Abstract
Oncogene-induced senescence represents a key tumor suppressive mechanism. Here, we show that Ras oncogene-induced senescence can be mediated by the recently identified haploinsufficient tumor suppressor apoptosis-stimulating protein of p53 (ASPP) 2 through a novel and p53/p19Arf/p21waf1/cip1-independent pathway. ASPP2 suppresses Ras-induced small ubiquitin-like modifier (SUMO)-modified nuclear cyclin D1 and inhibits retinoblastoma protein (Rb) phosphorylation. The lysine residue, K33, of cyclin D1 is a key site for this newly identified regulation. In agreement with the fact that its nuclear localization is required for its oncogenic activity, we show that nuclear cyclin D1 is far more potent than wild-type (WT) cyclin D1 in bypassing Ras-induced senescence. Thus, this study identifies SUMO modification as a positive regulator of nuclear cyclin D1, and reveals a new way by which cell cycle entry and senescence are regulated.
Similar content being viewed by others
Main
Cellular senescence is an irreversible cell cycle arrest and one of the major cellular processes that suppress tumor growth. Cyclin-dependent kinases (CDKs) are among the main regulators of cellular senescence, predominantly achieved through their phosphorylation of the retinoblastoma protein (Rb), preventing it from binding and inhibiting the E2F family of transcription factors that are involved in cell cycle progression. Deregulation of CDK pathways is common in the development of human cancers. Amplification and overexpression of cyclin D1, a regulatory subunit of CDK4/CDK6, has been observed in around 50% of human breast and esophageal cancers.1, 2 Interestingly, however, elevation of cyclin D1 alone is not sufficient to induce tumor growth in vivo,3, 4 partly owing to the fact that the cyclin D1/CDK4 or cyclin D1/CDK6 complex needs to locate in the nucleus to phosphorylate Rb during S-phase to promote cell proliferation.5, 6 Nuclear export, coupled with ubiquitin-dependent destruction of cyclin D1 in the cytoplasm during S-phase, is one of the best known mechanisms of cyclin D1/CDK4 kinase activity control. Hence, nuclear accumulation of cyclin D1 may be a key mechanism by which cyclin D1 exerts its oncogenic effects. In agreement with this, overexpression of mutant cyclin D1 (D1T286A), defective in phosphorylation-mediated nuclear export and subsequent proteolysis, induces cell transformation in vitro and triggers B-cell lymphoma in vivo.6, 7 Furthermore, transgenic mice overexpressing the same mutant, driven by mouse mammary tumor virus (MMTV) promoter (MMTV-D1T286A), developed mammary adenocarcinoma with a shorter latency relative to mice overexpressing wild-type (WT) cyclin D1 (MMTV-D1).8 These observations support the notion that the subcellular localization of cyclin D1 is critical in controlling its tumorigenicity. Identifying the molecular mechanisms that regulate the nuclear localization of cyclin D1 is, therefore, vital for our better understanding of tumor development.
SUMOylation is a form of post-translational modification that regulates the cellular localization of modified proteins. Small ubiquitin-like modifiers (SUMOs) are ubiquitin-like polypeptides that become covalently conjugated to cellular proteins in a manner similar to ubiquitylation.9 In vertebrates, three SUMO isoforms are expressed. SUMO-1 shares 43% identity with SUMO-2 and SUMO-3, whereas the latter two are closely related (sharing 97% identity). The most important differences between the mammalian SUMO paralogues are that the overall cellular concentration of SUMO-2/3 is greater than that of SUMO-1, as is the pool of free protein available for conjugation,10 and their conjugation pattern: only SUMO-2 and SUMO-3 can form conjugated chains through a single conserved acceptor lysine. Nonetheless, SUMO-1 may terminate chains that are elongated through serial conjugation of SUMO-2/3.11 The conjugation process involves an enzymatic cascade comprising the E1-activating enzyme activator of SUMO-1, the E2-conjugating enzyme ubiquitin-like protein SUMO-1 conjugating enzyme 9 and E3 ligases including the protein inhibitor of activated STAT family members RAN-binding protein 2 and human polycomb 2.9, 12
Oncogenic stress is one of the best studied cellular senescence-inducing signals and, of the oncogenes identified to date, Ras is the prototype. The Ras oncogene has been shown to predominantly use either p16ink4a/Rb or the alternate reading frame (Arf)/p21waf1/cip1/p53 pathway to mediate cellular senescence.13, 14, 15 However, it is now becoming clear that Ras oncogene can also induce senescence via p53-independent pathways such as CCAAT/enhancer-binding protein-β, or ones mediated by p63.16, 17
Apoptosis-stimulating protein of p53 (ASPP) 2 belongs to the evolutionarily conserved ASPP family of proteins that were initially identified through their ability to bind to, and regulate, the apoptotic function of p53 and its family members p63 and p73.18 Two different ASPP2 transgenic mouse models were generated, expressing either exon 3- or exons 10–17-deleted ASPP2 genes. These two mouse models are referred to here as ASPP2(Δ3/Δ3) and ASPP2(Δ10−17/Δ10−17), respectively. Importantly, heterozygous mice of ASPP2(Δ3/+) and ASPP2(Δ10−17/+) are both prone to developing spontaneous tumors, establishing ASPP2 as a new haploinsufficient tumor suppressor.19, 20 ASPP2 expression is frequently downregulated in human tumors, and reduced ASPP2 expression is tightly associated with patients' poor prognosis.21, 22 As ASPP2 can bind other cellular proteins including p65 reticuloendotheliosis viral oncogene homolog A, B-cell lymphoma 2, protein phosphatase 1, YES-associated protein, adenomatous polyposis coli-like and amyloid-β precursor protein binding protein 1,23, 24, 25, 26, 27, 28 it is possible that ASPP2 suppresses tumor growth in ways other than promoting p53-mediated apoptosis. As apoptosis and senescence are two major tumor suppressive pathways, we have investigated whether ASPP2 can exert its tumor suppressive effects by mediating cellular senescence.
