and Myogenin 48 h after HES1 silencing in whole-cell extracts. (b) Immunofluorescence analysis shows de novo expression of endogenous myosin heavy chain (MHC, red) in multinucleated fibers of Notch3, HES1 and Notch3 plus HES1 siRNA-transfected RD and RH30 cells after 6 days. Representative images of three assays
Then, we infected RMS cells with an adenovirus expressing both human HES1 and the GFP protein (AdHES1) 24 h after Notch3 silencing (see Supplementary Figure 3 for infection efficiency detection). A vector expressing only GFP (AdGFP) was the control vector. As shown in Figure 6a, p21Cip1 and Myogenin levels were downregulated 48 h after overexpression of HES1 in both Notch3 and control siRNA-treated cell lines compared with their AdGFP-infected counterparts. However, in Notch3 knocked down cells levels of both proteins remained higher than in control siRNA cells suggesting that (i) the efficiency of HES1 overexpression is not sufficient to completely block these molecular modulations and/or (ii) additional Notch3-dependent pathways collaborate with HES1.
HES1 overexpression in RMS cells abrogates the effects of Notch3 knockdown. RD and RH30 cells were infected with an Adenovirus expressing HES1 (AdHES1) or a control Adenovirus (Ad-GFP) 24 h after transfection with CTR or Notch3 siRNAs. (a) Western blotting showing levels of Notch3IC, HES1, Myogenin and p21Cip1 24 h and 48 h after Adenovirus infection. β-actin and α-tubulin were the loading controls. Representative of three independent experiments. (b) Cell cycle analysis by propidium iodide of RD and RH30 cells 48 h after adenovirus infection with AdHES1 and AdGFP. Left, representative diagrams. Right, histograms depict the fold change of the cell percentage in the different cell cycle phases for AdHES1 versus AdGFP infected RD and RH30 cells, after normalization for the efficiency of infection (Supplementary Figure 3). (c) Immunofluorescence analysis of AdHES1- and AdGFP-infected RD and RH30 cells (GFP, green) with anti-myosin heavy chain (MHC, red) antibody 6 days after infection. Representative images of three independent experiments
However, consistent with these results, after normalization for the percentage of GFP-positive RD and RH30 cell populations, cell cycle analysis showed an about 10 and 20% increase of the percentage of HES1-overexpressing cells (AdHES1) in S phase and 22 and 35% in G2 phase compared with AdGFP control cells, respectively (Figure 6b, both panels and Supplementary Figure 4).
Notably, RD cells overexpressing HES1 did not differentiate 6 days after infection (i.e., 7 days after siRNA treatment), as testified by the mutual exclusion of GFP and MHC expression (Figure 6c). Instead, a small number of Notch3 siRNA RH30 cells overexpressing HES1 are still able to differentiate even if they formed thinner myotubes compared with AdGFP control ones.
These data indicate that HES1 knockdown partially mimics Notch3 downregulation in RMS cell lines and, coherently, HES1 forced expression partially counteracts Notch3-depletion effects. Therefore, they are supportive for a role of HES1 in the maintenance of Notch3-dependent pro-tumour effects in RMS.
Short-hairpin (sh)RNA-mediated Notch3 reduction decreases RMS tumour growth rate in vivo
In the attempt to evaluate whether Notch3 downregulation affects tumour growth in vivo, RH30 cells were transfected with a GFP-expressing plasmid encoding an shRNA against Notch3. In vitro, a 3 weeks transfection with such Notch3 shRNA resulted in the formation of persistent RH30 multinucleated GFP-positive cells (Figure 7A, left), where Notch3 activation was strongly reduced, as assessed in lysates from cells sorted according to GFP expression (Figure 7A, right). In addition, a marked decrease in the proliferation rate of Notch3 shRNA cells compared with control shRNA ones was observed (data not shown). Then, nude mice were inoculated with suspensions of RH30 cells containing a sub-population (∼40%) of cells transiently transfected with Notch3 shRNA (Supplementary Figure 5). The reason for using a mixture of cells is that in preliminary experiments, when we inoculated mice with 100% Notch3 shRNA-expressing cells no xenograft growth was observed within 2 months from the date of injection (data not shown), thus no tumour growth was measurable. Analysis of xenografts from cell suspensions containing Notch3 shRNA (40%)/wild-type (wt) cells ∼12 days after tumours became palpable, that is, within the linear growth phase, showed significant reduction of tumour volume as compared with that obtained by inoculation of control shRNA (∼50%)/wt cell suspensions (Figure 7B). GFP-positive cells and cells expressing the proliferative marker Ki67 were reduced in xenografts derived from Notch3 shRNA injected mice compared with their control shRNA ones (Figures 7C and D). Western blotting analysis demonstrated reduction of Notch3 expression in Notch3 shRNA/wt compared with control shRNA/wt pooled xenograft samples (Figure 7E).
