Abstract
Neuroblastoma (NB) is a common childhood malignant tumor of the neural crest-derived sympathetic nervous system. In NB the frequent loss of heterozygosity (LOH) on chromosome 1p raises the possibility that this region contains tumor-suppressor genes whose inactivation contributes to tumorigenesis. The human homolog of the Drosophila neural fate determination gene CASZ1, a zinc-finger transcription factor, maps to chromosome 1p36.22, a region implicated in NB tumorigenesis. Quantitative real-time PCR analysis showed that low-CASZ1 expression is significantly correlated with increased age (≥18 months), Children's Oncology Group high-risk classification, 1p LOH and MYCN amplification (all P<0.0002) and decreased survival probability (P=0.0009). CASZ1 was more highly expressed in NB with a differentiated histopathology (P<0.0001). Retinoids and epigenetic modification agents associated with regulation of differentiation induced CASZ1 expression. Expression profiling analysis revealed that CASZ1 regulates the expression of genes involved in regulation of cell growth and developmental processes. Specific restoration of CASZ1 in NB cells induced cell differentiation, enhanced cell adhesion, inhibited migration and suppressed tumorigenicity. These data are consistent with CASZ1 being a critical modulator of neural cell development, and that somatically acquired disruption of normal CASZ1 expression contributes to the malignant phenotype of human NB.
Similar content being viewed by others
Main
Neuroblastoma (NB) is the most common extra-cranial childhood solid tumor and derives from the neural crest cells that give rise to the developing sympathoadrenal nervous system.1 Deletion of chromosome 1p (chr1p) is found in approximately 35% of NB1 and frequently occurs in other cancers, such as melanoma, oligodendroglioma and breast cancers.2 Evidence indicates that the region between D1S508 and D1S244 markers on chr1p36 is one of the commonly deleted regions in NB.3 There are 30 genes in this region and expression analysis of 4 of the 30 genes showed that two of them (CASZ1 and PIK3CD) are expressed at significantly lower levels in unfavorable NB as compared with favorable ones.3 However, the tumor-suppressor functions of CASZ1 and PIK3CD have not been investigated.3
CASZ1 maps to Chr1p36.22 and is a recently described zinc-finger (ZnF) transcription factor whose expression is upregulated during induction of NB and myoblast cell differentiation.4 In Drosophila, the CASZ1 homolog, Drosophila castor (dcas), is expressed at a late stage of neuroblast development that precedes the cessation of proliferation and initiation of neuronal differentiation.5, 6, 7 Dcas functions to regulate neural fate and loss of dcas in Drosophila results in impaired differentiation and alterations in glial cell number and migration.5, 7, 8 In Xeneopus CASZ1 is required for heart development and onset of cardiomyocytes differentiation at the ventral midline; the CASZ1-depleted midline cells overproliferate and remain a coherent population of nonintegrated cells positioned on the outer wall of the ventricle.9 Although functional studies have not been performed in mammals to date, murine studies indicate developmental regulation of CASZ1 in the neural crest-derived peripheral nervous system10 cells, which are thought to be the origin of NB tumors.
NB patients whose tumors are undifferentiated have poor prognoses while those whose tumors exhibit a more differentiated histopathology or gene expression profile have less malignant tumors and better prognoses.1 The finding that re-introduction of Chr1p into NB cells induces differentiation11 supports the concept that loss of genes regulating developmental processes contributes to NB tumorigenicity. Developmentally regulated transcription factors are implicated in tumorigenesis in several pediatric tumors: MYCN in NB,1, 2, 3, 4 RB gene in retinoblastoma,12 and REST/NRSF in medulloblastoma.13 These transcription factors are also important regulators of signal transduction pathways that control developmental programs. Thus, we propose that the ZnF transcription factor CASZ1 is a candidate NB tumor-suppressor gene based on the following observations: first, CASZ1 localizes to chr1p36 and it is underexpressed in unfavorable NB patients;3 second, CASZ1 is required for neuronal differentiation in Drosophila and is a neural fate determination gene;5 third, CASZ1 expression is developmentally regulated during neurogenesis in mouse and is upregulated when NB cells are induced to differentiate.4, 10
In this study, we examine the expression of CASZ1 in primary tumors from NB patients and determine how the reconstitution of CASZ1 expression in NB tumor cells alters their biologic functions. We provide evidence that CASZ1 has properties associated with tumor-suppressor gene function as it induces cell differentiation, inhibits tumor cell migration and tumor growth in vitro and in vivo in murine xenograft models.
Results
Decreased CASZ1 expression is clinically relevant and significantly associated with poor prognosis in NB
Using fluorescence in situ hybridization, CASZ1 cDNA maps to chr1p36 and loss of a CASZ1 allele occurs in a number of NB cell lines and primary NB tumor specimens (Supplementary Figures 1a and b). CASZ1 deletion in each of the four NB tumor samples tested, as represented by the percentage of cells with CASZ1 deletion versus all cells counted vary from 60 to 88%. To investigate CASZ1 expression in NB cell lines with and without 1p loss of heterozygosity (LOH), we performed real-time PCR to assess CASZ1 mRNA level. We found that low-CASZ1 is not always associated with 1p LOH in the eight NB cell lines tested, as shown in Supplementary Figure 1c, CASZ1 level is low in SY5Y and NGP cells, which have no 1p LOH, suggesting that in addition to 1p LOH, epigenetic alterations may contribute to low expression of CASZ1 in NB cells with or without 1p LOH. We treated AS, BE2, SY5Y, KCNR and NGP NB cell lines, which have representative genetic backgrounds characteristic of NB tumors (Supplementary Table 1) with the demethylating agent 5-Aza-20-deoxycytidine (5-Aza-dC) or the clinically relevant class I histone deacetylase inhibitor, depsipeptide (rompidepsin) and examined changes in CASZ1 expression. Consistent with a previous report,3 we did not see the induction of CASZ1 by 5-Aza-dC in the AS or BE2 cell lines but 5-Aza-dC induced a three to sevenfold increase in CASZ1 gene expression in SY5Y, KCNR and NGP (Supplementary Figure 1d). Unlike 5-Aza-dC, depsipeptide-induced CASZ1 gene expression in all five NB cell lines tested from ∼2- to 20-fold (Supplementary Figure 1e). Thus, changes in DNA methylation or histone acetylation are associated with increases in CASZ1 expression in NB cells either with or without 1p LOH.