Here, we show that Ras oncogene-induced senescence can be mediated by ASPP2 through its ability to prevent Ras from inducing nuclear accumulated cyclin D1. Importantly, ASPP2 mutant cells allowed us to identify SUMO modification as a novel regulation that leads to nuclear localization of cyclin D1. Lysine 33 (K33) of cyclin D1 is predominantly responsible for this newly identified modification. Nuclear localization is vital for the oncogenic properties of cyclin D1 in vitro and in vivo, and this is supported by the observation that nuclear cyclin D1 is able to bypass Ras-induced senescence.
Results
H-RasV12-induced senescence is mediated by ASPP2
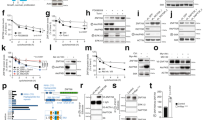
Mouse embryonic fibroblasts (MEFs) are a well-established experimental system used to study oncogene-induced senescence. To investigate whether ASPP2 has a role in mediating Ras oncogene-induced senescence, MEFs were generated from ASPP2(+/+) and ASPP2(Δ3/Δ3) embryos, where exon 3 of the murine ASPP2 gene was deleted as previously described,19 and infected with oncogenic H-RasV12. As expected, the expression of oncogenic H-RasV12-induced senescence in ASPP2(+/+) MEFs was detected by the presence of the senescence marker senescence-associated β-galactosidase (SA-β-Gal), but not in ASPP2(Δ3/Δ3) MEFs (Figure 1a and b).
ASPP2 mediates senescence induced by H-RasV12. MEFs generated from ASPP2(+/+) and ASPP2(Δ3/Δ3) embryos were infected with oncogenic H-RasV12 and stained for SA-β-Gal activity (a). Comparison of the number of colonies formed by ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs transduced with empty vector (Vector) or H-RasV12 (Ras)-expressing retroviruses (b). Anchorage-independent growth assay for ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs transduced with H-RasV12 (c). Colonies were photographed at × 20 magnification. Error bars represent S.D. of at least three independent experiments. H-RasV12-expressing ASPP2(Δ3/Δ3) MEFs were infected with retroviruses expressing full length ASPP2 or empty vector and subjected to SA-β-gal staining. The percentage of SA-β-gal-positive cells is shown in the right panel. Error bars represent the range from two independent experiments (d)
The ability of ASPP2 to suppress the transforming activity of H-RasV12 was further investigated using a soft agar assay to measure anchorage independence. Whereas ASPP2(Δ3/Δ3) MEFs formed few small soft agar colonies, H-RasV12-expressing ASPP2(Δ3/Δ3) MEFs formed at least 10 times more and bigger colonies than those seen in ASPP2(+/+) MEFs (Figure 1c). Retroviruses expressing full length ASPP2 were used to infect H-RasV12-expressing ASPP2(Δ3/Δ3) MEFs to investigate whether reintroduction of ASPP2 could mediate H-RasV12-induced senescence. SA-β-Gal activity was detected in around 30% of cells infected (Figure 1d). In MEFs, Ras-induced cellular senescence is the predominant phenomenon. p53 also predominantly induces senescence rather than apoptosis in Ras-expressing MEFs. In agreement with this, we failed to detect any impact of ASPP2 in inducing apoptosis (data not shown). These results illustrate that ASPP2 is a novel mediator of H-RasV12-induced senescence, which prevents H-RasV12 from transforming MEFs.
ASPP2 mediates Ras-induced senescence by preventing H-RasV12-induced nuclear accumulation of cyclin D1
As Ras oncogene can induce senescence through a p53-dependent pathway and ASPP2 was originally identified as a p53 activator, we checked whether ASPP2 could affect H-RasV12-induced expression of p19Arf, p53 and p21waf1/cip1. Interestingly, the expression of H-RasV12 caused a similar increase in the expression levels of p19Arf, p53 and p21waf1/cip1 in both ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs (Figure 2a), suggesting that ASPP2 mediates Ras oncogene-induced senescence independently of p53.
ASPP2 inhibits Rb phosphorylation and nuclear accumulation of cyclin D1 induced by Ras oncogene independently of p53. WBs showing the expression levels of endogenous ASPP2, p19Arf, p53 and p21waf1/cip1, as indicated in ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs, with or without H-RasV12 expression (a); phosphorylated Rb and total Rb (b); and cyclin D1, CDK4, p16ink4a, p27kip1, cyclin A, cyclin E and CDK2 in ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs, with or without H-RasV12 expression (c). In (b), the ratio of p-Rb to Rb was calculated by densitometry using the QuantityOne software (Bio-Rad Laboratories Ltd., Hemel Hempstead, Hertfordshire, UK) and error bars represent the range from two independent experiments. (d) Expression levels of p53, p19Arf, p21waf1/cip1 and cyclin D1 in p53(+/+) and p53(−/−) MEFs, with or without H-RasV12 expression. Tubulin expression levels confirm equal loading. (e) Indirect immunofluorescence of cyclin D1 in ASPP2(+/+) or ASPP2(Δ3/Δ3) MEFs, and p53(+/+) or (−/−) MEFs, with or without H-RasV12 expression. Graphs show the percentage of nuclear cyclin D1-expressing cells. Error bars represent the range from two independent experiments
To examine whether ASPP2 mediates Ras oncogene-induced senescence through the Rb pathway, we compared the phosphorylation status of Rb in ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs. Similar amounts of phosphorylated Rb were detected in both ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs. Interestingly, Ras oncogene only induced a clear reduction in Rb phosphorylation in ASPP2(+/+) MEFs, but caused a marked three–fourfold increase in the amount of phosphorylated Rb in H-RasV12-expressing ASPP2(Δ3/Δ3) MEFs (Figure 2b). These results suggest that ASPP2 is required for Ras oncogene to inhibit Rb phosphorylation, and ASPP2 may use this property to mediate Ras oncogene-induced senescence.