Notch3 downregulation in RMS cells reduces tumour growth in vivo. (A) RH-30 cells were transfected with a Notch3 short hairpin (sh)RNA- or control (CTR) shRNA-GFP-plasmids and images of GFP fluorescence (grey) were acquired after 3 weeks of puromycin selection (Left). Arrows depict nuclei of newly formed myofibers (200 × magnification). (Right) 48 h post-shRNAs transfection, positive GFP was separated from negative GFP cells by cell sorting and expression of Notch3IC, and β-actin (loading control) were analyzed by western blotting. Representative of two independent experiments. (B, Left) Mouse-bearing CTR shRNA (as a mixture of control (CTR) shRNA-GFP/wild-type cells) and Notch3 shRNA (as a mixture of Notch3 shRNA-GFP/wild-type cells) tumour xenografts (black arrows; left flank: a and right flank: f, respectively). (Middle) xenografts from nude mice injected with Notch3 shRNA RH30 cells (wild-type/Notch3 shRNA cell ratio ∼60%/40%; f, g, h, i and l). Xenografts from CTR shRNA cells (wild-type/CTR shRNA cell ratio ∼50%/50%; a, b, c, d and e) were controls. (Right) Histogram reports tumour volumes of each xenograft. (C, Left) Haematoxylin/eosin staining and immunolabeling of Ki67 and GFP of 5 μm serial sections from xenografts of mice injected with CTR shRNA and Notch3 shRNA RH30 cells (from d and i samples) (400 × magnification). GFP right panels are a higher magnification of left panels (600 × magnification). Representative of three xenografts per condition. (D) Histogram depicts the average of the percentage of GFP-positive cells per field in five fields per tumour section. Bars, S.D. *P<0.05. (E) The expression of Notch3IC was analysed by western blotting in lysates of samples from mice injected with CTR shRNA and Notch3 shRNA RH30 cell suspensions (three xenografts were pooled per group)
In summary, we have shown that Notch3 knockdown induces myogenic differentiation and decreases tumourigenicity of RMS cells in vitro and in vivo, and that Notch3 controls cell cycle progression and the ERK and Akt pathways in RMS cells. Moreover, we have provided evidence that Notch3 effects in RMS cells are mediated, at least in part, by HES1. Our data indicate that Notch3 is pivotal to the choice between proliferation and differentiation in RMS cells.
Discussion
In this study, we investigated the involvement of Notch signalling in the inability of RMS cells to complete the myogenic program. Consistently with data on primary RMS tumours,15 we showed that Notch3 is overexpressed in RMS cell lines and abnormally activated in a ligand-dependent manner. We also provided evidences that there is a causal relationship between Notch3 expression and the tumourigenic properties of RMS cells in vitro, as Notch3 silencing is sufficient in restoring myogenesis and inhibiting cell cycle progression and anchorage-independent growth. Moreover, lowering Notch3 expression, even only in a fraction of cells, decreased tumour growth in vivo.