We evaluated CASZ1 mRNA expression in 59 primary NB patients’ tumors by TaqMan real-time PCR. All samples had detectable levels of CASZ1 mRNA. The proportion of patients by known risk factors was similar to those of the NB population in general (Supplementary Table 2). The median level of CASZ1 expression, 0.77 was selected as the optimal cut-off in this cohort (in terms of event-free survival, EFS) to categorize each sample as either a high-CASZ1 or a low-CASZ1 expresser. Although the sample size is small, there is statistical evidence to support any choice of CASZ1 expression cut-off between 0.77 and 1.5.
Low levels of CASZ1 were detected in the majority (77%) of tumor samples in patients ≥18 months, only 27% of the samples in patients <18 months samples expressed low-CASZ1 levels (P=0.0002, Table 1). The relative risk for an event in a patient ≥18 months expressing low-CASZ1 is 2.9-fold higher than that for a patient <18 months. Similarly, low-CASZ1 expression was statistically significantly associated with unfavorable Shimada histology, 1p LOH, no 11q LOH and MYCN amplification, and having a Children's Oncology Group (COG) high-risk profile14 (all P<0.004, Table 1). Although CASZ1 mRNA level is higher in stage 1/2/3/4S than in stage 4 disease, the difference is not statistically significant (P=0.0692, Table 1). NB patients whose tumors expressed low-CASZ1 generally had a poor overall survival rate (P=0.0009) (Figure 1a). Consistent with this finding, NB patients whose tumors express low-CASZ1 have a poor overall survival rate (P=0.0000027) was also found in NB Prognosis Database from Dr. Javed Khan’s lab (http://home.ccr.cancer.gov/oncology/oncogenomics/), which was based on microarray analysis using 56 patient tumor samples (Figure 1b). Similarly, in Dr. Rogier Versteeg’s R2 database (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi), in a study of 88 NB patients, those patients whose tumors express low-CASZ1 have a poor overall survival rate (P=0.0011) (Supplementary Figure 2).
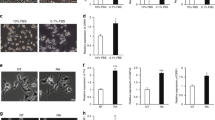
Loss of CASZ1 expression is correlated with poor prognosis in NB. (a) Kaplan–Meier curves of overall patient survival for low (<0.77) versus high (>0.77) CASZ1 expression in 59 NB patients. The association of high-CASZ1 expression with good prognosis is highly statistically significant (P=0.0009). CASZ1 mRNA level was determined on Taqman real-time PCR. (b) Kaplan–Meier curves of overall patient survival for low versus high-CASZ1 expression in 56 other NB patients (using median CASZ1 level as cutoff) from NB Prognosis Database (http://home.ccr.cancer.gov/oncology/oncogenomics/). The association of high-CASZ1 expression with good prognosis is highly statistically significant (P=0.00000207). CASZ1 mRNA level was determined on microarray
CASZ1 regulates the expression of genes that are important for cell growth and developmental processes
Retinoids are known to induce growth arrest and differentiation of NB tumor cells, and in the KCNR cell line treated with 5 μM all-trans retinoic acid (RA), CASZ1 expression increased within 6 h, peaking at 24 h (Figure 2a, top). At this time, the KCNR cells showed obvious morphological changes marked by the loss of cellular aggregation, increased cell adherence and neurite extension (Figure 2a, bottom). The induction of CASZ1 by RA suggests a role for CASZ1 in the regulation of NB cell differentiation.
Transcriptome regulated by CASZ1. (a) Northern analysis of CASZ1 expression in RA-induced KCNR differentiation. Top: the CASZ1 expression in KCNR after RA treatment at the indicated hours (shown as h), 18S RNA stained with ethidium bromide before transfer was used as loading control. Middle: the CASZ1 expression level as normalized to 18S RNA. The CASZ1 level in the control cells (shown as c) is defined as one unit. Bottom: the morphological differentiation of KCNR after RA treatment of indicated amount of time. (b) Full-length FLAG-tagged CASZ1 in Tet-on vector was stably transfected into SY5Y, NGP and LAN5 NB cell lines that express Tet repressor. The stable clones obtained were named SY5YtetCASZ1, NGPtetCASZ1 and LAN5tetCASZ1. Induction of CASZ1 expression by Tet (1 μg/ml) at 24 h was visualized by immunoblotting whole cell lysate with anti-FLAG antibody. (c) IPA of 125 common targets of CASZ1 in SY5YtetCASZ1 replicates, NGPtetCASZ1 and LAN5tetCASZ1 determined by microarray. Threshold represents P=0.05
As the transcriptome regulated by CASZ1 has not been investigated in any species to date, we evaluated the global transcriptome regulated by CASZ1 to gain insight into potential targets and mechanisms of action. We used Agilent whole human genome oligo microarray analyses (G4112F, Agilent, Santa Clara, CA, USA) to investigate the genome-wide transcriptional consequences of expression of CASZ1 in three NB cell lines in which CASZ1 gene expression is regulated by tetracycline (Tet) (SY5YtetCASZ1, NGPtetCASZ1 and LAN5tetCASZ1). Tet-induced FLAG-CASZ1 expression was visualized by immunoblotting (Figure 2b) and immunocytochemical staining revealed that CASZ1 predominantly localizes in the nucleus within 12 h of Tet treatment (Supplementary Figure 3). After Tet-induction (24 h) 125 genes were commonly regulated by CASZ1 in SY5YtetCASZ1 replicates, the NGPtetCASZ1 and LAN5tetCASZ1 cell line (Supplementary Table 3, increased/decreased >1.5-fold in all cell lines). Bio-function assay of these 125 differently expressed genes using Ingenuity Pathways Analysis (IPA) tool, indicated that the top 8 molecular and cellular function classes of the altered genes involved cell growth and proliferation, cell death, cellular movement, as well as cellular development (Figure 2c).
There are 11 common CASZ1 target genes involved in cell growth and proliferation, the upregulated genes include HOPX, INHA, CD9, GRAP2, NGFR, CNN1, CLU and EDNRA; and the downregulated genes include Kit ligand (KITLG), hepatocyte growth factor (HGF) and PTGER2. Among those upregulated genes, CLU is a recently reported NB tumor- and metastasis-suppressor gene,15 while NGFR also suppresses NB tumorigenicity.16 To validate the microarray data, we performed real-time PCR focus on those genes that involved in tumor growth and developmental processes. CASZ1 induces a 2- to 40-fold increase in mRNA level of these genes including tumor-suppressor gene CD9, CLU and NGFR (Figure 3a). This induction is not due to Tet because there is no change in the level of mRNA expression of these genes after Tet treatment of the empty vector-transfected NB cell lines (unpublished data).