To understand how H-RasV12 affects Rb phosphorylation in the absence of functional ASPP2, we investigated the expression levels of CDKs and their inhibitors in ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs, with or without H-RasV12 infection. The expression levels of CDK4, p16ink4a, p27kip1, cyclin E, cyclin A and CDK2 were not affected by ASPP2 status (Figure 2c). Interestingly, however, ASPP2 status did affect the expression pattern of cyclin D1. Although cyclin D1 protein levels were increased in both H-RasV12-expressing ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs, increased cyclin D1 was often associated with the expression of higher molecular weight (MW) protein bands in H-RasV12-expressing ASPP2(Δ3/Δ3) MEFs, detected by at least two different anti-cyclin D1 antibodies (Figure 2c and data not shown). This suggests that ASPP2 may mediate the post-translational modification of cyclin D1, induced by Ras oncogene. It is important to note that we failed to detect similar changes in p53(+/+) and p53(−/−) MEFs, irrespective of Ras expression (Figure 2d). Hence only ASPP2, but not p53, may influence cyclin D1 expression in response to Ras oncogene. As cyclin D1 is a regulatory subunit of CDKs that phosphorylate Rb, this observation suggests that modified cyclin D1 may be responsible for the observed increase in Rb phosphorylation.
It has also been reported that only nuclear cyclin D1 confers tumor growth.29 Remarkably, ASPP2(Δ3/Δ3) MEFs had a prominent, fourfold higher number of nuclear cyclin D1-expressing cells in response to H-RasV12 induction compared with ASPP2(+/+) MEFs (Figure 2e). In contrast, no difference was observed in the nuclear localization of p21waf1/cip1 between ASPP2(+/+) and ASPP2(Δ3/Δ3) MEFs (Supplementary Figure 1). Furthermore, H-RasV12-induced nuclear accumulation of cyclin D1 was specifically prevented by ASPP2, but not p53, as similar numbers of nuclear cyclin D1 were detected in Ras-expressing p53(+/+) and p53(−/−) MEFs (Figure 2e, middle panel). This confirms that the pathway regulated by Ras and ASPP2 is p53-independent. Together, the results show that ASPP2 mediates H-RasV12-induced senescence by preventing H-RasV12 from inducing nuclear cyclin D1, Rb phosphorylation and cell cycle entry.
H-RasV12 induces SUMOylation of cyclin D1
Owing to the large change in gel mobility, we hypothesized that cyclin D1 modification was likely caused by molecules that can form large chains, such as ubiquitin or SUMO. As cyclin D1's ubiquitination may be influenced by Ras,30 we tested whether the presence of H-RasV12 could influence the expression pattern of ubiquitinated cyclin D1 in ASPP2(Δ3/Δ3) cells. The high MW cyclin D1 observed in Figure 2c was highly insoluble and only detected when 8 M urea buffer was used to lyse the cells. Thus, 8 M urea cell lysates were first diluted 10-fold in NP-40 buffer to refold the solubilised and denatured high MW cyclin D1, to allow an anti-cyclin D1 antibody to specifically immunoprecipitate cyclin D1. The presence of ubiquitinated cyclin D1 was analyzed using an anti-ubiquitin antibody. Interestingly, H-RasV12 expression did not significantly alter the expression pattern of ubiquitinated proteins in ASPP2(Δ3/Δ3) cells (Figure 3a, lanes 1–4). Furthermore, the ubiquitinated cyclin D1 expression pattern was similar between vector or H-RasV12-expressing ASPP2(Δ3/Δ3) cells (Figure 3a, lanes 5 and 6). Treatment with the proteasome inhibitor MG132 also had a minimal impact on the expression pattern of ubiquitinated cyclin D1 (Figure 3a, lanes 8 and 9). Importantly, ubiquitinated cyclin D1 had a completely different mobility pattern than that observed in Figure 2c, suggesting that the high mobility cyclin D1 bands detected in H-RasV12-expressing ASPP2(Δ3/Δ3) cells were not caused by altered ubiquitination activity.
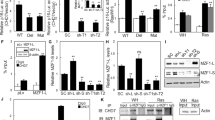
H-RasV12 induces accumulation of SUMO-modified cyclin D1 in ASPP2(Δ3/Δ3) MEFs. (a) H-RasV12 expression does not affect cyclin D1's ubiquitination pattern. Total cell lysates (6 mg) derived from ASPP2(−/−) MEFs with or without H-RasV12 expression, treated with proteasomal inhibitor MG132 where indicated, were immunoprecipitated with anti-cyclin D1 or control (IgG) antibodies. Immunoprecipitates and an aliquot (50 μg) of total cell lysate were subjected to WB analysis using anti-ubiquitin antibody. Ubiquitin-cyclin D1 is labelled. (b) Total cell lysates (6 mg) derived from ASPP2(Δ3/Δ3) MEFs, with or without H-RasV12 expression, were immunoprecipitated and subjected to WB analysis as in (a) to detect SUMO-modified cyclin D1 as labelled. (c) ASPP2(Δ3/Δ3) MEFs, with or without H-RasV12 expression, were transfected with 1 μg of cyclin D1-expressing plasmid, with or without 1 μg of plasmid expressing 6His-tagged SUMO-1 or 6His-tagged SUMO-2 and 6His-tagged SUMO-3. 6His-tagged SUMOs were trapped on a nickel column, and SUMOylated products separated and analyzed with anti-cyclin D1 antibody to detect SUMO-modified cyclin D1 as indicated
SUMO modification, particularly by SUMO-2 and SUMO-3, often alters a protein's cellular localization. Knowing that modified cyclin D1 is tightly associated with the accumulation of nuclear cyclin D1, we tested whether cyclin D1 could be SUMO-modified and whether this modification was altered in H-RasV12-expressing ASPP2(Δ3/Δ3) cells. Interestingly, we detected an enrichment of SUMO-2/SUMO-3-modified protein in H-RasV12-expressing ASPP2(Δ3/Δ3) cells, in comparison with ASPP2(Δ3/Δ3) cells expressing vector only (Figure 3b, compare lanes 1 and 2). We also detected a distinct expression pattern of high MW bands in cyclin D1 immunoprecipitates derived from H-RasV12-expressing ASPP2(Δ3/Δ3) MEFs compared with those expressing vector only (Figure 3b, lanes 3 and 4). In particular, we detected an enrichment of SUMO-modified cyclin D1 with MWs raging from 35 to 50 kDa (e.g., labelled band 2 and 3). These results suggest that cyclin D1 may be SUMO-modified, and that H-RasV12 enhances this modification in ASPP2(Δ3/Δ3) MEFs.