All these observations are consistent with recent studies demonstrating that (i) Notch3 is highly expressed in the most immature subpopulation of satellite stem cells that gives rise to skeletal muscle-committed precursors,21 (ii) the expression of Notch3 in satellite cells is mutually exclusive with that of early skeletal muscle markers such as MyoD and Myogenin,21, 22 (iii) the expression of Notch3 in proliferating myoblasts is lost during the induction of differentiation22 and, conversely, (iv) the induction of Notch3 expression in myoblasts correlates with the loss of skeletal muscle features blocking myogenesis.17
In vitro Notch3 knockdown resulted in both alveolar and embryonal cells in an early significant overexpression of the p21Cip1 protein, along with an increase of Myogenin and the hypo-phosphorylated form of pRb, as reported during physiological skeletal muscle differentiation.23, 24 Interestingly, alveolar cells showed lower induction of p21Cip1 and needed sustained levels of Notch3 silencing compared with embryonal ones. Consistent with the notorious context-dependence of Notch signalling, this finding is possibly related to high Notch1 and/or fusion proteins expression in alveolar RMS.25
Notch3 knockdown also increased phosphorylation of p38 MAPK, previously shown to be inhibited by Notch in myoblasts,26 Akt and Akt-target mTOR, which all cooperate to myogenesis.18, 19, 20, 23, 27 Although the difference between the molecular mechanisms underlying Akt/mTOR-dependent mitogenesis or myogenesis in RMS needs to be fully elucidated, these results provide insights on Notch-Akt/mTOR pathway cross-talk in this soft-tissue sarcoma suggesting potential therapeutic applications.28 We also noticed an increase in PTEN, reported to be regulated by Notch pathway in several cancer types,29, 30 and that might act as a negative feedback associated with prolonged Akt activation.31 However, the concomitant increase in the less active Ser380-phosphorylated PTEN form could control the PTEN-PI3K-Akt axis to avoid suppression of the myogenic process.32
Notably, Notch3 depletion was consistently associated with strong inhibition of HES1 expression, suggesting that Notch3 regulation of RMS cells differentiation and proliferation occurs, at least in part, through HES1. Indeed, (i) forcing the HES1 expression in Notch3 knocked down RMS cells partially overcame, whereas (ii) co-downregulation of Notch3 and HES1 facilitated Notch3-depletion effects. These observations demonstrate that HES1 lowering is necessary for the Notch3 silencing-dependent anti-proliferative and pro-myogenic effects, and support HES1 as a downstream effector of Notch3 signalling in RMS.
This feature is specific of the Notch3 paralog as neither the downregulation of Notch1, also overexpressed and activated in RMS cell lines compared with myoblasts, nor that of Notch2 resulted in HES1 protein level reduction.
Such differential effects of Notch receptors have been reported in several types of cancer and appear context-dependent. Among these, in breast cancer, medulloblastoma, multiple myeloma and mesothelioma, some Notch paralogs have an oncogenic role whereas others behave as tumour suppressors modulating differentially downstream targets.33 The mechanism regulating these differential functions is under intense investigation and could be related to a diverse transcriptional activity but also to the ability to bind the RBP-jk/CBF-1 complex and/or to displace corepressors from the transcriptional complex.14
Increasing evidence suggests that oncogenesis in RMS involves multistage dysregulation of several signalling proteins.34 It is likely that other pathways cross-talk with Notch3 during RMS development. The cross-talk of different Notch-receptors pathways in tumours has been previously reported.35 In this regard, we cannot exclude that Notch1 cooperates with Notch3, at least in the embryonal cells. However, recent data showed no significant differences in the expression of Notch1 mRNA levels in primary RMS tumours compared with normal muscle tissues,15 suggesting that this Notch paralog could have a minor role in RMS pathogenesis. In this regard, it is conceivable that Notch1 upregulation in RMS cell lines could be ascribed to the molecular features of tumour tissues from which these cells derive. On the basis of this possibility, in RMS tumours with high Notch1 expression/activation, Notch1 and Notch3 signalling pathways could cooperate to sustain the tumourigenic phenotype.
Strikingly, Myogenin expression was severely impaired after Notch2 downregulation in RMS cell lines. This observation is consistent with findings showing similar Notch2 mRNA and activation levels in RMS cell lines as compared with normal myoblasts, and with the evidence that Notch receptors can have differential and even opposite roles in the same type of cancer.30 Future studies could dissect the specific role of each Notch paralog in RMS and Notch3-dependent direct and indirect effects.
Accordingly to the in vitro results, after injection of tumour cells (i.e., 2 × 106 cells per injection), we noticed that xenografts formed by the cell population including Notch3 shRNA cells grew more slowly compared with control ones. Moreover, the lower percentage of GFP-positive cells found in Notch3 shRNA/wt xenografts suggests that they resulted mostly from wild-type untrasfected cells with respect to control shRNA/wt ones. The evidence that xenografts containing Notch3-depleted population showed GFP cells that remain viable could be explained by the polyclonal character of the injected cell population, in which the level of Notch3 knockdown was heterogeneous and, possibly, in certain cells could not be sufficient to block cell engraftment and proliferation. However, Notch3 downregulation even in a fraction of cells affects the in vivo growth of alveolar RMS cells that are highly resistant to treatments.
