Abstract
Overexpression of BMI1 correlates with cancer development, progression, and therapy failure; however, the underlying molecular mechanisms remain to be fully elucidated. Using the C666-1 nasopharyngeal cancer (NPC) model, the role of BMI1 in mediating response of NPC cells to radiation therapy (RT) was investigated. The results showed a novel radioresistance function for BMI1 in NPC, wherein BMI1 depletion sensitized NPC cells to RT. Cell cycle analysis and transmission electron microscopy (TEM) showed apoptosis as the major mode of cell death, and the mitochondria as a primary targeted cellular organelle. Genome-wide microarray and pathway analyses revealed that the P53 pathway is a critical mediator of this process. Cotransfection with siP53 rescued C666-1 cells from cytotoxicity upon BMI1 depletion and RT, thereby corroborating the role for P53. Pretreatment with the antioxidant, Trolox, inhibited apoptosis, indicating that production of reactive oxygen species (ROS) is also mediating cytotoxicity. In vivo, BMI1 depletion combined with RT abrogated tumor-forming capacity in SCID mice, showing the relevance of this process in a more complex tumor environment. Hence, we show a novel role for BMI1 in conferring radioresistance in cancer cells through the downregulation of p53-mediated apoptosis. These results suggest a potential strategy of BMI1 depletion combined with RT for tumors wherein BMI1 appears to be driving disease progression.
Similar content being viewed by others
Main
Over the past several years, the polycomb group (PcG) family of genes has emerged as key regulators of cancer development and progression.1 PcG proteins were first described in Drosophila, with subsequent identification of their mammalian homologs,2 which function in several multimeric polycomb repressive complexes, such as PRC1, PRC2, PRC3, or PRC4.3 PRC1, also known as the ‘maintenance complex’, contains the BMI1 proto-oncogene.4 In embryonic and adult stem cells, PcG genes maintain cellular identity by repressing transcription of key developmental genes, such as homeobox (Hox) genes, and by preventing senescence through repression of the INK4A locus.5, 6 In addition to their role during development, several members of the PcG gene family are overexpressed in different cancer types. In particular, several studies correlated BMI1 overexpression with disease progression and poor clinical outcome.7, 8, 9 The precise molecular mechanism by which BMI1 contributes to cancer development and therapy failure, however, remains to be fully elucidated; some studies have suggested repression of the INK4A locus and induction of hTERT as possible mechanisms.10, 11
Nasopharyngeal carcinoma (NPC) represents a subset of head-and-neck epithelial cancers, which is treated by radiation therapy (RT) for early disease, or combined with chemotherapy for locally advanced NPC. Despite progress in NPC management, a significant proportion of NPC patients (43%) will still develop distant metastases within 2 years,12 underscoring the need for an improved understanding of the molecular mechanisms underlying therapy failure.
Overexpression of BMI1 has been previously associated with reduced survival in NPC patients treated with RT.7 In this study, we evaluated the expression of BMI1 in primary NPC tissues and xenograft tumor models, and also investigated the role of BMI1 in resistance of NPC cells to RT. Our data showed the overexpression (⩾2-fold) of BMI1 transcript in 70% of primary NPC specimens, and 100% of xenograft tumors. Importantly, a novel role for BMI1 in conferring radioresistance in NPC was identified, wherein BMI1 depletion significantly increased radiation sensitivity of NPC cells. Genome-wide microarray analysis of cells, depleted of BMI1 and exposed to RT, combined with experimental validation showed that the p53 pathway and reactive oxygen species (ROS) production are critical mediators of this process. Thus, our data are the first to elucidate a molecular mechanism by which BMI1 confers radioresistance in NPC.
Results
BMI1 is overexpressed in primary NPC tissues
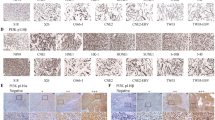
BMI1 expression was evaluated in 17 primary human NPC biopsy specimens, and in 3 NP xenograft tumors, showing the overexpression (⩾2-fold) of BMI1 transcript in 12 out of 17 primary biopsies, and in all 3 xenograft tumors (Figure 1a). Overexpression of BMI1 protein in primary NPC samples was confirmed by immunohistochemistry (Figure 1b), whereas adjacent normal cells showed negative or low expression of BMI1. Using immunofluorescent microscopy, the C666-1 NPC cells showed nuclear expression of BMI1 (Figure 1c).