To provide further evidence that cyclin D1 can be SUMO-modified, we cotransfected His-tagged SUMO-expressing plasmids together with cyclin D1 into ASPP2(Δ3/Δ3) MEFs, with or without H-RasV12. Expressed His-tagged SUMO was pulled down by nickel beads and the presence of cyclin D1 was detected by an anti-cyclin D1 antibody. Under the conditions used, the nickel beads pulled down the cotransfected cyclin D1 non-specifically (Figure 3c, lanes 1 and 2). However, when cotransfected with SUMO-1, they pulled down more cyclin D1 with a high MW, similar to that seen in Figure 3b. Furthermore, the presence of H-RasV12 enriched the amount of SUMO-modified cyclin D1 (Figure 3c, lanes 3 and 4, in particular band 3 in lane 4). The amount of high MW cyclin D1 was further increased when SUMO-1–3 were coexpressed with cyclin D1 (Figure 3c, lanes 5 and 6, in particular bands 2 and 3 in lane 6). The pattern of high MW cyclin D1 resembled that detected by probing immunoprecipitated endogenous cyclin D1 with anti-SUMO antibody. These results illustrate that cyclin D1 is SUMO-modified, and that this modification is induced by H-RasV12 in ASPP2(Δ3/Δ3) MEFs.
SUMO modification leads to nuclear accumulation of cyclin D1
SUMO modification is often associated with the specific cellular localization of modified proteins. Thus, we checked the localization of exogenously expressed cyclin D1 in a human osteosarcoma cell line (U2OS), which expresses endogenous Ras and ASPP2, in the presence of SUMO-1-, SUMO-2- and SUMO-3-expressing plasmids. Nuclear cyclin D1 was detected in more than 60% of U2OS cells coexpressing cyclin D1 and SUMO-2 and 3 (but not SUMO-1; Figure 4a). To confirm the role of SUMOylation in the nuclear localization of cyclin D1, we infected H-RasV12-expressing ASPP2(Δ3/Δ3) MEFs with adenovirus-expressing sentrin-specific protease 2 (SENP2), a protease that specifically removes SUMO modifications. We observed that nuclear cyclin D1 disappeared from cells infected by SENP2-expressing adenovirus, but not from cells infected by control virus, (Figure 4b), suggesting that SUMO modification is required for the accumulation of nuclear cyclin D1.
SUMO modification leads to nuclear accumulation of cyclin D1. (a) Indirect immunofluorescence of cyclin D1 and SUMO in U2OS cells cotransfected with cyclin D1, SUMO-1-, or SUMO-2- and SUMO-3-expressing plasmids. Graphs show the percentage of nuclear cyclin D1-expressing cells over the total number of cyclin D1-expressing cells. Error bars represent the range from three independent experiments. (b) Indirect immunofluorescence showing expression of endogenous cyclin D1 in H-RasV12-expressing ASPP2(Δ3/Δ3) MEFs, infected with control or SENP2-expressing adenovirus for 24 h. The percentage of nuclear cyclin D1-expressing cells is indicated in the bottom/right corner of each panel
SUMO modification on K33 is critical for the nuclear localization of cyclin D1
Sequence analysis performed with the SUMOsp web server31 revealed the existence of four putative SUMOylation sites on cyclin D1: a canonical one at K33, and three type II sites at K72, K95 and K96 (Figure 5a). To identify the lysine residue important for cyclin D1 modification and nuclear localization, residues K33, K72, K95 and K96 were individually mutated to alanine to generate non-modifiable cyclin D1 mutants K33A, K72A, K95A and K96A, respectively. The cellular localizations of exogenously expressed cyclin D1 and its mutants in the presence of SUMO-2- and SUMO-3-expressing plasmids (Figure 5b) were then examined. Interestingly, nuclear cyclin D1 was detected in 60–70% of the cells coexpressing cyclin D1, cyclin D1-K72A, cyclin D1-K95A and cyclin D1-K96A with SUMO-2 and SUMO-3, but only in 15% of the cells coexpressing cyclin D1-K33A and SUMO-2 and SUMO-3. These results demonstrate that SUMO is likely to modify cyclin D1 on K33, and that K33 is critical for cyclin D1's nuclear localization.