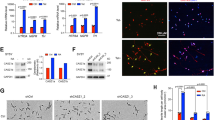
CASZ1 regulates tumor-suppressor and developmental gene expression. (a) The genes upregulated by CASZ1 in SY5YtetCASZ1 cells discovered by microarray were confirmed by real-time PCR (all P<0.05). (b) The upregulation of NGFR protein expression in SY5YtetCASZ1 cells by CASZ1 was detected by FACS analysis using FITC-labeled NGFR antibody. (c) Knock down of CASZ1 expression by CASZ1 siRNA but not control siRNA downregulated NGFR mRNA expression in SK-N-BE2 cells (detected by real-time PCR, data are shown as mean±S.D., *P<0.05). (d) Two regions of the NGFR promoter representing −3724 to +13 bp (+1 marks transcription start site) (p-3724luc) and −207 to +13 bp (p-207 luc) coupled to a luciferase reporter gene expression vector 38 were transiently transfected into SY5YtetCASZ1 cells. Transfected SY5YtetCASZ1 cells were kept in the presence or absence of Tet for 2 days, and proteins were harvested to evaluate NGFR promoter activity. Relative activity of NGFR promoter analyzed using dual-luciferase assay kit show that induction of CASZ1 expression significantly increases p-3724 luc activity (*P<0.0002)
The following studies support NGFR being a transcriptional target of CASZ1. Consistent with the CASZ1-induced increases in NGFR mRNA, induction of CASZ1 increases membrane NGFR expression in SY5YtetCASZ1 cells, as detected by fluorescence-activated cell sorting (FACS) analysis (Figure 3b). Conversely, in the SK-N-BE2 NB cell line that expresses relatively high endogenous CASZ1 mRNA, siRNAs targeted to CASZ1 cause an 80% decrease in CASZ1 that is accompanied by a 50% decrease in NGFR mRNA expression (Figure 3c). The finding that induction of CASZ1 expression increased NGFR promoter reporter constructs p-3724 luciferase activity (3.5-fold) is consistent with CASZ1 transcriptionally regulating NGFR promoter (Figure 3d). The upregulation of NGFR by CASZ1 is consistent with a role for CASZ1 in inducing NB cell differentiation and suppressing tumor growth.
CASZ1 induces NB cell differentiation
Consistent with our finding that CASZ1 regulates developmental programs and neural differentiation, our biologic assessment of CASZ1 function indicated that within 3 days of induction of CASZ1, neurite extensions are detected in SY5YtetCASZ1 (Figure 4a top, left panel). By day 12, there is more overt morphological differentiation with the formation of dense cellular aggregates and the anastamoses of neuritic extensions to form thick fasciles (Figure 4a bottom, left panel). This morphologic differentiation is accompanied by increases in MAP2 and GAP43 levels, biochemical markers of neural differentiation (Figure 4a, right panel). CASZ1 strongly upregulates expression of tyrosine hydroxylase (TH) at protein level (Figure 4a, right panel), implying CASZ1 induces dopaminergic differentiation in SY5Y. Moreover, CASZ1 induction of SY5YtetCASZ1 cell differentiation under serum-free conditions is even more dramatic with marked neurite extensions and growth cone varicosities, and increases in the growth cone associated protein GAP43 (Supplementary Figure 4, green arrow). CASZ1-induced differentiation is accompanied by downregulation of c-Myc expression (Supplementary Figure 4), which frequently occurs on induction of differentiation in a number of cell lineages. Induction of CASZ1 also induces differentiation in LAN5tetCASZ1 cells (Figure 4b). Although CASZ1 does not induce morphologic differentiation in NGPtetCASZ1, analysis of the 125 CASZ1 common target genes using BibiloSphere Pathway Edition tool, revealed that CASZ1 induced 22 genes that are also expressed in mature neurons (Supplementary Table 4) including HTR3A, NGFR and TH mRNA (Figures 3a and 4c) in all three cell lines. Thus CASZ1 induces neural gene expression in all tested cell line models and morphologic differentiation in a subset of NB cell line models.
CASZ1 induces NB cell differentiation. (a) In SY5YtetCASZ1 cells (left, top panel), neurite extension is seen within 3 days after induction of CASZ1 expression with more overt morphological characteristics of differentiation including dense cellular aggregates and fasciculation of neurites after 12 days (left, bottom panel). GAP43, MAP2 and TH, biochemical markers of neural differentiation were upregulated by CASZ1. The upregulation was visualized by immunoblotting with antibodies against GAP43, MAP2 and TH. The induction of CASZ1 expression was visualized by immunoblotting with anti-Flag antibody (right panel). (b) In LAN5tetCASZ1 cells, morphological characteristics of differentiation including dense cellular aggregates and fasciculation of neurites after 7 days induction of CASZ1 expression. (c) The upregulation of neural genes expression in LAN5tetCASZ1 and NGPtetCASZ1 cells by CASZ1 were confirmed by real-time PCR (all P<0.05)
CASZ1 enhances adhesion and inhibits motility
CASZ1 upregulates gene expression of a number of cell–cell adhesion molecules, such as COL13A1, CD9, ICAM2 and PCDH1 in all of the NB cell lines tested (Supplementary Table 3). So we functionally evaluated the effect of CASZ1 expression on cell adhesion and motility in NB cells. Cells expressing increased levels of CASZ1 are 2–3-fold more adherent based on their relative resistance to trypsinization (Figure 5a). Moreover increased expression of CASZ1 results in inhibition of cell movement in a scratch wound assay. We found a significant inhibition of cell movement on induction of CASZ1 expression in both SY5YtetCASZ1 (2.7-fold) and NGPtetCASZ1 (3.2-fold) (Figures 5b and c). The inhibition of wound healing was not a consequence of slower proliferation rates in the CASZ1 overexpressing cells, because CASZ1 inhibited cell movement in cells, which had been growth inhibited by the cell proliferation inhibitor mitomycin C before the scratch assay. These data indicate that CASZ1 expression increases cell adhesion and decreases cell motility function.