BMI1 is overexpressed in primary NPC specimens and xenograft tumors. (a) Quantitative real-time PCR analysis of BMI1 transcript in 17 primary NPC specimens, three xenograft tumors (C666-1, C15, and C17), compared with that of pooled control RNA, derived from three patients' normal nasopharyngeal epithelial tissues. Data are presented as box and whiskers, with the mean and error bars indicated; *P<0.05; ***P<0.0005. (b) Immunohistochemistry of BMI1 expression in a representative formalin-fixed and paraffin-embedded NPC biopsy. Black arrows indicate nuclear expression of BMI1; T, tumor; N, normal. (c) Immunofluorecence overlay of BMI1 (green) and nuclear (blue) staining, showing nuclear expression of BMI1 in C666-1 cells
BMI1 knockdown sensitizes C666-1 cells to radiation
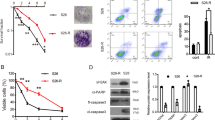
BMI1 expression has been reported to be associated with significantly reduced patient survival after RT with or without chemotherapy,7 implying that BMI1 overexpression might be mediating radioresistance. To address this possibility, C666-1 cells were transfected with small interfering RNA targeting BMI1 (siBMI1), or control siRNA (siCtrl). SiBMI1 showed >90% reduction in BMI1 transcript level, observed at both 48 and 72 h post-transfection (Figure 2a), which resulted in undetectable BMI1 protein expression (Figure 2b). Persistent knockdown of BMI1 transcript was also observed on day 4 and day 6 post-transfection (Supplementary Figure 1). Under those experimental conditions, C666-1 cells were equally transfectable with siCtrl and siBMI1 (Supplementary Figure 2). BMI1 knockdown alone was associated with only a modest reduction in cell viability (Figure 2c), but when combined with RT (6 Gy), a significant increase in cytotoxicity was observed, with 32% viability for siBMI1 plus RT versus 65% viability for siCtrl plus RT (P=0.0002). A dose-dependent increase in radiation cytotoxicity was observed following transfection with increasing concentration of siBMI1 (Figure 2d), which was determined to be a synergistic interaction on further analysis (Supplementary Figure 3).
BMI1 knockdown combined with RT reduces C666-1 viability. (a) Representative qRT-PCR analysis of BMI1 mRNA expression level in C666-1 cells at 48- and 72 h post-transfection with siCtrl, or siBMI1 (30 nM). The expression level is normalized to that of siCtrl cells, presented as the mean±S.D. from two replicates. (b) Western blot (WB) analysis of BMI1 protein expression in C666-1 cells 72 h post-transfection with 30 nM siBMI1 or siCtrl. GAPDH was used as loading control. (c) MTS viability assay for C666-1 cells transfected with 30 nM siBMI1 or siCtrl, followed by 0 or 6 Gy RT (at 72 h), measured at 4 days (open bar) or 6 days (solid bar) post-transfection. The data are normalized to viability of siCtrl-transfected cells, presented as mean±S.E. from three independent experiments, each conducted in triplicate; ***P<0.0005. (d) Viability of C666-1 cells transfected with increasing concentration (0–60 nM) of siBMI1 or siCtrl (for 72 h), then exposed to 0 (open symbols) or 6 Gy (half-filled symbols) RT, with the MTS assay conducted 6 days post-transfection. Viability is normalized to that of mock-transfected cells; the data are presented as mean±S.E.; n=10 from two independent experiments
BMI1 knockdown in combination with RT induces apoptosis
Cell cycle analysis was then conducted on C666-1 cells, which showed a 15% sub-G0 population (apoptosis) in siBMI1, compared to 4% in siCtrl-treated cells, 4 days post-transfection (Figure 3a). When BMI1 knockdown was combined with RT, the proportion of apoptotic cells increased to 22%, while mock or siCtrl treated cells combined with RT showed no significant change in the apoptotic fraction. This increase in apoptosis was associated with induction of caspase-3–7 activity by >1.5-fold, when BMI1 depletion was combined with RT, observed as early as 24 h post-RT (Figure 3b). Induction of apoptosis was also confirmed by DAPI staining and visualization of cells showing morphologic features of apoptosis (data not shown). Consistent with these data, overexpression of BMI1 in normal oral epithelial (NOE) cells led to an increase in radiation resistance compared with cells transfected with control plasmid (Supplementary Figure 4).
BMI1 knockdown combined with RT induces apoptosis and γ-H2AX expression. (a) Cell cycle analysis was conducted on day 4 for C666-1 cells transfected with 30 nM siBMI1 or siCtrl (for 72 h), then exposed to RT (0 Gy (upper panel) or 6 Gy (lower panel). The proportion of cells in the sub-G0 is indicated on each plot. The data are representative of at least three independent experiments. (b) Caspase-3–7 activity was measured in cells transfected with 30 nM siCtrl or siBMI1 for 48 h, and then exposed to 0 or 4 Gy RT. The data are presented as the ratio of caspase activation of siBMI1/siCtrl; with (hatched bar) or without RT (open bar). Each data point represents the mean±S.E. from three independent experiments; n=4–6. (c) The percentage of γ-H2AX-positive C666-1 cells transfected with 30 nM siBMI1 or siCtrl (for 48 h), then exposed to 4 Gy RT, were determined, tracked for up to 24 h post-RT. The data are presented as mean±S.E. from two independent experiments, each performed in duplicate. (d) Fluorescent microscopy for γ-H2AX staining (left panel) in siBMI1-treated cells from (c), at 30 min and 24 h post-RT. Cells were costained with Hoechst 33342 to visualize nuclear DNA (middle panel); the overlay of γ-H2AX with Hoechst 3342 is shown in the merged panel (far right), showing that γ-H2AX was only observed in the cell with the nuclear fragmentation (white arrow)
BMI1 depletion induced increase in phosphorylated histone H2AX (γ-H2AX) staining following RT
γ-H2AX is commonly utilized to assess DNA double strand breaks (DSBs) inflicted by RT.13 When C666-1 cells were depleted of BMI1, 15 min after RT exposure, γ-H2AX staining (Figure 3c) was observed in 18% of cells, versus 11% for the siCtrl plus RT population (P=0.05). Recovery kinetics of DNA repair, as indicated by loss of γ-H2AX staining over time, was similar between the siBMI1- and siCtrl-treated cells except at 24 h post-RT, wherein a second peak of γ-H2AX staining was observed only in the siBMI1-treated cells (17 versus 7% for the siCtrl-treated cells; P=0.01). When visualized in greater detail using fluorescent microscopy, it was evident that at 24 h post-RT, γ-H2AX expression was only observed in the cells undergoing apoptosis, as shown by chromatin condensation and DNA fragmentation (Figure 3d). This phenomenon has been reported by others wherein γ-H2AX staining can be observed during the early stages of apoptosis.14
Transmission electron microscopy (TEM) showed apoptosis and mitochondrial death in BMI1-depleted cells, and exposed to RT
To acquire greater cellular detail on the effects of these treatments, TEM was conducted, showing normal subcellular morphology and healthy mitochondria for the siCtrl plus RT cells at both 24 and 48 h post-RT (Figure 4a and c). In contrast, siBMI1 plus RT-treated cells exhibited early stages of apoptosis manifested by nuclear and mitochondrial condensation, although the nuclear membrane remained intact (Figure 4b). There were progressive aberrations observed at 48 h, whereby siBMI1 plus RT cells showed a marked increase in chromatin condensation, nuclear and mitochondrial swelling, and formation of intracytoplasmic vacuoles, associated with loss of nuclear membrane integrity (Figure 4d).