K33 is critical for cyclin D1's nuclear localization. (a) Cyclin D1 protein sequence with SUMOylation sites highlighted (with lysine residues in blue and flanking residues in red). (b) indirect immnofluorescence of cyclin D1 and SUMO-2/3 in U2OS cells contrasfected with cyclin D1, cyclin D1 mutants and SUMO-2 and SUMO-3-expressing plasmids as indicated. (c) Bar graph shows the percentage of cyclin D1 positive cells expressing nuclear cyclin D1 as that seen in (b). Error bars represent the range from three independent experiments. In each experiment 100 cyclin D1 positive cells per sample were counted
Nuclear cyclin D1 is more potent than WT cyclin D1 in rescuing Ras-induced senescence
Cyclin D1 is phosphorylated by glycogen synthase kinase 3β (GSK-3β) on a conserved C-terminal threonine, T-286, during the G1/S transition.5 By mutating T-286 to A-286, the resulting mutant cyclin D1 T286A is refractory to GSK-3β-dependent phosphorylation and CRM export, so is constitutively nuclear. Importantly, transgenic mouse model studies have shown that the nuclear cyclin D1 T286A mutant, but not WT cyclin D1, promotes cell transformation in vitro and tumor growth in nude mice when overexpressed.3, 4, 6, 32
We hypothesized that ASPP2 mediates H-RasV12-induced senescence by preventing the nuclear accumulation of cyclin D1. Cyclin D1 is, therefore, a downstream effecter of ASPP2, and nuclear cyclin D1 should be able to bypass H-RasV12-induced senescence in ASPP2(+/+) MEFs. ASPP2(+/+) MEFs were infected with oncogenic H-RasV12 alone, or in combination with cyclin D1 or cyclin D1 T286A. Protein expression levels and cellular localization of WT and cyclin D1 T286A were then examined via immunoblotting and immunofluuresence staining (Figure 6a and b). The expression of oncogenic H-RasV12 was found to induce endogenous cyclin D1 expression (Figure 6a, compare lanes 1 and 4). This induction was at the mRNA level (Reference 33, and data not shown) and agreed with previous findings. However, H-RasV12 expression did not affect exogenously expressed cyclin D1 or cyclin D1 T286A (Figure 6a, compare lanes 2 and 5 and lanes 3 and 6), allowing us to test whether nuclear cyclin D1 was more potent than WT cyclin D1 in bypassing H-RasV12-induced senescence.
Overexpression of the constitutive nuclear cyclin D1 mutant is sufficient to overcome Ras-induced senescence. WB (a) and indirect immunofluorescence (b) showing expression of exogenous (Ex) and endogenous (End) cyclin D1 and Ras in ASPP2(+/+) MEFs, infected with control, cyclin D1- or cyclin D1-T286A-expressing retrovirus, with or without H-RasV12-expressing retrovirus. ASPP2(+/+) MEFs infected as in (a) were stained for SA-β-Gal activity (c) and BrdU incorporation (d), after 24 h pulse, 1 week after infection. Graphs show the percentage of SA-β-gal-positive cells over the total number cells (e) and the percentage of BrdU-positive cells over the total number of cells (f). Error bars represent the range from three independent experiments
At 7 days after infection, SA-β-Gal staining or 24 h 5-bromo-2-deoxyuridine (BrdU) labelling was used to identify senescent cells. Very few ASPP2(+/+) MEFs underwent senescence when infected with retroviruses expressing either cyclin D1 or cyclin D1 T286A. Under the same conditions, however, around 90% of the cells infected with H-RasV12 alone stained positive for SA-β-Gal. When cells were co-infected with H-RasV12 and WT cyclin D1, the number of SA-β-Gal-positive cells reduced to around 65%. Perhaps most importantly, when H-RasV12 was co-infected with nuclear cyclin D1 T286A, only 25% of the cells stained positive for SA-β-Gal. Interestingly, the SA-β-Gal staining pattern was almost a mirror image of that of BrdU-labelled cells (Figure 6d and f). These results demonstrate that nuclear cyclin D1 is more potent than the WT in bypassing Ras oncogene-induced cellular senescence.
Discussion
By investigating the mechanisms that ASPP2 uses to mediate Ras oncogene-induced senescence, we identified SUMOylation as a novel mechanism that regulates the nuclear localization of cyclin D1. We also showed that nuclear cyclin D1 is more effective than WT cyclin D1 in bypassing Ras-induced cellular senescence.
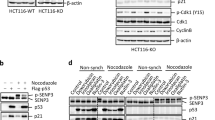
Existing studies have demonstrated that cyclin D1 is exported from the nucleus to the cytoplasm. At G1 phase, cyclin D1 complexes with CDK4 and translocates to the nucleus, to phosphorylate Rb following CDK-activating kinase activation. GSK-3β kinase then enters the nucleus during G1/S transition and phosphorylates cyclin D1 at T-286,5, 34 triggering CRM1-mediated nuclear export of cyclin D1.35 Once T286-phosphorylated cyclin D1 is exported to the cytoplasm, it is ubiquitinated and degraded by the proteosome.32 In contrast, however, very little is known about the mechanism that regulates cyclin D1's nuclear entry. In our attempts to understand how ASPP2 mediates Ras oncogene-induced senescence, we have identified one mechanism by which cyclin D1's nuclear accumulation is regulated. Cyclin D1 is SUMO-modified by SUMO-2/3 (but not SUMO-1), and ASPP2 inhibits Ras oncogene-induced SUMO modification of endogenous cyclin D1 in vivo. Interestingly, SUMO-modified cyclin D1 was nuclear and insoluble, as SUMO-modified cyclin D1 could only be detected when cells were lysed with 8 M urea. This may explain why SUMO-modified cyclin D1 has not previously been identified by conventional immunoprecipitation or kinase assays. The insoluble nature of SUMO-modified cyclin D1 is in agreement with its nuclear localization. To detect SUMO-modified endogenous cyclin D1 in Ras-expressing ASPP2 mutant cells, SUMOylated cyclin D1 was denatured with 8 M urea and the protein was refolded in diluted buffer for immunoprecipitation. This refolding process is likely to only work with a proportion of the population of SUMO-modified cyclin D1, and may be a reason why the mobility shifts of cyclin D1 observed in Figures 2c and 3b are similar but not identical. Furthermore, we do not yet know the molecular mechanisms by which ASPP2 prevents Ras oncogene from inducing SUMO cyclin D1's SUMO modification.