Altogether, these results offer new insights into the deregulation of the myogenic program in the pathological context of RMS. Our results appear also relevant under a clinical translational point of view, as increasing evidence point out on the identification of small molecules and inhibitors specific to each Notch receptor.36 Notch3-selective inhibition, as reported for a preclinical model of lung cancer,37 could be, therefore, evaluated as potential adjuvant strategy in the treatment of chemotherapy-resistant RMS. Notch3-targeted therapy could potentially reduce possible toxic effects of complete Notch signalling pathway blockade.37
In conclusion, our data identify Notch3 as an inducer of the dysregulated differentiative program in RMS providing evidence that its blockade restores myogenesis and prevents tumourigenic potential of tumour cells, thus suggesting novel therapeutic approaches.
Materials and Methods
Cell lines
RH30 (alveolar RMS, ARMS) and RD (embryonal RMS, ERMS) cell lines were obtained from American Type Culture Collection (Rockville, MD, USA). RH41 (ARMS), A204 and RH18 (ERMS) cell lines were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). Normal Human Skeletal Muscle cells (SkMC; myoblasts) were obtained from PromoCell (Heidelberg, Germany). Supplementary Methods report cell lines and culture details.
Western blotting
Western blotting was performed on whole-cell lysates as previously described.38 See Supplementary Methods for antibodies and additional details.
Nuclear fraction-enrichment
Cells were lysed in cytoplasm lysis buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 0.2 mM EDTA and 1 mM DTT), containing protease inhibitors, 0.5 mM phenylmethylsulfonylfluoride (PMSF) and 0.6% Nonidet P-40 (Sigma Chemical Co., St. Louis, MO, USA). Lysates were centrifuged at 10 000 r.p.m. for 10 min at 4 °C, and the supernatants (cytoplasmic fractions) were split into aliquots and rapidly frozen. The nuclear pellet was washed in buffer A without Nonidet P-40 and finally resuspended in nuclear lysis buffer B (20 mM HEPES pH 7.9, 0.4 M NaCl, 2 mM EDTA, 1 mM DTT), containing protease inhibitors and 1 mM PMSF (Sigma Chemical Co.). Samples were incubated on ice for 30 min and centrifuged at 13 000 r.p.m. for 10 min at 4 °C; the supernatants (nuclear fractions) were split into aliquots and rapidly frozen. The remaining pellets, containing DNA as well as proteins tightly associated with DNA, were washed in reticulocyte buffered buffer (RSB: 10 mM NaCl, 10 mM Tris-HCl pH 7.5 and 1.5 mM MgCl2) and finally resuspended in RSB and HCl 0.4N to obtain HCl-soluble proteins (Castiglia et al., Neurochem Res (1994); Cestelli et al., Cell Mol Neurobiol. 1992). The samples were incubated at least 1 h at 4 °C and centrifuged at 10 000 r.p.m. for 20 min at 4 °C. The acid-soluble proteins were recovered from the supernatants by precipitation with 10 volumes of acetone at −20 °C and centrifugation at 10 000 r.p.m. for 20 min at 4 °C. HCl-soluble proteins were finally resuspended in distilled water, split into aliquots and frozen.
Cleaved Notch1 (Val1744) was obtained from Cell Signaling (Beverly, MA, USA) and the anti-acetyl-Histone H3 was obtained from Millipore (Temecula, CA, USA). The other antibodies were already described in the revised version of the manuscript.
Real time RT-PCR
Total RNA was extracted using TRizol (Invitrogen, Carlsbad, CA, USA) and analyzed by real-time RT-PCR as described.39 See Supplementary Methods for primer sequences and details.
Transient RNA interference
Cells were transfected with double-stranded synthetic 21-mer RNA oligonucleotides (siRNA) against Notch3 (Hs01_00101285; Sigma Chemical Co.) or fluorescein-labeled non-targeting siRNA control (100 nM final concentration) using Oligofectamine (Invitrogen) according to manufacturer's recommendations. The efficacy of siRNA transfection was assessed by visualization of fluorescent cells under an Eclipse TE200 microscope (Nikon, Sesto Fiorentino, Firenze, Italia) and images were acquired through Metamorph software version 7.1 (Crisel, Roma, Italia). JAG1 silencing was performed at 100 nM (final concentration) using Silencer Select siRNA reagents (Applied biosystems, CA, USA/Ambion, TX, USA).