CASZ1 regulates NB cell adhesion and migration. (a) Detachment assay was performed by trypsin treatment of cells for 2–3 min; the ratio of the number of detached cells to the total cell number was calculated. The NB cell lines adhered more strongly to the substratum after induction of CASZ1 expression for 3 days (*P<0.005). (b) Wound confluence was graphed over time to quantitatively evaluate the characteristics of wound closing. The IncuCyte scratch wound assay software was used to analyze the data. Induction of CASZ1 expression caused a statistically significant reduction in migration in SY5YtetCASZ1 cells compared with the migration of control cells in a scratch wound assay (all time point P<0.00001 after wounds generated for 2.5 h). The cells photographed under phase-contrast microscopy at 22 h were showed at the right. (c) Wound confluence on induction of CASZ1 expression caused a statistically significant reduction in migration in NGPtetCASZ1 cells compared with the migration of control cells in a scratch wound assay (all time point P<0.0005 after wounds generated 2.5 h). The cells photographed under phase-contrast microscopy at 22 h were showed at the right
CASZ1 suppresses NB tumorigenicity
Upon CASZ1 induction, NB cell growth was inhibited 80% in SY5Y cells and 50% in NGP cells at day 8 (Figure 6a). MTS assays showed similar results while the growth of the empty vector control cell lines were not affected by Tet treatment (Figure 6b). FACS analysis showed that in NBtetCASZ1 cell lines, overexpression of CASZ1 leads to a modest increase in the percentage of cells in the G1 phase of the cell cycle (5–8%) at day 4 (Supplementary Figure 5a, b). The overexpression of CASZ1 functions to delay progression of SY5YtetCASZ1 cells through the cell cycle (Supplementary Figure 5c).
CASZ1 inhibits NB cell growth in vitro. (a) Cell number was counted at different time point using hemocytometer (all time point P<0.05 since day 3). (b) Cell proliferation was assessed using MTS assay after NB cells treated with or without Tet for 7 days (*P<0.005). (c) SY5YtetCASZ1 and NGPtetCASZ1 cell soft agar clonogenicity decreased when CASZ1 was induced to express by Tet (*P<0.005; **P<0.05)
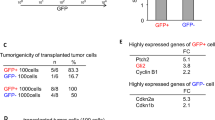
To test the effects of CASZ1 on tumorigenicity of NB cells, anchorage-independent growth was first assessed by soft agar clonogenicity. CASZ1-expressing cells showed a 90–50% decrease in soft agar clonogenicity compared with the controls in SY5YtetCASZ1 cells and NGPtetCASZ1 cells (Figure 6c).
To assess in vivo tumorigenicity, SY5YtetCASZ1 and NGPtetCASZ1 cells were implanted at a subcutaneous site into mice that had received placebo or Tet in the drinking water during the previous week. Tet-treated animals showed decreases in tumor growth and increased CASZ1 expression was detected in their tumors (Figures 7a and b). A nearly complete inhibition of tumor growth was detected by eight weeks when CASZ1 was expressed in the SY5Y cell background compared with placebo-treated mice (Figure 7a). A 75% decrease in tumor growth was detected by four weeks in NGP cell background compared with placebo-treated mice (Figure 7b). CASZ1-expressing xenograft tumors did not lead to a more differentiated histology (data not shown). However, in CASZ1-expressing tumors there is an increase in expression of NGFR (1.9-fold for SY5Y; 5.8-fold for NGP) as well as TrkA (2-fold for SY5Y; 3.4-fold for NGP) (Figure 7c and d), which are highly expressed in differentiated NB tumors and markers of good prognosis in NB.16, 17
CASZ1 inhibits NB cell growth in vivo. (a) SY5YtetCASZ1 cells were subcutaneously injected into nude mice. The induction of CASZ1 expression significantly inhibited tumor growth in the xenograft model (n=8 mice per group, total 16 mice; Volume=long × short2/4; data are shown as mean±S.E.M., all time point P<0.02 since day 24). (b) NGPtetCASZ1 cells were subcutaneously injected into nude mice. The induction of CASZ1 expression significantly inhibited tumor growth in the xenograft model (n=10 mice per group, total 20 mice; data are shown as mean±S.E.M., all P<0.02 since day 16). (c) TrkA and NGFR mRNA levels were significantly upregulated in SY5Y xenograft tumors expressing CASZ1 (*P<0.05). (d) TrkA and NGFR mRNA levels were significantly upregulated in NGP xenograft tumors expressing CASZ1 (*P<0.05)
Discussion
This study demonstrates a role for CASZ1 as a candidate NB tumor-suppressor gene based on: its localization to the chromosome 1p36 region;1, 2, 3 the association of low-CASZ1 expression with poor prognosis and overall survival (Table 1, Figure 1); its regulation of genes involved in tumor growth and developmental processes (Figure 2, Supplementary Table 3); and its functional ability to inhibit NB tumor cell growth in vitro and in vivo (Figures 6 and 7).
Previous studies showed that there are at least three discrete regions at chr1p are commonly deleted in NB, which indicates that these regions harbor putative tumor-suppressor genes.1 Recent evidence indicates that CHD5 (1p36.31) a chromatin remodeling gene,18, 19 KIF1β (1p36.21) a pro-apoptotic gene20, 21 and miR-34a, a micro RNA known to regulate MYCN expression22, 23 are candidate 1p36 NB tumor-suppressor genes. CASZ1 (1p36.22) localizes to one of the commonly deleted region in NB on chr1p between the markers D1S508 and D1S244.3 Although the recent study in 184 primary NB with 1p LOH showed that 1p36.3 (minimally between D1S2795 and D1S252) is the shortest region of overlap (SRO),24 we found in their study that loss of CASZ1 occurs in 180 of the 184 (98%) 1p LOH primary NB tumors. In this study, we provide functional evidence that CASZ1 is a tumor-suppressor gene. The identification of multiple loci on 1p36 with tumor suppressor-like activity is consistent with the hypothesis that alterations at multiple loci within this region contribute to tumorigenesis.
Classical tumor suppressors have altered expression of both alleles resulting in a loss of gene function. Although no coding nonsynonymous tumor-specific CASZ1 mutations have been found by sequencing 46 NB patient DNA samples,3 we cannot exclude the possibility that one allele of CASZ1 is deleted and the other allele is mutated in a larger NB population. Currently, we are sequencing the 160 kb regions encompassing the CASZ1 gene to assess mutational status in NB patient samples. With respect to CASZ1, our unpublished studies combined with others3 showed that although a demethylating agent induces CASZ1 expression, bisulfite sequencing results indicate that methylation of CpG rich regions 5′ and 3′ of the CASZ1 gene does not account for low-CASZ1 expression. We found that depsipeptide, a clinically relevant HDAC inhibitor upregulated CASZ1 expression even in NB tumors with an intact Chr1p. Although HDAC inhibitors result in activation of hundreds of genes, treatment with TSA induces only CASZ1 and 3 other genes in the 30 genes that localize to the Chr1p36 SRO between the markers D1S508 and D1S244.15 This indicates that only a subset of Chr1p36 genes may be silenced by histone deacetylation. Taken together, the above information suggests that epigenetic silencing either directly or indirectly contributes to the low expression of CASZ1 in NB cells.