Previous studies have shown that SUMO-modified proteins tend to locate in the nucleus.36 In some proteins, nuclear accumulation by SUMO modification is achieved by interference with nuclear export signal (NES) functions. By modifying a site in close proximity to a NES, SUMO modification may inhibit nuclear export by masking the NES signal. Although the identified K33 of cyclin D1 is not in close proximity to any known cyclin D1 NES, the two lysine residues (K95 and K96) in close proximity to the first NES of cyclin D1 failed to influence its nuclear localization when coexpressed with SUMO-2 and SUMO-3. How SUMO modification at K33 can influence the nuclear localization of cyclin D1, therefore, remains unknown. Future studies are needed to test whether SUMO modification on K33 can mask an NES or expose a nuclear localization signal (if one exists) of cyclin D1, or whether it causes nuclear retention by facilitating cyclin D1's association with nuclear factors or structures. Regardless of how K33 SUMO modification causes cyclin D1's nuclear localization, we show here that SUMOylated nuclear cyclin D1 is active in phosphorylating Rb, as the amount of phosphorylated Rb detected in H-RasV12-expressing ASPP2 mutant MEFs is much higher than that in ASPP2 WT MEFs. This also agrees with the finding that nuclear cyclin D1 is far more potent than the WT in bypassing Ras oncogene-induced senescence.
Mutation of Ras oncogene and overexpression of cyclin D1 occur frequently in human cancers. In colon and breast cancers, for example, mutation of Ras and overexpression of cyclin D1 can be as high as 50%.1, 33, 37 It is also interesting to note that downregulation of ASPP2 in human breast cancers was also observed in over 20% of mutant p53-expressing tumors.18 Furthermore, a large percentage of human tumors overexpress nuclear cyclin D1, and nuclear localization is required for the oncogenic properties of cyclin D1. Thus, it will be interesting to identify whether nuclear cyclin D1 expression is associated with Ras mutation and/or a reduced expression of ASPP2. Importantly, inhibiting cyclin D1's SUMO modification may be beneficial in the treatment of patients expressing high levels of nuclear cyclin D1.
Materials and Methods
Cell culture and genotyping
MEFs were prepared from day 13.5 embryos from XTV-ASPP2(+/Δ3) intercrosses. Genotyping was performed by PCR using primers a (5′-CCTCTCACAAAAGGAAATAACCTG-3′), b (5′-AGGAAAACCACCACCTTCAC-3′) and c (5′-TACCCGCTTCCATTGCTCAG-3′). Primers a and b amplify a 900 base pair –(bp) fragment from the WT allele, and primers b and c amplify a 1100-bp fragment from the knockout allele. p53 MEFs were prepared from day 13.5 embryos from p53 (+/−) intercrosses. All MEFs and U2OS cells were cultured in DMEM supplemented with 10% fetal bovine serum and penicillin-streptomycin (Gibco-BRL, Invitrogen Ltd, Paisley, UK).
Plasmids
pBabe-Puro Ha-RasV12 and pBabe-Bleo Ha-RasV12 were obtained from Julian Downward (Cancer Research UK, London, UK). pBabe-Puro cyclin D1 and pBabe-Puro cyclin D1-T286A were obtained from Alan Diehl (University of Pennsylvania, Philadelphia, PA, USA). pBabe-Puro cyclin D1-K33A, K72A, K95A and K96A were obtained by site-directed mutagenesis. The following primers were used: K33A (GCGGGCCATGCTGGCGGCGGAGGAGACCTGC); K72A (GGTCTGCGAGGAACAGGCGTGCGAGGAGGAGG); K95A (CGCTGGAGCCCGTGGCAAAGAGCCGCCTGCAGC); K96A (CGCTGGAGCCCGTGAAAGCGAGCCGCCTGCAGC). The retroviral vector for ASPP2 was generated by cloning ASPP2 cDNA fragments into pLPCX-Puro using XhoI and NotI restriction enzymes. 6His-tagged SUMO-1, SUMO-2 and SUMO-3 were cloned in pcDNA3 using BamHI and EcoRI restriction enzymes.
Western blot (WB) analysis
WB analysis was performed with lysates prepared 6–8 days after transduction of Ras from subconfluent cells, with urea buffer (8 M urea, 1 M thiourea, 0.5% 3-((3-cholamidopropyl)dimethylammonium)-1-propanesulfonate (CHAPS), 50 mM dithiothreitol (DTT) and 24 mM Spermine) or NP40 lysis buffer (1%NP40, 0.5% sodium deoxycholate in phosphate-buffered saline (PBS)) as described.38 Primary antibodies were from: Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA (p21waf1/cip1(F-5), cyclin E, cyclin A, CDK2), Abcam Inc., Cambridge, MA, USA (tubulin, cyclin D1(DCS-6), p16ink4a, p19Arf), Novocastra/Leica Microsystems GmbH, Wetzlar, Germany (p53(CM-5)), BD Biosciences, Franklin Lakes, NJ USA (Ras, Rb), Upstate Millipore Corporate Headquarters, Billerica, MA, USA (Ras clone 10), Cell Signaling Technology, Inc., Danvers, MA, USA (cyclin D1, CDK4, ubiquitin) and Invitrogen (SUMO-2/3). Anti-ASPP2 antibodies were generated in rabbits and the serum was named S32. To detect p27kip1, we used the mouse monoclonal SX53.G8. Signals were detected using the ECL detection system (GE Healthcare, Pollards Wood, Chalfont, Buckinghamshire, UK).