Transmission electron microscopy (TEM) images of C666-1 cells treated with siCtrl,or siBMI1 with RT. C666-1 cells were transfected with siCtrl (a, c) or siBMI1 (b, d) for 48 h, then exposed to 4 Gy RT. TEM was utilized to visualize these cells at 24 (a, b), or 48 h (c, d) post-RT. (a) siCtrl plus RT cells exhibited healthy cellular and subcellular morphology (left panel) with normal appearing mitochondria (right panel, arrow), and intact nuclear membrane (right panel, arrowhead). (b) At 24 h post-RT, siBMI1 plus RT cells exhibited early stages of apoptosis manifested by nuclear (middle panel), and mitochondrial (right panel, arrow) chromatin condensation, although the nuclear membrane remained intact (right panel, arrowhead). (c) At 48 h, siCtrl plus RT showed normal appearing cells (left and middle panels), normal mitochondria (right panel, arrow), and intact nuclear membrane (right panel, arrowhead). (d) At 48 h post-RT, most siBMI1-treated cells showed late stages of apoptosis manifested by nuclear chromatin condensation, nuclear swelling, and decreased density of cytoplasm (left and middle panels). Mitochondrial swelling (right panel, arrow), formation of intracellular vacuoles (right panel, asterisk), and loss of nuclear membrane integrity (right panel, arrowhead) were also observed
Microarray analysis revealed significant enrichment for the p53 pathway in siBMI1 plus RT cells
To elucidate the molecular mechanisms underlying this process, wherein BMI1 depletion enhanced radiation sensitivity, microarray analyses were conducted on BMI1-depleted cells (for 48 h), with or without RT (4 Gy). Total RNA was collected from such treated cells on day 3, then hybridized onto the Affymetrix array (HG-U133 plus 2.0). Using a twofold cutoff compared with control cells, several hundred genes were differentially expressed upon BMI1 depletion, with or without the addition of RT (Supplementary Tables 1 and 2). As BMI1 is a transcriptional repressor, we focused on transcripts that were upregulated in either the siBMI1 or siBMI1 plus RT cells (Figure 5a). BMI1 depletion alone upregulated 14 transcripts by ⩾2-fold, compared with control cells, which upregulated 505 genes upon the addition of RT (Figure 5a, right circle); in contrast, RT alone upregulated only 23 genes. Quantitative real-time PCR (qRT-PCR) was conducted to validate the expression level of four selected transcripts (TP53I3, CCNG1, LAMP2, and CRI1) identified from the microarray data; corroborating their respective over or underexpressions (Figure 5b). Pathway analysis of the differentially expressed genes in the siBMI1 plus RT group revealed significant enrichment for genes in the P53 signaling pathway (P=7.7 × 10−7, Figure 5c). Several of these P53 targets are known to be involved in induction of apoptosis and cell cycle regulation (Figure 5d). We sought to determine whether expression of these P53 target genes was critically dependent on the presence of P53. To that end, C666-1 cells were transfected with siCtrl, siBMI1, or siBMI1 plus siP53, then exposed to RT to examine for expression of representative P53 targets (DKK1 and TP53I3). Figure 5e shows that co-transfection of siBMI1 and siP53 with RT led to significant reduction in DKK1 and TP5313 expression level (below basal), compared with siBMI1 plus RT. Knockdown of p53 is corroborated by undetectable P53 protein level using western blotting (Figure 5f). Taken together, these data clearly indicate that P53 is a critical mediator of cytotoxicity in NPC cells treated with siBMI1 plus RT.