ON-TARGETplus SMART pool siRNA (Thermo Fisher Scientific, Lafayette, CO, USA), were used for downregulating HES1.
Immunofluorescence
Immunofluorescence for visualizing MHC was performed as previously described38 using the MF-20 antibody (Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA, USA). Briefly, cells were washed three times in PBS, fixed for 10 min in 4% PFA and permealized for 5 min with 0.2% Triton X-100 in PBS. After 30 min in PBS containing 3% bovine serum albumin, slides were incubated for 1 h at room temperature with the MF-20 antibody against myosin heavy chain (MHC; DSHB, USA). After two washing in PBS, cells were treated with a fluorescein-linked secondary antibody. After being stained for 2 min with 200 ng/ml DAPI in PBS, chamber slides were mounted in GelMount (Biomeda, Foster City, CA, USA). Images were acquired with an Eclipse E600 fluorescence microscope, through LUCIA software version 4.81 (Nikon, Sesto Fiorentino, Firenze, Italy).
Cell proliferation and cell cycle assay
Cells were transfected at 24 h after seeding (Day 0) with siRNAs, harvested and counted at the reported time points. For cell cycle assay, the 5-ethynyl-2′-deoxyuridine (EdU) incorporation was performed using Click-iT EdU Flow Cytometry assay Kit (Invitrogen), and cells were analyzed with flow cytometry using a FACSCantoII and a FACSDiva 6.1 software (BD Biosciences, San Josè, CA, USA). For adenoviruses infections, GFP-positive cells’ cell cycle was evaluated using propidium iodide staining and flow cytometry (FCM) analysis. Cells were harvested by trypsinization, washed in cold phosphate buffered saline (PBS) 1 × , fixed in 50% PBS and 50% acetone/methanol (1 : 4 v/v) for at least 1 h, and, after removing alcoholic fixative, stained with a solution containing 50 μg/ml PI and 100 μg/ml RNase (Sigma Chemical Co.) for 30 min at room temperature, in dark. Samples were then measured by using a FACScan cytofluorimeter. Twenty thousand events per sample were acquired.
Soft-agar colony formation assay
After 48 h of transfection with Notch3 siRNA or control siRNA, RMS cells were assayed for their capacity to form colonies in soft-agar as previously described.38 Briefly, a total of 2 × 104 or 3 × 104 cells were suspended in DMEM (10% FCS) containing 0.35% Noble agar (Sigma Chemical Co.). Cells were plated on a layer of 0.7% Noble agar in DMEM (10% FCS) onto a 35-mm petri dish. Medium was refreshed every 5 days. On week 4, the number of colonies per field was counted under the contrast-phase Eclipse E600 microscope (Nikon). Two independent experiments were carried out in triplicate.
Adenovirus infection
A recombinant, replication-deficient adenoviral CMV bicistronic vector expressing both human HES1 (AdHES1) and the green fluorescent protein (GFP) (a gift of GP Dotto) was used to infect RD and RH30 cells (100 plaque-form units per cell) as described.40 A vector expressing only the reporter gene GFP (AdGFP) (a gift of G Toietta) was used as control. The efficiency of infection was determined analysing the percentage of GFP-positive cells with flow cytometry using a FACSCantoII and a FACSDiva 6.1 software (BD Biosciences).
Prolonged RNA interference with shRNA and cell sorting
Short hairpin RNA (sh)RNA-mediated silencing was performed by transfecting cells with a pGFP-V-RS vector expressing 29 nucleotide shRNAs against Notch3 and a green fluorescent protein (GFP) as reporter gene (TG311139: Notch3 target sequence 5′-GTGAGAGCTGCAGAATATCGATGAC-3′; Origene, Rockville, MD, USA) using Lipofectamine 2000 (Invitrogen). A non-targeting shRNA was used as control. Notch3 silencing was assessed by western blotting after cell sorting and 48 h after transfection. Living cells identified by Forward and Side Scatters light were sorted as GFP-positive and GFP-negative populations by a FACSAriaII and with a FACS DIVA 6.1 software (BD Biosciences). After 3 weeks of selection with puromycin (1 μg/ml), images of fluorescent cells were acquired under an Eclipse TE200 fluorescence microscope (Nikon) through Metamorph software version 7.1 (Crisel, Roma, Italia).