Indirect immunofluorescence
Cells were fixed in 4% PBS-paraformaldeyde for 15 min, incubated in 0.2% Triton-X-100 for 5 min, then in 0.2% Fish Skin Gelatine in PBS for 10 min and stained for 30 min with an anti-cyclin D1 antibody (Abcam), anti-His (Santa Cruz), anti-BrdU fluorescein isothiocyanate (BD bioscience) or anti-p21waf1/cip1 (SX118). Antibodies were used at 1 : 50 dilution in 0.2% Fish Skin Gelatine-PBS. Staining with the secondary antibody was performed as previously described,39 followed by visualization under a fluorescence microscope. All the procedures were performed at 20°C.
Analysis of senescence
Analysis of senescence in cell culture was performed using a Senescence β-galactosidase staining kit (Cell Signaling) as described previously.38 To quantify SA-β-gal positives, at least 200 cells were counted in random fields in each of the duplicated wells.
Transformation assays
A total of 105 ASPP2(Δ3/Δ3) and ASPP2(+/+) MEFs were infected with pBABE-Puro H-RasV12 or empty vector retroviruses. Cells were selected with puromycin for 4 days, and plates were then fixed and stained with Giemsa 4 weeks later. For anchorage-independent growth, 5 × 104 cells transduced with pBABE-Puro H-RasV12 were resuspended in medium containing 0.3% low-melting-point agarose and plated onto solidified bottom layer medium containing 0.5% agarose. Colonies were photographed and counted after 3–4 weeks.
Immunoprecipitations
Cells were lysed in urea buffer (8 M urea, 1 M thiourea, 0.5% CHAPS, 50 mM DTT and 24 mM Spermine), after which extracts were diluted 10-fold in NP40 lysis buffer (1%NP40, 0.5% sodium deoxycholate in PBS) and precleared with 60 μl of protein G beads (50% slurry) for 60 min at 4°C. For each immunoprecipitation, 25 μl anti-cyclin D1 mAb (Abcam) or IgG was used, 25 μl of 50% slurry of protein G beads were added and then samples were rotated at 4°C overnight. Beads were washed five times with 1 ml of 10-fold diluted urea buffer and 20 μl of 2 × SDS loading buffer was added. Immunoprecipitations were analyzed by WB as indicated.
Purification of 6His-tagged SUMO-cyclin D1 conjugates
At 36 h post-transfection, cells were washed twice with PBS and scraped in 1 ml of PBS. In all, 20% of the cell suspension was used for direct WB analysis. The remainder was lysed in 6 ml of 6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 8.0, 5 mM imidazole and 10 mM β-mercaptoethanol. A total of 75 μl of Ni2+-NTA-agarose beads (Qiagen, Fleming Way, Crawley, West Sussex, UK) was then added and lysates were rotated at 20°C for 4 h. The beads were successively washed for 5 min in each step at 20°C with 750 μl of each of the following buffers: 6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 8.0, 10 mM β-mercaptoethanol; 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH8.0, 10 mM β-mercaptoethanol; 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 6.3, 10 mM β-mercaptoethanol (buffer A) plus 0.2% Triton X-100; buffer A and then buffer A plus 0.1% Triton X-100. After the last wash, 6His-tagged SUMOylated products were eluted by incubating beads in 75 μl of 200 mM imidazole, 0.15 M Tris/HCl pH6.7, 30% glycerol, 0.72 M β-mercaptoethanol, 5% SDS for 20 min at 20°C. Eluates were mixed at a 1 : 1 ratio with 2 × Laemmli-buffer and analyzed by WB.
Abbreviations
- Arf:
-
alternate reading frame
- ASPP:
-
apoptosis-stimulating protein of p53
- bp:
-
base pair
- BrdU:
-
5-bromo-2-deoxyuridine
- CDK:
-
cyclin-dependent kinase
- CHAPS:
-
3-((3-cholamidopropyl)dimethylammonium)-1-propanesulfonate
- DTT:
-
dithiothreitol
- GSK-3β:
-
glycogen synthase kinase 3 β
- h:
-
hours
- K:
-
lysine
- MEFs:
-
mouse embryonic fibroblasts
- MMTV:
-
mouse mammary tumor virus
- MW:
-
molecular weight
- NES:
-
nuclear export signal
- PBS:
-
phosphate-buffered saline
- Rb:
-
retinoblastoma protein
- SA-β-Gal:
-
senescence-associated β-galactosidase
- SENP2:
-
sentrin-specific protease 2
- SUMO:
-
small ubiquitin-like modifier
- T:
-
threonine
- WB:
-
western blot
- WT:
-
wild type
- YAP:
-
YES-associated protein
References
Barnes DM, Gillett CE . Cyclin D1 in breast cancer. Breast Cancer Res Treat 1998; 52: 1–15.
Shamma A, Doki Y, Shiozaki H, Tsujinaka T, Yamamoto M, Inoue M et al. Cyclin D1 overexpression in esophageal dysplasia: a possible biomarker for carcinogenesis of esophageal squamous cell carcinoma. Int J Oncol 2000; 16: 261–266.
Quelle DE, Ashmun RA, Shurtleff SA, Kato JY, Bar-Sagi D, Roussel MF et al. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes & Dev 1993; 7: 1559–1571.
Resnitzky D, Gossen M, Bujard H, Reed SI . Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol 1994; 14: 1669–1679.
Diehl JA, Cheng M, Roussel MF, Sherr CJ . Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes & Dev 1998; 12: 3499–3511.
Alt JR, Cleveland JL, Hannink M, Diehl JA . Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes & Dev 2000; 14: 3102–3114.
Gladden AB, Woolery R, Aggarwal P, Wasik MA, Diehl JA . Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene 2006; 25: 998–1007.
Lin DI, Lessie MD, Gladden AB, Bassing CH, Wagner KU, Diehl JA . Disruption of cyclin D1 nuclear export and proteolysis accelerates mammary carcinogenesis. Oncogene 2008; 27: 1231–1242.
Johnson ES . Protein modification by SUMO. Annu Rev Biochem 2004; 73: 355–382.