Genome-wide microarray analysis reveals enrichment for genes in the P53 pathway in BMI1-depleted C666-1 cells and exposed to RT. (a) Treeview representation (left panel) of microarray expression data filtered on a twofold change in siBMI1 plus RT sample compared with control cells showing upregulation (red color) in the majority of transcripts. Venn diagram (right panel) depicting the overlap in genes that were upregulated ⩾2.0-fold in siBMI1, siBMI1 plus RT (4 Gy), and Ctrl plus RT (4 Gy) cells. (b) Validation of microarray data were conducted using qRT-PCR to measure the expression levels of TP53I3, CCNG1, LAMP2, and CRI1, between the siBMI1 plus RT, relative to siCtrl plus RT cells after 72 h transfection. (c) Pathway enrichment analysis for genes upregulated in the siBMI1 plus RT group as a function of specific signaling cascade, the percentage of genes, and the respective P-value, ranked in decreasing statistical significance. (d) Heatmap showing genes in the P53 pathway that were upregulated in cells treated with siBMI1 plus RT, and were significantly enriched (from c); * Additional P53 transcriptional targets which are not part of the KEGG P53 pathway.40 (e) C666-1 cells were treated with optiMEM, optiMEM plus RT (6 Gy), or transfected with 30 nM of siCtrl, siBMI1, or siBMI1 plus siP53, all with RT (6 Gy) delivered after 48 h. qRT-PCR for the GAPDH, DKK1, TP53I3, or p53 was performed on day 3 post-transfection. (f) C666-1 cells were treated as in (e), followed by total protein extraction, and P53 expression was detected using western blotting on day 3. GAPDH was used as loading control. O, optiMEM; C, siCtrl; B, siBMI1; B+P, siBMI1 plus siP53
Induction of cell death in BMI1 knockdown plus RT is dependent on P53, and involves production of ROS
To further assess the involvement of P53 in mediating cytotoxicity in cells depleted of BMI1 and exposed to RT, siRNA was utilized to simultaneously knockdown the expression of BMI1 and P53 before exposure to RT. As shown in Figure 6a, transfection of C666-1 cells with siP53 followed by RT led to a slight increase in viability compared to transfection with siCtrl. Importantly, siP53 co-knockdown with siBMI1 before RT rescued C666-1 cells, thereby corroborating the role of P53 in mediating cytotoxicity. To further investigate the role of P53 in mediating cell death or apoptosis, further experiments were conducting using the human UT-SCC-8 laryngeal squamous cancer cell line, which does not express P53 by western blot (Figure 6b). Transfection of UT-SCC-8 cells with siBMI1 led to a substantial reduction in BMI1 transcript level (Figure 6c); yet, no difference in apoptosis was observed with siCtrl or siBMI1-treated cells plus RT (Figure 6d). Concordantly, no difference in cell viability was observed (data not shown). Taken together, these data supported the conclusion that induction of apoptosis or cell death in siBMI1 plus RT C666-1 cells is dependent upon an intact P53.
Cytotoxicity in BMI1-depleted C666-1 cells plus RT is mediated through P53 and ROS production. (a) Viability was assessed for C666-1 cells treated with optiMEM (O), optiMEM plus RT (O+RT), or transfected with 30 nM of siCtrl, siBMI1, or siBMI1 plus siP53 for 72 h, then followed by RT (6 Gy). The MTS viability assay was conducted 6 days post-transfection. The data are presented as mean±S.E. from two independent experiments, n=6. (b) Western blot for basal P53 expression in C666-1 and UT-SCC-8 cells. GAPDH served as loading control. (c) qRT-PCR analysis for BMI1 expression in UT-SCC-8 cells transfected with 30 nM siCtrl or siBMI1 at 48 h post-transfection. Data are presented as mean±S.D.; n=3 (d) Percent apoptosis in UT-SCC-8 cells transfected with 30 nM siCtrl or siBMI1, then exposed to RT (6 Gy) after 48 h. Apoptosis was assayed using flow cytometry at 24 h post-RT. Data are presented as mean±S.D.; n=3. (e) C666-1 cells were transfected with 30 nM siBMI1 or siCtrl, then on days 2 and 3; cells were treated with the indicated doses of Trolox or DMSO before exposure to 6 Gy RT. Apoptosis was measured on day 4 using flow cytometry as described previously (Figure 3a). Data are presented as mean±S.D. from two independent experiments, n=2. (f) C666-1 cells were transfected with 30 nM siBMI1 or siCtrl (for 72 h), exposed to 0 or 6 Gy, and then implanted into the left gastrocnemius muscle of SCID mice on day 6 post-transfection. Tumor growth was monitored by measuring leg plus tumor diameter over time; the data are presented as mean±S.E. for each group of mice, each arm consisted of five mice per group. (g) Mice in (f) were monitored for survival for up to 6 months. All mice were killed upon reaching the humane end point, and not from any other reason. (h) A proposed model for induction of cell death in C666-1 cells depleted of BMI1 and exposed to RT. RT exposure leads to activation of P53, transcription of several P53 proapoptotic target genes, production of ROS, and induction of apoptosis. In the presence of BMI1 and the PRC1 complex, transcription of those proapoptotic genes, and production of ROS is inhibited, thus rendering cells resistant to RT-induced apoptosis; *P<0.05; **P<0.005; n.s, not significant
One major mechanism of RT-induced cytotoxicity is mediated through production of ROS.15 Indeed, several upregulated genes in siBMI1 plus RT-treated cells are involved in oxidative stress (PXDN, TP53I3, SESN2, SESN1, CLN8, and HMOX1). To assess whether induction of apoptosis in the C666-1 system is mediated through the production of ROS, the ability of Trolox,16 a widely used antioxidant, to inhibit apoptosis was examined. As shown in Figure 6e, treating C666-1 cells with 0.2 or 1.0 mM Trolox before RT significantly reduced apoptosis in siBMI1 plus RT-treated cells to 18 and 15%, respectively, in contrast to 22% for pre-RT exposure to DMSO, showing that ROS production also mediated apoptosis in this process.