Xenograft experiments
Athymic 6-week-old female BALB/c nude mice (nu+nu+) were purchased from Charles River (23885 Calco (Lecco), Italy). Procedures involving animals and their care were conformed to institutional guidelines that comply with national and international laws and policies (EEC Council Directive 86609, OJ L 358, 12 December 1987). RH30 cells transfected with Notch3 or CTR shRNA plasmids were transfected again 24 h later and allowed to grow for additional 24 h. The efficiency of transfection was determined by flow cytometry at this time. Cells (2 × 106) were injected subcutaneously into the posterior flanks of nude mice in a cold mixture of PBSMatrigel (ratio 1 : 1). At 2 weeks after the tumours became palpable, mice were sacrificed, xenografts were removed and tumour volume was measured as described.35 Western blotting on total lysates from RMS xenografts were performed by pulling three Notch3 or CTR shRNA-derived specimens from snap frozen tissues.
Immunohistochemistry of xenograft samples
Serial 5 μm sections from paraffin-embedded tumours and control tissues were deparaffinized and antigen retrieval was performed for 25 min at 98 °C with citrate pH6. After endogenous peroxidase blocking at room temperature (RT), 3–5% BSA or 5% goat serum in Tris-buffered saline was applied for non-specific site blocking. Additional blocking was performed using Biotin Blocking System (DAKO Carpintera, CA, USA). Sections were incubated with anti-GFP (Clontech, Mountain View, CA, USA), anti-Ki67 (Novocastra Laboratories Ltd, Newcastle upon Tyne, UK) antibodies and the appropriate secondary antibodies. Positive reactions were visualized by incubation with diaminobenzidine (DAB chromogen kit, DAKO Carpinteria, CA, USA). Negative controls were done by treating serial cross-sections simultaneously with isotype non-specific IgG or omitting the primary antibody. Section were then dehydrated and mounted in non-aqueous mounting medium. Levels of GFP expression were semi-quantitatively quantified at 400 × magnification by scoring the percentage of GFP-stained cells per field in at least five fields per section. Scoring was done by two blinded independent observers. Sections were scored using an Eclipse E600 microscope (Nikon). Images were acquired through LUCIA software, version 4.81 (Nikon) with a Nikon Digital Camera DXM1200F.
Statistical analysis
Continuous variables were analyzed by Mann-Whitney U-test or Kruskal-Wallis test for pairwise or multiple comparisons, respectively. Statistical significance was set at a two-tailed P-value <0.05. All analyses were performed with SPSS 11.5.1 for Windows Package (SPSS, Inc., 1989–2002 and LEADTOOLS 1991–2000, LEAD Technologies Inc., Berks, UK).
Abbreviations
- RMS:
-
rhabdomyosarcoma
- Notch1–4:
-
type I transmembrane Notch receptor paralog 1–4
- NotchIC:
-
Notch intracellular domain
- HES:
-
hairy and enhancer of split
- HEY:
-
HES-related repressor
- RBP-Jk:
-
recombination signal-binding protein for immunoglobulin kappa J region
- CBF1:
-
C promoter-binding factor
- CSL or CBF1/Su(H)/Lag1:
-
CBF1/suppressor of hairless/Lag-1
- SkMC:
-
skeletal muscle cells
- PAX3-FKHR:
-
paired box 3-FOXO1 forkhead box
- PAX7-FKHR:
-
paired box 7-FOXO1 forkhead box
- siRNA:
-
small interfering RNA
- (sh)RNA:
-
short-hairpin RNA
- IC:
-
intracellular cleaved domains
- mRNAs:
-
messenger RNAs
- ERK:
-
extracellular signaling regulated kinases
- Akt:
-
v-akt murine thymoma viral oncogene homolog
- MHC:
-
myosin heavy chain
- MAPK:
-
mitogen-activated protein kinase
- CDK:
-
cyclin-dependent kinase
- mTOR:
-
mammalian target of rapamycin
- PTEN:
-
phosphatase and tensin homolog deleted on chromosome ten
- pRB:
-
retinoblastoma
- PI3K:
-
phosphatidylinositol-3-kinase
- GFP:
-
green fluorescent protein
- MyoD:
-
myogenic differentiation 1
References
Loeb DM, Thornton K, Shokek O . Pediatric soft tissue sarcomas. Surg Clin North Am 2008; 88: 615–627 vii.