Low-CASZ1 expression is predictive of poor outcome and is associated with other unfavorable clinical and biological features: age ≥18 months, unfavorable histology, 1p LOH and MYCN amplification. CASZ1 expression and grade are not associated in this study, yet this may be due to the fact that in 14 of the 59 samples tumor grade was unknown (Table 1). It was somewhat unexpected that high-CASZ1 expression would be associated with 11q LOH, which is a poor prognostic indicator (Table 1). This could be because CASZ1 levels are only relevant in the 1p LOH subset of NB tumors. However, the high levels of CASZ1 associated with 11q LOH may be due to the loss of upstream regulators of CASZ1 on 11q that may directly or indirectly suppress CASZ1 levels. Conversely, high levels of CASZ1 may also be due to loss of gene(s) on 11q, which are CASZ1 downstream targets that CASZ1 regulates to control neuroblast cell growth and tumorigenicity, and this loss causes a negative feedback circuit. Such a model has been described in lung cancer in which inactivation of the tumor-suppressor Rb leads to increased expression of the tumor-suppressor p16INK4a (see ref. 25). In fact, in Drosophila, CASZ1 (dcas) is expressed at a specific time and in specific cells by both feedback and feed-forward regulation with other temporal transcription factor Hunchback, Kruppel, Pdm and Grainyhead to keep the proper neuroblast sublineage development.26 It is unknown if a similar network occurs in mammals. Clearly the elucidation of the CASZ1-regulated signal transduction pathways will be important in understanding its association with 11q LOH. A larger study is also needed in order to determine if CASZ1 expression is independently prognostic in comparison with other prognostic factors.
The level of expression or CASZ1 gene dosage is associated with NB patient prognosis. Haploinsufficiency for tumor suppression has been demonstrated in the case of p27KIP1, p53, NF1 and PTEN.27 With respect to CASZ1, HDAC inhibitors upregulate CASZ1 expression in the cell lines with 1p LOH, suggesting the remaining allele is epigenetic silenced. However, we also found that CASZ1 mRNA expression was detectable in all NB patient samples and NB cell lines tested. These findings suggest epigenetic silencing of CASZ1 leads to its low expression, instead of complete depletion of CASZ1 in 1p LOH lines. Thus, CASZ1 may function like haploinsufficient tumor-suppressor genes, where a partial loss of CASZ1 may be sufficient to result in a cellular phenotype that leads to NB tumorigenesis.
Our study represents the first functional evidence in mammalian models that the highly evolutionarily conserved CASZ1 gene regulates the expression of genes important for cell growth and neural and muscle developmental processes as proposed in Drosophila and Xenopus models.5, 7, 8, 9 Our study provides compelling functional evidence that the CASZ1 suppresses tumor cell growth in vitro and in vivo (Figures 6 and 7). CASZ1 induces NB cells differentiation in vitro; and in CASZ1-expressing xenograft tumors there is an increase in expression of NGFR and TrkA (Figures 7c and d), which are highly expressed in differentiated NB tumors and markers of good prognosis in NB. Although the CASZ1-expressing xenograft tumors did not have a more differentiated histology (data not shown), this may be due to the subcutaneous transplantation of the NB cell lines. Additional studies in an orthotopic setting will be needed to determine whether CASZ1 restoration enables differentiation of NB tumors in vivo.
The transcriptome regulated by CASZ1 in NB cell lines reveals a number of induced targets whose increased expression has been associated with tumor suppression or with metastasis suppression (NGFR, CLU, CD9 and ICAM2).15, 28, 29, 30, 31 Among the targets that CASZ1 suppresses are the metastasis enhancers HGF and KITLG.32, 33 Preliminary studies have not identified a single critical target gene that mediates CASZ1 functions and current studies are aimed at delineating the targets and signaling pathways by which CASZ1 suppresses tumor growth and metastasis.
CASZ1 is expressed in a number of different tissues besides neuronal cells.4 It is interesting that CASZ1 in Xenopus is required for cardiomyocytes differentiation and loss of CASZ1 causes over-proliferation of cardiac progenitor cells.9 Inspection of data from available expression profiling data sets (http://www.oncomine.org),34 reveals that in neural crest-derived melanoma, lower levels of CASZ1 expression are found in cutaneous melanoma compared with normal skin (P=2.64E–5, Supplementary Figure 6). Low expression of CASZ1 is also found in gastric intestinal type adenocarcinoma, head and neck squamous cell carcinoma, and clear cell renal cell carcinoma (all P<0.0001, Supplementary Figure 6). Our functional studies support a role for CASZ1 as a tumor-suppressor in NB but it is possible that loss of CASZ1 may interrupt the normal developmental program of other tissues types and have a role in their tumorigenesis.
Materials and Methods
Cell culture
NB cell lines LAN5, SMS-KCNR (KCNR), SK-N-AS (AS), SK-N-BE2 (BE2), NGP and SH-SY5Y (SY5Y) were cultured in RPMI 1640 containing 10% fetal bovine serum, 2 mM glutamine and antibiotics as previously described.35 Background of these NB cell lines is shown in Supplementary Table 1. Cells were transfected using Nucleofector (Amaxa Biosystems, Gaithersburg, MD, USA) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol.
Cell treatment
When noted, cells were treated with 5 μM of RA dissolved in 95% ethanol. 5-Aza-dC (Sigma-Aldrich Co., St. Louis, MO, USA) was dissolved to 20 mM in 50% acetic acid in distilled water. Depsipeptide, provided by Fujisawa Pharmaceuticals (Osaka, Japan), 5 mg/ml was dissolved in 4 : 1 propylene glycol/ethanol, diluted to100 μg/ml in dimethyl sulfoxide and stored at −20 °C. For experiments, 5-Aza-dC and depsipeptide were diluted to indicate concentrations in cell culture media.
Stable clones
The full-length human CASZ1 transcription factor gene spans 160 kb, encodes an 8.0 kb transcript with 21 exons (Genbank accession number: NM_001079843) with 11 class C2H2 ZnF motifs.4 Full-length Flag-tagged CASZ1 cDNA4 was cloned in the Tet inducible vector pT-REx-DEST30. The control vector pDest-30 with an out-of-frame CAT gene with no start codon served as the empty vector control.