BMI1 depletion and RT completely abrogates tumor-forming potential in C666-1 cells
To assess the long-term consequences of BMI1 knockdown in NPC, C666-1 cells were transfected with siBMI1 or siCtrl, and then exposed to 0 or 6 Gy RT, followed by implantation into SCID mice, to monitor the tumor growth. Cells depleted of BMI1 showed no difference in tumor-forming capacity compared with siCtrl treated cells (Figure 6f), consistent with the modest in vitro effect (Figure 2c). Radiation delayed tumor formation by approximately 40 days; remarkably, BMI1 depletion combined with RT completely prevented tumor formation in C666-1 cells, followed for more than 6 month, translating into a 100% survival rate (Figure 6g).
Discussion
Despite recent advances in cancer management and therapy, local or distant tumor recurrences remain a major clinical challenge. Several studies have correlated the overexpression of various members of the polycomb gene family with cancer progression and therapy failure; however, the precise molecular mechanisms by which PcG members mediate their role in cancer remains to be elucidated. Given that >80% of NPC worldwide are EBV-positive, these experiments were conducted using the C666-1 NPC line, the only EBV-positive NPC line available. Our data showed a novel function for the BMI1 polycomb gene in conferring radioresistance in NPC, through repression of p53-mediated apoptosis upon DNA damage. NPC cells normally do not undergo apoptosis when exposed to RT (Figure 3a); however, when BMI1 was depleted from the C666-1 cells, this led to significant increase in cytotoxicity and apoptosis, particularly upon RT exposure (Figure 2c and 3a). Thus, our data are consistent with recent reports of apoptosis induction in tumor cells following BMI1 knockdown and 5-FU treatment.17, 18 Some of the proposed mechanisms include re-expression of p16 or signaling through PI3K. To that end, we examined P16 expression in C666-1 cells depleted of BMI1. No induction of p16 transcript was observed (data not shown); thus, p16 is unlikely to be involved in mediating cell death in NPC. This was not surprising given that several tumor cell lines (including the C666-1 NPC cells) have p16 silenced through promoter hypermethylation.19 Consistent with this notion, quantitative methylation analysis showed no alteration in p16 promoter methylation status following BMI1 depletion (data not shown). Similarly, no induction of the P14arf transcript was observed under these experimental conditions.
The genome-wide microarray analysis was undertaken to acquire insight into downstream effects of BMI1 depletion in C666-1 cells. To date, only a few studies have examined global gene expression following BMI1 knockdown;20, 21 none has conducted such experiments in combination with DNA-damaging agents such as RT. When human embryonic fibroblasts were depleted of PcG genes, only a small fraction (∼10%) of PcG targets were transcriptionally activated,20 consistent with the current report wherein only 14 transcripts were upregulated (⩾2-fold) in BMI1-depleted C666-1 cells (Figure 5a, b, and Supplementary Table 1). In contrast, when cells were additionally exposed to RT, a significantly greater number of transcripts (505) were induced (Figure 5a, b, and Supplementary Table 2), with several being well-known transcriptional targets of p53, integral to the p53 response induced by DNA damage (Figure 5c and d). Thus, our data support a model in which depletion of PcG genes on its own is not sufficient to induce the re-expression of PcG-repressed genes, whereas an induction stimulus appears to be necessary for the active transcription of some PcG target genes. Several of the differentially expressed genes in our study (ATF3, BMP2, and so on) were also induced following BMI1 depletion in human embryonic cells, corroborating the published microarray data.20 The presence of P53 and induction of oxidative stress appeared to be important mediators of cell death inflicted by BMI1 depletion combined with RT (Figure 6a-e). It has previously been reported that signaling through the P53 pathway can lead to apoptosis under stress conditions (such as RT) through transcription of stress response genes, such as TP53I3, PUMA, SESN1, and SESN2.22 Several of those genes were also induced in our study, corroborating the role for p53 in inducing cytotoxicity through oxidative damage. Interestingly, BMI1 knockdown did not induce any increase in P53 expression at either the transcriptional or protein levels (Figure 5e and f); thus, it is likely that BMI1 is modulating P53 function post-transcriptionally, or possibly by repressing P53 targets through the PRC1 complex (Figure 6h). Whether overexpression of BMI1 will lead to similar repression of those targets in normal cells, however, remains to be investigated. Nonetheless, our data are in agreement with a recent report documenting a similar function for BMI1 in repressing P53-dependent apoptosis in neuronal cells, corroborating our current findings in a different system.23
Several studies have documented the overexpression of BMI1 in the tumor-initiating cells (TI-C) of leukemia, brain, breast, and head-and-neck cancers.24, 25, 26, 27 The C666-1 cell line has an undifferentiated phenotype, and expresses several stem cell-related genes, such as CD133 (PROM1), CD44, BMI1, and maternal embryonic leucine zipper kinase (MELK; data not shown). Thus, it could be postulated that BMI1 might be conferring radiation resistance in the subset of TI-Cs. In fact, it has recently been shown that the TI-C in a brain tumor model are the subpopulation that survives RT by undergoing transient cell cycle arrest, whereas the remaining bulk population undergoes P53-dependent apoptosis.28 Similarly, C666-1 cells undergo G1 and G2M arrest in response to RT; yet, only a small fraction, if any, undergoes apoptosis (Figure 3a). In contrast, BMI1-depleted cells were more prone to apoptotic cell death in response to RT. Similarly, the TI-C from breast cancer cells are also resistant to RT through reduction in ROS levels.29 In our system, ROS also appears to be a component of the underlying mechanism leading to cell death when depleted of BMI1 then exposed to RT. Thus, our data corroborate the published reports in the literature, and provide a possible mechanism leading to radiation resistance in the TI-C cells, although this remains to be proven. Importantly, the combination of BMI1 depletion and RT completely abrogated tumor-forming capacity of C666-1 cells (Figure 6f and g), which is not often observed in this tumor model using an siRNA approach.30 Although these experiments were conducted using transient siRNA transfection experiments, our data showed that the interaction between BMI1 depletion with RT occurs as early as 24 h post-RT (Figure 3a), which might therefore explain the impressive in vivo data, despite using a transient transfection approach.