Williamson D, Missiaglia E, de Reynies A, Pierron G, Thuille B, Palenzuela G et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol 2010; 28: 2151–2158.
Tapscott SJ, Thayer MJ, Weintraub H . Deficiency in rhabdomyosarcomas of a factor required for MyoD activity and myogenesis. Science 1993; 259: 1450–1453.
Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 2008; 14: 369–381.
Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest 2009; 119: 2366–2378.
Artavanis-Tsakonas S, Rand MD, Lake RJ . Notch signaling: cell fate control and signal integration in development. Science 1999; 284: 770–776.
Kopan R . Notch: a membrane-bound transcription factor. J Cell Sci 2002; 115 (Pt 6): 1095–1097.
Iso T, Kedes L, Hamamori Y . HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 2003; 194: 237–255.
Conboy IM, Conboy MJ, Smythe GM, Rando TA . Notch-mediated restoration of regenerative potential to aged muscle. Science 2003; 302: 1575–1577.
Vasyutina E, Lenhard DC, Wende H, Erdmann B, Epstein JA, Birchmeier C . RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proc Natl Acad Sci USA 2007; 104: 4443–4448.
Kitzmann M, Bonnieu A, Duret C, Vernus B, Barro M, Laoudj-Chenivesse D et al. Inhibition of Notch signaling induces myotube hypertrophy by recruiting a subpopulation of reserve cells. J Cell Physiol 2006; 208: 538–548.
Dahlqvist C, Blokzijl A, Chapman G, Falk A, Dannaeus K, Ibanez CF et al. Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development 2003; 130: 6089–6099.
Sang L, Coller HA, Roberts JM . Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 2008; 321: 1095–1100.
Radtke F, Raj K . The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer 2003; 3: 756–767.
Roma J, Masia A, Reventos J, Sanchez de Toledo J, Gallego S . Notch pathway inhibition significantly reduces rhabdomyosarcoma invasiveness and mobility in vitro. Clin Cancer Res 2011; 17: 505–513.
Andersson ER, Sandberg R, Lendahl U . Notch signaling: simplicity in design, versatility in function. Development 2010; 138: 3593–3612.
Ono Y, Sensui H, Okutsu S, Nagatomi R . Notch2 negatively regulates myofibroblastic differentiation of myoblasts. J Cell Physiol 2007; 210: 358–369.
Wu Z, Woodring PJ, Bhakta KS, Tamura K, Wen F, Feramisco JR et al. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol Cell Biol 2000; 20: 3951–3964.
Wilson EM, Rotwein P . Selective control of skeletal muscle differentiation by Akt1. J Biol Chem 2007; 282: 5106–5110.
Park IH, Chen J . Mammalian target of rapamycin (mTOR) signaling is required for a late-stage fusion process during skeletal myotube maturation. J Biol Chem 2005; 280: 32009–32017.
Kuang S, Kuroda K, Le Grand F, Rudnicki MA . Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 2007; 129: 999–1010.
Kitamoto T, Hanaoka K . Notch3 null mutation in mice causes muscle hyperplasia by repetitive muscle regeneration. Stem Cells 2010; 28: 2205–2216.
Ciccarelli C, Marampon F, Scoglio A, Mauro A, Giacinti C, De Cesaris P et al. p21WAF1 expression induced by MEK/ERK pathway activation or inhibition correlates with growth arrest, myogenic differentiation and onco-phenotype reversal in rhabdomyosarcoma cells. Mol Cancer 2005; 4: 41.
Guo K, Wang J, Andres V, Smith RC, Walsh K . MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol Cell Biol 1995; 15: 3823–3829.
Hecker RM, Amstutz RA, Wachtel M, Walter D, Niggli FK, Schafer BW . p21 Downregulation is an important component of PAX3/FKHR oncogenicity and its reactivation by HDAC inhibitors enhances combination treatment. Oncogene 2010; 29: 3942–3952.
Kondoh K, Sunadome K, Nishida E . Notch signaling suppresses p38 MAPK activity via induction of MKP-1 in myogenesis. J Biol Chem 2007; 282: 3058–3065.
Puri PL, Wu Z, Zhang P, Wood LD, Bhakta KS, Han J et al. Induction of terminal differentiation by constitutive activation of p38 MAP kinase in human rhabdomyosarcoma cells. Genes Dev 2000; 14: 574–584.