Saitoh H, Hinchey J . Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 2000; 275: 6252–6258.
Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH et al. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics 2008; 7: 132–144.
Anckar J, Sistonen L . SUMO: getting it on. Biochem Soc Trans 2007; 35 (Pt 6): 1409–1413.
Lloyd AC . Ras versus cyclin-dependent kinase inhibitors. Curr Opin Genet Dev 1998; 8: 43–48.
Pantoja C, Serrano M . Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene 1999; 18: 4974–4982.
Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW . Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997; 88: 593–602.
Sebastian T, Malik R, Thomas S, Sage J, Johnson PF . C/EBPbeta cooperates with RB:E2F to implement Ras(V12)-induced cellular senescence. EMBO J 2005; 24: 3301–3312.
Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW et al. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol 2009; 11: 1451–1457.
Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 2001; 8: 781–794.
Vives V, Su J, Zhong S, Ratnayaka I, Slee E, Goldin R et al. ASPP2 is a haploinsufficient tumor suppressor that cooperates with p53 to suppress tumor growth. Genes & Dev 2006; 20: 1262–1267.
Kampa KM, Acoba JD, Chen D, Gay J, Lee H, Beemer K et al. Apoptosis-stimulating protein of p53 (ASPP2) heterozygous mice are tumor-prone and have attenuated cellular damage-response thresholds. Proc Natl Acad Sci USA 2009; 106: 4390–4395.
Liu ZJ, Lu X, Zhang Y, Zhong S, Gu SZ, Zhang XB et al. Downregulated mRNA expression of ASPP and the hypermethylation of the 5′-untranslated region in cancer cell lines retaining wild-type p53. FEBS Letters 2005; 579: 1587–1590.
Lossos IS, Natkunam Y, Levy R, Lopez CD . Apoptosis stimulating protein of p53 (ASPP2) expression differs in diffuse large B-cell and follicular center lymphoma: correlation with clinical outcome. Leuk Lymphoma 2002; 43: 2309–2317.
Yang JP, Hori M, Takahashi N, Kawabe T, Kato H, Okamoto T . NF-kappaB subunit p65 binds to 53BP2 and inhibits cell death induced by 53BP2. Oncogene 1999; 18: 5177–5186.
Naumovski L, Cleary ML . The p53-binding protein 53BP2 also interacts with Bc12 and impedes cell cycle progression at G2/M. Mol Cell Biol 1996; 16: 3884–3892.
Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D . Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J 1997; 16: 1876–1887.
Espanel X, Sudol M . YES-associated protein and p53-binding protein-2 interact through their WW and SH3 domains. J Biol Chem 2001; 276: 14514–14523.
Nakagawa H, Koyama K, Murata Y, Morito M, Akiyama T, Nakamura Y . APCL, a central nervous system-specific homologue of adenomatous polyposis coli tumor suppressor, binds to p53-binding protein 2 and translocates it to the perinucleus. Cancer Res 2000; 60: 101–105.
Chen Y, Liu W, McPhie DL, Hassinger L, Neve RL . APP-BP1 mediates APP-induced apoptosis and DNA synthesis and is increased in Alzheimer's disease brain. J Cell Biol 2003; 163: 27–33.
Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, Nakagawa H et al. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell 2008; 14: 68–78.
Okabe H, Lee SH, Phuchareon J, Albertson DG, McCormick F, Tetsu O . A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PloS One 2006; 1: e128.
Xue Y, Zhou F, Fu C, Xu Y, Yao X . SUMOsp: a web server for SUMOylation site prediction. Nucleic Acids Res 2006; 34 (Web Server issue): W254–W257.
Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell 2006; 24: 355–366.
Downward J . Targeting RAS signalling pathways in cancer therapy. Nat Rev 2003; 3: 11–22.
Diehl JA, Zindy F, Sherr CJ . Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev 1997; 11: 957–972.
Benzeno S, Diehl JA . C-terminal sequences direct cyclin D1-CRM1 binding. J Biol Chem 2004; 279: 56061–56066.
Rodriguez MS, Dargemont C, Hay RT . SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 2001; 276: 12654–12659.
Bartkova J, Lukas J, Muller H, Lutzhoft D, Strauss M, Bartek J . Cyclin D1 protein expression and function in human breast cancer. Int J Cancer 1994; 57: 353–361.
Wang W, Chen JX, Liao R, Deng Q, Zhou JJ, Huang S et al. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol Cell Biol 2002; 22: 3389–3403.
Slee EA, Gillotin S, Bergamaschi D, Royer C, Llanos S, Ali S et al. The N-terminus of a novel isoform of human iASPP is required for its cytoplasmic localization. Oncogene 2004; 23: 9007–9016.
Acknowledgements
We would like to thank the Ludwig Institute for Cancer Research, EU and AICR for supporting this work and Dr Claire Beveridge for her help in preparing the paper. EL was partially supported by an FIRC fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Knight
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, X., Lapi, E., Sullivan, A. et al. SUMO-modified nuclear cyclin D1 bypasses Ras-induced senescence. Cell Death Differ 18, 304–314 (2011). https://doi.org/10.1038/cdd.2010.101
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2010.101
Keywords
This article is cited by
-
Senolytic Effect of Cerium Oxide Nanoparticles (CeO2 NPs) by Attenuating p38/NF-кB, and p53/p21 Signaling Pathways
Journal of Cluster Science (2022)
-
ASPP1 deficiency promotes epithelial-mesenchymal transition, invasion and metastasis in colorectal cancer
Cell Death & Disease (2020)
-
PIK3R3 inhibits cell senescence through p53/p21 signaling
Cell Death & Disease (2020)
-
SUMO and the robustness of cancer
Nature Reviews Cancer (2017)
-
Nuclear iASPP may facilitate prostate cancer progression
Cell Death & Disease (2014)