In summary, our data are the first to show the role of BMI1 in radioresistance of NPC cells, mediated by repression of P53 signaling and reduction of ROS production. This result provides a broad molecular framework for this process; opportunities for more detailed mechanistic studies remain. The in vivo data support the development of small molecule inhibitors of BMI1, which could be utilized as either a radiosensitizer or a cancer stem cell target in the clinical management of NPC.
Materials and Methods
Patient samples
Seventeen snap-frozen biopsies were obtained from patients with NPC, stored in liquid nitrogen, and analyzed using qRT-PCR. Formalin-fixed and paraffin-embedded (FFPE) NPC patient biopsies were randomly selected for immunohistochemical analysis from an archival bank of materials, which have been evaluated previously.31, 32, 33
qRT-PCR
Supplementary Table 3 lists all the primer sequences used in this study. To measure BMI1 mRNA expression in primary NPC specimens, qRT-PCR was performed using the ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). To assess the efficiency of BMI1 knockdown, C666-1 cells were transfected with 30 nM siBMI1 or siCtrl, and at indicated time points, cells were harvested and total RNA was isolated using the RNeasy Mini Kit (Qiagen, Chatsworth, CA, USA). Reverse transcription was performed using SuperScript III Reverse Transcriptase (Invitrogen Corp., Carlsbad, CA, USA) according to the manufacture's recommendations. The relative fold change in RNA expression was calculated using the 2−ΔΔCt method, where the average of ΔCt values for the amplicon of interest was normalized to that of endogenous control genes (GAPDH or β-actin), compared with control specimens.
Cloning and transfection experiments
Full length BMI1 cDNA was amplified from C666-1 cells using the primers indicated in Supplementary Table 3, and AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA, USA), and was then ligated into the pcDNA3.1 TOPO vector according to the manufacture's recommendations (Invitrogen).
Immunohistochemical detection of human BMI1 expression
BMI1 immunohistochemistry analysis was evaluated on 4-μm FFPE tumor sections using microwave antigen retrieval, in combination with the Level-2 Ultra Streptavidin system (Signet Laboratories, Dedham, MA, USA). Detection of BMI1 utilized a monoclonal anti-BMI1 antibody (Clone F6, 1 : 50 dilution, Millipore Inc., Billerica, MA, USA).
Cell culture and transfection
The EBV-positive C666-1 NPC cell line34 was maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 100 mg/l penicillin, and 100 mg/l streptomycin (RPMI-10) at 37°C 5% CO2. C15 and C17 NP xenograft tumors were passaged in vivo, as described previously.35 UT-SCC-8 cells were grown in DME H21 medium supplemented with 10% fetal bovine serum, 100 mg/l penicillin, and 100 mg/l streptomycin (RPMI-10) at 37°C 5% CO2. BMI1 SMARTpool (siBMI1),9 and P53 SMARTpool (siP53) were purchased from Dharmacon (Dharmacon Inc., Lafayette, CO, USA). Control siRNAs were purchased from either Dharmacon or Ambion (Ambion, Austin, TX, USA). C666-1 cells were transfected with the indicated siRNA using the Lipofectamine RNAiMAX (Invitrogen) reverse transfection protocol, according to the manufacturer's instructions. At 48 or 72 h post-transfection, cells were exposed to RT (4 or 6 Gy). UT-SCC-8 cells were transfected using lipofectamine 2000. NOE cells36 were seeded in a 24-well plate for 24 h before transfection with 1 μg of pcDNA3.1 or pcDNA3.1 BMI1 using lipofectamine 2000. Media was changed 48 h post-transfection, then cells were exposed to 6 Gy RT. Cells were harvested 4 days later, and the proportion of apoptotic cells were quantified using propidium iodine (PI) staining.