Tet-inducible expression is dependent on expression of the Tet repressor in the recipients. Recipient cells were prepared by stable transfection of the pcDNA6/TR (Tet repressor) plasmid (Invitrogen) into NGP and LAN5 cells and selection with blasticidin. Clones, LAN5tet and NGPtet, were isolated and expanded in media containing 5 μg/ml blasticidin (Invitrogen). Full-length Flag-tagged CASZ1 in pT-REx-DEST30 or an empty vector control were transfected into SY5Ytet,36 LAN5tet and NGPtet cells selected with G418 (300–500 μg/ml) and blasticidin (3–5 μg/ml). Antibiotic-resistant transfectants were isolated and evaluated for Tet (1 μg/ml) regulated CASZ1 expression. Stable clones expressing CASZ1, are labeled SY5YtetCASZ1, NGPtetCASZ1 and LAN5tetCASZ1 while the stable clones with empty vector are labeled SY5Ytetemv, NGPtetemv and LAN5tetemv.
RNAi
RNAi was performed as described previously.4 To target CASZ1 expression we designed a CASZ1 siRNA to the target sequence 5′-AACGGACTGCCCACAGATAAA-3′. Non-silencing siRNA (Qiagen, Valencia, CA, USA; cat. no. 1022076100) was used as negative control. SiRNA was transfected into NB cells by using Nucleofector (Amaxa Biosystems) following the manufacturer's protocol.
Real-time quantitative PCR
The primer sets for the CASZ1, NGFR and TH genes are shown in Supplementary Table 5. Expression of β-actin was used for normalization. Quantitative real-time PCR was performed on ABI Prism 7000 (PE Applied Biosystems, Poster City, CA, USA) using SYBR Green SuperMix as described previously.4 The PCR was performed in duplicate or triplicate and repeated at least once. Taqman real-time PCR was applied for primary tumor study.
Primary tumor study
Total RNA from NB tumors from 59 patients before chemotherapy was provided by the COG NB Biology Group. RNA (2 μg) was used as template for cDNA synthesis using Superscript III (Invitrogen), and CASZ1 levels assessed by Taqman real-time PCR. CASZ1 (part no. 4331182, assay ID: Hs00214901_m1) and β-actin (endogenous control) (part no. 4333762F) real-time PCR primers were designed and synthesized by PE Applied Biosystems. PCR reactions were performed according to the manufacture's protocol and standard curves were made during each PCR reaction.
Northern analysis
Total RNA was isolated with RNeasy kit (Qiagen) following the manufacturer's protocol. Forty micrograms of total RNA was run on 1% formaldehyde gel blotted to Nytran filters and hybridized purified inserts of CASZ1 DNA. The probes were labeled with 32P by Rediprime II (Amersham Biosciences, Buchinghamshire, UK).
FACS analysis for the determination of NGFR expression
SY5YtetCASZ1 cells treated with Tet (1 μg/ml) for 2 days were washed with PBS twice and detached by trypsin-free detachment agents, then washed once with 5 ml of PBS. Two million cells were incubated with anti-NGFR-FITC antibody (Cedarlane Laboratories Limited, Burlington, NC, USA) or normal mouse IgG-FITC for 45 min at 4 °C and washed with PBS. FACS analysis was performed using a flow cytometer (FACScan; Becton Dickinson, Franklin Lakes, NJ, USA). NGFR expression was measured using the FlowJo software (Ashland, OR, USA).
Cell cycle analysis
Cell cycle analysis was performed using propidium iodide staining. NBtetCASZ1 cells treated with Tet (1 μg/ml) for 4 days were harvested, washed with PBS twice and stained with 50 μg/ml propidium iodide containing 5 μg/ml RNase A for 30 min at room temperature, and analyzed by flow cytometry (FACScan; Becton Dickinson) and FloJo software.
Western blot and indirect immunofluorescence assay
Nuclear protein extraction, whole cell protein extraction, western blot analysis and indirect immunofluorescence assay were performed as descried previously.4 For western blot analyses, an anti-Flag (1 : 1000) antibody from Sigma (St. Louis, MO, USA) was used to detect Flag-CASZ1 expression. Antibodies to MAP2 (1 : 1000) (Chemicon International, Billerica, MA, USA), GAP43 (1 : 1000) (Abcam, Cambridge, MA, USA), α-tubulin (1 : 2000), β-actin (1 : 2000), Histone H3 (1 : 200) and TH (1 : 1000) (Santa Cruz, Santa Cruz, CA, USA) was used to detect each protein expression. For indirect immunofluorescent cell staining, cells were incubated with anti- GAP43 (1 : 250) or anti- α-tubulin (1 : 200) (Molecular Probes, Carlsbad, CA, USA) for 1 h at 23 °C, washed three times, followed by incubation with goat anti-rabbit IgG-Rhodamine or goat anti-mouse IgG-FITC (1 : 200) for 45 min at 23 °C.
Cell adhesion and scratch wound assay
Cell adhesion was performed as described by Su et al.37 with some modification. Briefly, cell adhesion was measured using a trypsinization assay. SY5YtetCASZ1 and NGPtetCASZ1 cells were plated onto a six-well plate overnight and then treated with Tet (1 μg/ml) for 3 days, then treated with 0.5 ml of 0.05% trypsin-EDTA for 2–3 min to stimulate cell detachment. The number of detached cells was counted using a hemocytometer and expressed as a percentage of the total number of cells on the plate. For scratch wound assay, SY5YtetCASZ1 and NGPtetCASZ1 cells were plated into Essen ImageLock 24-well plates (for NGPtetCASZ1 cells, the plate was coated with collagen I), and after 24 h incubation, the cells were treated with or without Tet (1 μg/ml) for 24 h, then the medium was changed from 10% FBS containing complete medium to 0.5% FBS containing low serum medium with or without Tet (1 μg/ml). After overnight incubation, the cells were incubated with mitomycin-C (10 μg/ml) for 2 h to inhibit cell proliferation, after three times washing with low serum medium, the confluent monolayer of the cells was scratched using the Essen 24-well WoundMaker (Ann Arbor, MI, USA) following their protocols and the cells were cultured in the complete medium with or without Tet (1 μg/ml). The wound confluence was obtained and analyzed by using the IncuCyte phase-contrast imaging and scratch wound assay system and software (ESSEN INSTRUMENTS, Ann Arbor, MI, USA).
Promoter activity assay
The NGFR promoter constructs were generously provided by Philip Barker.38 The constructs were transiently transfected into SY5YtetCASZ1 cell line and after 24 h, cells were treated with Tet. After 2 days, cells were harvested, protein isolated and relative luciferase activity assessed using a dual-luciferase reporter assay system (Promega, Madison, WI, USA), the experiments were repeated for three times.