Petricoin 3rd EF, Espina V, Araujo RP, Midura B, Yeung C, Wan X et al. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res 2007; 67: 3431–3440.
Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 2007; 13: 1203–1210.
Graziani I, Eliasz S, De Marco MA, Chen Y, Pass HI, De May RM et al. Opposite effects of Notch-1 and Notch-2 on mesothelioma cell survival under hypoxia are exerted through the Akt pathway. Cancer Res 2008; 68: 9678–9685.
Crackower MA, Oudit GY, Kozieradzki I, Sarao R, Sun H, Sasaki T et al. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell 2002; 110: 737–749.
Salmena L, Carracedo A, Pandolfi PP . Tenets of PTEN tumor suppression. Cell 2008; 133: 403–414.
Ranganathan P, Weaver KL, Capobianco AJ . Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer 2011; 11: 338–351.
Naini S, Etheridge KT, Adam SJ, Qualman SJ, Bentley RC, Counter CM et al. Defining the cooperative genetic changes that temporally drive alveolar rhabdomyosarcoma. Cancer Res 2008; 68: 9583–9588.
Rizzo P, Miao H, D’Souza G, Osipo C, Song LL, Yun J et al. Cross-talk between Notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res 2008; 68: 5226–5235.
Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y et al. Therapeutic antibody targeting of individual Notch receptors. Nature 2010; 464: 1052–1057.
Lin L, Mernaugh R, Yi F, Blum D, Carbone DP, Dang TP . Targeting specific regions of the Notch3 ligand-binding domain induces apoptosis and inhibits tumor growth in lung cancer. Cancer Res 2010; 70: 632–638.
Ciarapica R, Annibali D, Raimondi L, Savino M, Nasi S, Rota R . Targeting Id protein interactions by an engineered HLH domain induces human neuroblastoma cell differentiation. Oncogene 2009; 28: 1881–1891.
Clement N, Gueguen M, Glorian M, Blaise R, Andreani M, Brou C et al. Notch3 and IL-1beta exert opposing effects on a vascular smooth muscle cell inflammatory pathway in which NF-kappaB drives crosstalk. J Cell Sci 2007; 120 (Pt 19): 3352–3361.
Sriuranpong V, Borges MW, Ravi RK, Arnold DR, Nelkin BD, Baylin SB et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res 2001; 61: 3200–3205.
Acknowledgements
We thank E Giorda and L Cifaldi for FACS analysis and D Doreau for RT-PCR. We acknowledge GP Dotto and G Toietta for providing AdHES1 and AdGFP vectors, respectively. Notch1, Notch2 and Jagged1 (Artavanis-Tsakonas S), Myogenin (Wright WE) and MHC (Fishman DA) antibodies were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. SS is a Chercheur National of the Fonds de la Recherche en Santé du Quebec. Grant support: Italian Ministry of Health, AIRC (10338) and ISS (70BF/8) (R. Rota); FIRB-MIUR grant (G Cortese); Universitè Pierre et Marie Curie (I Limon); AIRC 5 per mille (FL).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by P Mehlen
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Raimondi, L., Ciarapica, R., De Salvo, M. et al. Inhibition of Notch3 signalling induces rhabdomyosarcoma cell differentiation promoting p38 phosphorylation and p21Cip1 expression and hampers tumour cell growth in vitro and in vivo. Cell Death Differ 19, 871–881 (2012). https://doi.org/10.1038/cdd.2011.171
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2011.171
Keywords
This article is cited by
-
Pharmacological targeting of the ephrin receptor kinase signalling by GLPG1790 in vitro and in vivo reverts oncophenotype, induces myogenic differentiation and radiosensitizes embryonal rhabdomyosarcoma cells
Journal of Hematology & Oncology (2017)
-
PAX7 is a required target for microRNA-206-induced differentiation of fusion-negative rhabdomyosarcoma
Cell Death & Disease (2016)
-
Targeting hedgehog signaling reduces self-renewal in embryonal rhabdomyosarcoma
Oncogene (2016)
-
MicroRNA-101 is repressed by EZH2 and its restoration inhibits tumorigenic features in embryonal rhabdomyosarcoma
Clinical Epigenetics (2015)
-
Pharmacological inhibition of EZH2 as a promising differentiation therapy in embryonal RMS
BMC Cancer (2014)