Cell viability and measurement of apoptosis
Cell viability was assessed using the soluble tetrazolium salt (MTS) cell proliferation assay as described previously,36 as these cells are not amenable to clonogenic assays. Briefly, 20 μl of the MTS reagent (Promega, Madison, WI, USA) were added to each well in a 96-well plate at the indicated time points, and absorbance was measured at λ492. To measure the fraction of cells in the sub-G0-G1 phase of the cell cycle, cells were harvested and washed twice in FACS buffer (PBS/0.5% BSA). Cells were resuspended in 1 ml of FACS buffer, and then 3 ml of ice-cold 70% ethanol was added to fix the cells for 1 h on ice. Cells were washed once, before resuspending in 500 μl of FACS buffer containing 40 μg/ml RNAse A (Sigma, St. Louis, MO, USA) and 50 μg/ml of PI. Cells were incubated at room temperature for 30 min in the dark before being analyzed in BD FACScalibur (Becton Dickinson, San Jose, CA, USA) using FL-2A and FL-2W channels. FACS data were analyzed using FlowJo 7.5 software (Tree Star, San Carlos, CA, USA).
Caspase activation was measured using Promega Apo-ONE Homogeneous Caspase-3–7 Assay. Briefly, cells were transfected with siCtrl or siBMI1, and were exposed to 4 Gy RT after 48 h. At the indicated time points, caspase 3–7 substrates were added according to the manufacturer's specifications. Plates were analyzed using SPECTRAFluor Plus Fluorometer (Tecan US, Inc., Durham, NC, USA) at excitation and emission wavelengths of 485 and 535 nm, respectively.
Inhibition of ROS
C666-1 cells were transfected as described above and at 48 and 72 h post-transfection, the antioxidant, Trolox, (Sigma) was added at 0, 15, 100, and 500 μ M (stock 500 mM in DMSO) concentrations. Two hours following the second Trolox treatment on day 3, cells were exposed to 6 Gy RT. Cells were collected on day 4, fixed in 70% ethanol, then stained and analyzed to measure the apoptotic fraction as described above.
Quantitation of γ-H2AX
Fifty thousand C666-1 cells were transfected with 30 nM siBMI1 or siCtrl and were exposed to 4 Gy RT after 48 h. Staining for γ-H2AX was conducted as described previously.13 Both attached and detached C666-1 cells were collected at the indicated times post-RT, pelleted by centrifugation, washed in FACS buffer (PBS/0.5% BSA), fixed with 3 ml 70% ethanol, and maintained at −20°C. After collecting all samples, cells were pelleted by centrifugation, and rehydrated with 1 ml cold TBS (pH 7.4) plus 4% FBS and 0.1% Triton X-100 (TST) on ice for 10 min. Cells were then centrifuged and resuspended in 200 μl of FITC-labeled mouse monoclonal antiphosphorylated (Ser139) histone H2AX antibody (1 : 500 dilution in TST, clone JBW301, Millipore Inc., Bedford, MA, USA) for 2 h at room temperature. Cells were then washed twice in TBS, and analyzed using BD FACScalibur.
TEM
C666-1 cells were transfected with siCtrl or siBMI1, then exposed to 4 Gy as described above. Cells from five transfection replicates were harvested at the indicated time points, and TEM was then performed (University of Toronto Microscopy Imaging Laboratory, Toronto, ON, USA), as described previously.37 Briefly, harvested cells were fixed using a mixture of 4% paraformaldehyde and 2.5% glutaraldehyde in a 0.1 mol/l Sorensen's phosphate buffer, pH 7.2. Cells were post-fixed with 1% osmium tetroxide, dehydrated with ethanol, washed with propylene oxide, treated with epoxy resin, polymerized at 60°C for 48 h, sectioned on a Reichert Ultracut E microtome (Leica Inc., Deerfield, IL, USA) to 80-nm thickness, collected on 300 mesh copper grids, and counterstained with uranyl acetate and lead citrate. Analysis was performed using a Hitachi H7000 transmission electron microscope (Hitachi, Tokyo, Japan) at an accelerating voltage of 75 kV.
Immunoblotting
C666-1 cells were transfected with 30 nM siBMI1 or siCtrl and 72 h post-transfection; cells were harvested and lysed in 1 M Tris-HCl (pH 8), 5 M NaCl, and 1% NP40 plus protease inhibitor cocktail (Roche Diagnostics, Quebec, Canada). Protein concentration was determined using the Bio-Rad Detergent-Compatible Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA). In al, 20 μg of protein were loaded onto 10% Tris-glycine protein gels (Invitrogen) for electrophoresis. The protein was then transferred onto a nitrocellulose membrane using a Trans-Blot S.D. Semi-Dry Transfer Cell (Bio-Rad) and blocked using TBST (0.1% Tween-20 plus 5% fat-free dry milk). The membrane was probed with rabbit polyclonal anti-BMI1 antibody (1 : 100 dilution, Abcam, Cambridge, MA, USA), and anti-GAPDH antibody (Abcam, Cambridge, MA, USA). Immunoblotting for P53 was conducted using the anti-P53 monoclonal antibody (clone 1C12, 1 : 1000 dilution; Cell Signaling Technology, Danvers, MA, USA)
Microarray and bioinformatic analyses
C666-1 cells were transfected with 30 nM siBMI1 or siCtrl, and at 48 h post-transfection, half of the transfected cells were exposed to 4 Gy RT. Total RNA was extracted from five transfection replicates of irradiated and nonirradiated cells, 17 h post-RT, using the Qiagen RNeasy kit (Qiagen) according to the manufacturer's instruction, and tested for quality using the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Palo Alto, CA, USA). Extracted RNA was labeled and hybridized onto the Affymetrix GeneChip Human Genome U133 Plus 2.0 array (Affymetrix Inc., Santa Clara, CA, USA), which includes over 47 000 human transcripts. All procedures were conducted at the University Health Network Microarray center (Toronto, ON, USA).