Cell proliferation and clonogenicity assays
To assess the effect of CASZ1 on NB cell proliferation, SY5YtetCASZ1 and NGPtetCASZ1 cells were plated into 10 cm dishes. The next day Tet was added to the RPMI-1640 containing 10% fetal calf serum. Trypan blue diluted cell suspension was counted using hemocytometer at indicated time points. MTS assay (Promega) was also performed to assess effects of CASZ1 on NB cell proliferation. IncuCyte (ESSEN BioSCIENCE, Ann Arbor, MI, USA) was used to monitor kinetic SY5YtetCASZ1 cell proliferation in 24-well plate using the integrated confluence algorithm as a surrogate for cell number. To assess effects of CASZ1 on anchorage-independent cell growth, 1–2 × 104 cells were cultured in 0.7% top agarose in media containing G418 and Blasticidin (± Tet) plated on a layer of 1.4% bottom agar/RPMI to prevent the adhesion of cells to the culture plates. Medium was changed twice a week and visible colonies counted after 4 weeks.
In vivo tumorigenesis
Suspensions of SY5YtetCASZ1 (2 × 106) or NGPtetCASZ1 (5 × 105) cells were mixed with an equal volume of Matrigel solution (Trevigen, Gaithersburg, MD, USA) and implanted subcutaneously in the dorsal flank of 16–20 SCID mice (6–8 weeks female mice) per cell line. The animal studies were approved by the Animal Care and Use Committee of the National Cancer Institute (PB-023). Control mice received 5% sucrose in their drinking water while the other half received Tet (2 mg/ml) in 5% sucrose for 1 week before tumor implantation and during the course of the experiment. The tumors were measured thrice weekly, and the mice were killed once the tumor diameter reached 20 mm.
The oligo microarray
Agilent whole human genome oligo microarray kit (G4112F, Agilent) were used to determine the CASZ1-induced transcriptional profiles in SY5YtetCASZ1, NGPtetCASZ1 and LAN5tetCASZ1. Gene expression after Tet induction (24 h) was analyzed in the three different NB cell lines, as well as a biologic replicate for SY5Y cell line. Array hybridization, chemiluminescence detection and image acquisition and analysis were performed by GenUs Biosystems, Northbrook, IL, USA. The differentially expressed gene list was uploaded to IPA (Ingenuity http://www.ingenuity.com/) and BiblioSphere Pathway Edition tool (Genomatix http://www.genomatix.de/), and these were used to generate unbiased molecular and cellular functional analyses and physiological system development and functions.
Statistical analyses
The method of Kaplan–Meier was used to generate survival curves, with standard errors per Peto,39 and differences between the curves were tested with a log rank test. For EFS, time to event was calculated from the time of diagnosis until the first occurrence of relapse, progression, secondary malignancy or death from any cause, or until last contact if no event occurred. An event for overall patient survival was death. P-values <0.05 were considered statistically significant.
To identify an CASZ1 expression cut-off that maximized the difference in EFS rate between two groups (low versus high expression), 13 Cox proportional hazards models40 were run, one at each of 13 different CASZ1 expression cut-offs. Each model included only one term, a binary indicator for high-CASZ1 expression; the assumption of proportional hazards was tested, and was not violated. The value for every sixth patient (i.e, at each decile) was tested as a cut-off, and additional cut-offs was tested to gain resolution in the area of interest. A combination of minimum P-value and maximum relative risk were used to identify the cut-off that was the most prognostic of outcome.
The study was not powered to determine if CASZ1 expression was independently predictive of outcome, so multivariable modeling was not performed.
IPA of microarray data was analyzed by Fisher's exact test (http://www.ingenuity.com/). Additional statistical analyses of continuous data were performed using a t-test with P<0.05 considered significant. Values in the graphs are expressed as means±S.E.M. or S.D. The statistical tests were two-sided.
Accession codes
Abbreviations
- NB:
-
neuroblastoma
- LOH:
-
loss of heterozytosity
- Chr1p:
-
Chromosome 1p
- ZnF:
-
zinc-finger
- EFS:
-
event-free survival
- RA:
-
retinoic acid
- Tet:
-
tetracycline
- FACS:
-
fluorescence-activated cell sorting
- SRO:
-
shortest region of overlap
References
Brodeur GM . Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 2003; 3: 203–216.
Bagchi A, Mills AA . The quest for the 1p36 tumor suppressor. Cancer Res 2008; 68: 2551–2556.
Caren H, Fransson S, Ejeskar K, Kogner P, Martinsson T . Genetic and epigenetic changes in the common 1p36 deletion in neuroblastoma tumours. Br J Cancer 2007; 97: 1416–1424.
Liu Z, Yang X, Tan F, Cullion K, Thiele CJ . Molecular cloning and characterization of human castor, a novel human gene upregulated during cell differentiation. Biochem Biophys Res Commun 2006; 344: 834–844.
Mellerick DM, Kassis JA, Zhang SD, Odenwald WF . Castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron 1992; 9: 789–803.
Isshiki T, Pearson B, Holbrook S, Doe CQ . Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 2001; 106: 511–521.
Cui X, Doe CQ . Ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development 1992; 116: 943–952.
von Hilchen CM, Beckervordersandforth RM, Rickert C, Technau GM, Altenhein B . Identity, origin, and migration of peripheral glial cells in the Drosophila embryo. Mech Dev 2008; 125: 337–352.
Christine KS, Conlon FL . Vertebrate castor is required for differentiation of cardiac precursor cells at the ventral midline. Dev Cell 2008; 14: 616–623.
Vacalla CM, Theil T . Cst, a novel mouse gene related to Drosophila castor, exhibits dynamic expression patterns during neurogenesis and heart development. Mech Dev 2002; 118: 265–268.
Bader SA, Fasching C, Brodeur GM, Stanbridge EJ . Dissociation of suppression of tumorigenicity and differentiation in vitro effected by transfer of single human chromosomes into human neuroblastoma cells. Cell Growth Differ 1991; 2: 245–255.
Chen D, Livne-bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R . Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 2004; 5: 539–551.
Weissman AM . How much REST is enough? Cancer Cell 2008; 13: 381–383.
Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 2009; 27: 289–297.
Chayka O, Corvetta D, Dews M, Caccamo AE, Piotrowska I, Santilli G et al. Clusterin, a haploinsufficient tumor suppressor gene in neuroblastomas. J Natl Cancer Inst 2009; 101: 663–677.
Schulte JH, Pentek F, Hartmann W, Schramm A, Friedrichs N, Ora I et al. The low-affinity neurotrophin receptor, p75, is upregulated in ganglioneuroblastoma/ganglioneuroma and reduces tumorigenicity of neuroblastoma cells in vivo. Int J Cancer 2009; 124: 2488–2494.
Nakagawara A, Arima-Nakagawara M, Scavarda NJ, Azar CG, Cantor AB, Brodeur GM . Association between high levels of expression of the TRK gene and favorable outcome in human neuroblastoma. N Engl J Med 1993; 328: 847–854.
Fujita T, Igarashi J, Okawa ER, Gotoh T, Manne J, Kolla V et al. CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J Natl Cancer Inst 2008; 100: 940–949.
Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D et al. CHD5 is a tumor suppressor at human 1p36. Cell 2007; 128: 459–475.
Schlisio S, Kenchappa RS, Vredeveld LC, George RE, Stewart R, Greulich H et al. The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Develop 2008; 22: 884–893.
Munirajan AK, Ando K, Mukai A, Takahashi M, Suenaga Y, Ohira M et al. KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J Biol Chem 2008; 283: 24426–24434.
Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P et al. The MYCN oncogene is a direct target of miR-34a. Oncogene 2008; 27: 5204–5213.
Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res 2008; 6: 735–742.
White PS, Thompson PM, Gotoh T, Okawa ER, Igarashi J, Kok M et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene 2005; 24: 2684–2694.
Shapiro GI, Edwards CD, Kobzik L, Godleski J, Richards W, Sugarbaker DJ et al. Reciprocal Rb inactivation and p16INK4 expression in primary lung cancers and cell lines. Cancer Res 1995; 55: 505–509.
Brody T, Odenwald WF . Cellular diversity in the developing nervous system: a temporal view from Drosophila. Development 2002; 129: 3763–3770.
Fodde R, Smits R . Cancer biology. A matter of dosage. Science 2002; 298: 761–763.
Krygier S, Djakiew D . Neurotrophin receptor p75(NTR) suppresses growth and nerve growth factor-mediated metastasis of human prostate cancer cells. Int J Cancer 2002; 98: 1–7.
Jin H, Pan Y, He L, Zhai H, Li X, Zhao L et al. p75 neurotrophin receptor inhibits invasion and metastasis of gastric cancer. Mol Cancer Res 2007; 5: 423–433.
Takeda T, Hattori N, Tokuhara T, Nishimura Y, Yokoyama M, Miyake M . Adenoviral transduction of MRP-1/CD9 and KAI1/CD82 inhibits lymph node metastasis in orthotopic lung cancer model. Cancer Res 2007; 67: 1744–1749.
Yoon KJ, Phelps DA, Bush RA, Remack JS, Billups CA, Khoury JD . ICAM-2 expression mediates a membrane-actin link, confers a nonmetastatic phenotype and reflects favorable tumor stage or histology in neuroblastoma. PLoS One 2008; 3: e3629.
Hecht M, Papoutsi M, Tran HD, Wilting J, Schweigerer L . Hepatocyte growth factor/c-Met signaling promotes the progression of experimental human neuroblastomas. Cancer Res 2004; 64: 6109–6118.
Timeus F, Crescenzio N, Valle P, Pistamiglio P, Piglione M, Garelli E et al. Stem cell factor suppresses apoptosis in neuroblastoma cell lines. Exp Hematol 1997; 25: 1253–1260.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004; 6: 1–6.
Thiele CJ, Reynolds CP, Israel MA . Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature 1985; 313: 404–406.
Lovat PE, Oliverio S, Ranalli M, Corazzari M, Rodolfo C, Bernassola F et al. GADD153 and 12-lipoxygenase mediate fenretinide-induced apoptosis of neuroblastoma. Cancer Res 2002; 62: 5158–5167.
Su LT, Agapito MA, Li M, Simonson WT, Huttenlocher A, Habas R et al. TRPM7 regulates cell adhesion by controlling the calcium-dependent protease calpain. J Biol Chem 2006; 281: 11260–11270.
Ramos A, Ho WC, Forte S, Dickson K, Boutilier J, Favell K et al. Hypo-osmolar stress induces p75NTR expression by activating Sp1-dependent transcription. J Neurosci 2007; 27: 1498–1506.
Peto R, Peto J . Asymptotically efficient rank invariant test procedures. J Royal Stat Soc A 1972; 135: 185–198.
Cox D . Regression models and life tables (with discussion). J Royal Stat Soc B 1972; 34: 187–220.
Acknowledgements
We would like to thank the Children s Oncology Group (COG) Neuroblastoma Biology Group Committee for providing us with total RNA from neuroblastoma tumors from patients prior to chemotherapy. We thank Dr. Cris Q. Doe of Howard Hughes Medical Institute, University of Oregon for the critical reading of the manuscript. We would like to thank Drs. Chris Redfern, and Quentin Campbell Hewson of the Northern Institute for Cancer Research, Newcastle University, UK for providing us with the SY5Ytet (SY5Ytet12) cell line. The NGFR promoter – pGL3 basic vectors were generously provided by Philip Barker (Centre for Neuronal Survival, Montreal Neurological Institute, McGill University, Canada). The control vector pDest-30 with an out-of-frame CAT gene with no start codon was generously provided by Dominic Esposito (Protein Expression Laboratory, SAIC-Frederick, Inc.). We thank the CCR's Office of Science and Technology Partnerships, Drs. Shoshana Segal and David Goldstein for their coordination of the strategic partnerships that were key for microarray services. The authors thank Ms. Lauren Marks for her excellent support of the Cell and Molecular Biology Section, POB, CCR, NCI. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The study of NB tumor samples was done through the COG NB Biology Project 2004-01, with COG funding from grant number U10 CA98413.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by V De Laurenzi
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Liu, Z., Yang, X., Li, Z. et al. CASZ1, a candidate tumor-suppressor gene, suppresses neuroblastoma tumor growth through reprogramming gene expression. Cell Death Differ 18, 1174–1183 (2011). https://doi.org/10.1038/cdd.2010.187
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2010.187
Keywords
This article is cited by
-
Role of the CASZ1 transcription factor in tissue development and disease
European Journal of Medical Research (2023)
-
CASZ1: a promising factor modulating aldosterone biosynthesis and mineralocorticoid receptor activity
Hypertension Research (2023)
-
Allele-specific expression reveals genes with recurrent cis-regulatory alterations in high-risk neuroblastoma
Genome Biology (2022)
-
The expression changes of transcription factors including ANKZF1, LEF1, CASZ1, and ATOH1 as a predictor of survival rate in colorectal cancer: a large-scale analysis
Cancer Cell International (2022)
-
Loss of CASZ1 tumor suppressor linked to oncogenic subversion of neuroblastoma core regulatory circuitry
Cell Death & Disease (2022)