Data normalization was performed using GeneSifter software (GeneSifter.Net, VizX Laboratories, Seattle, WA, USA). Only genes with ‘present’ cell showing a ⩾2-fold change in expression over lipofectamine and siCtrl-transfected cells were included in the analysis. Heatmaps were generated using heatmap builder version 1.1 software (http://ashleylab.stanford.edu/tools_scripts.html) or Microsoft excel 2007. Gene overlap was assessed using the gene list Venn diagram tool (http://mcbc.usm.edu/genevenn/genevenn.htm). Differentially expressed genes were mapped onto KEGG pathways using DAVID Bioinformatics Database functional annotation tools (http://david.abcc.ncifcrf.gov/).38
Tumor formation
All animal experiments were conducted in accordance with the guidelines of the Animal Care Committee, University Health Network. For tumor formation assays, 5 × 104 C666-1 cells were transfected with 30 nM siBMI or siCtrl. After 72 h, where indicated, cells were exposed to 6 Gy RT using a 137Cs unit (Gamma-cell 40 Extractor; Nordion International, Inc., Ontario, Canada) at a dose rate of 1.1 Gy/min. On day 6, cells were harvested, resuspended in 100 μl PBS, and then injected intramuscular (IM) into the left gastrocnemius of 6–8-week-old female SCID mice. Tumor growth was monitored by measuring tumor plus leg diameter as described previously.36, 39
Statistical analysis
Statistical analyses and graphing were performed using Microsoft excel and Graphpad Prism 4.0 software (Graphpad software, San Diego, CA, USA).
Abbreviations
- NPC:
-
nasopharyngeal cancer
- RT:
-
radiation therapy
- TEM:
-
transmission electron microscopy
- PcG:
-
polycomb group
- ROS:
-
reactive oxygen species
References
Sparmann A, van Lohuizen M . Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 2006; 6: 846–856.
Marx J . Developmental biology. Combing over the polycomb group proteins. Science 2005; 308: 624–626.
Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE . The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol 2002; 22: 6070–6078.
Lund AH, van Lohuizen M . Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol 2004; 16: 239–246.
Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 2003; 423: 302–305.
Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C et al. The polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 2007; 21: 525–530.
Song LB, Zeng MS, Liao WT, Zhang L, Mo HY, Liu WL et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res 2006; 66: 6225–6232.
Mohty M, Yong AS, Szydlo RM, Apperley JF, Melo JV . The polycomb group BMI1 gene is a molecular marker for predicting prognosis of chronic myeloid leukemia. Blood 2007; 110: 380–383.
Glinsky GV, Berezovska O, Glinskii AB . Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 2005; 115: 1503–1521.
Dimri GP, Martinez JL, Jacobs JJ, Keblusek P, Itahana K, Van Lohuizen M et al. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res 2002; 62: 4736–4745.
Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M . Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 1999; 13: 2678–2690.
Lee N, Xia P, Quivey JM, Sultanem K, Poon I, Akazawa C et al. Intensity-modulated radiotherapy in the treatment of nasopharyngeal carcinoma: an update of the UCSF experience. Int J Radiat Oncol Biol Phys 2002; 53: 12–22.
Banath JP, Olive PL . Expression of phosphorylated histone H2AX as a surrogate of cell killing by drugs that create DNA double-strand breaks. Cancer Res 2003; 63: 4347–4350.
Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM . Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem 2000; 275: 9390–9395.
Leach JK, Van Tuyle G, Lin PS, Schmidt-Ullrich R, Mikkelsen RB . Ionizing radiation-induced, mitochondria-dependent generation of reactive oxygen/nitrogen. Cancer Res 2001; 61: 3894–3901.
McClain DE, Kalinich JF, Ramakrishnan N . Trolox inhibits apoptosis in irradiated MOLT-4 lymphocytes. FASEB J 1995; 9: 1345–1354.
Qin L, Zhang X, Zhang L, Feng Y, Weng GX, Li MZ et al. Downregulation of BMI-1 enhances 5-fluorouracil-induced apoptosis in nasopharyngeal carcinoma cells. Biochem Biophys Res Commun 2008; 371: 531–535.
Liu L, Andrews LG, Tollefsbol TO . Loss of the human polycomb group protein BMI1 promotes cancer-specific cell death. Oncogene 2006; 25: 4370–4375.
Kwong J, Lo KW, To KF, Teo PM, Johnson PJ, Huang DP . Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin Cancer Res 2002; 8: 131–137.
Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K . Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006; 20: 1123–1136.
Dutton A, Woodman CB, Chukwuma MB, Last JI, Wei W, Vockerodt M et al. Bmi-1 is induced by the Epstein–Barr virus oncogene LMP1 and regulates the expression of viral target genes in Hodgkin lymphoma cells. Blood 2007; 109: 2597–2603.
Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM . The antioxidant function of the p53 tumor suppressor. Nat Med 2005; 11: 1306–1313.
Chatoo W, Abdouh M, David J, Champagne MP, Ferreira J, Rodier F et al. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J Neurosci 2009; 29: 529–542.
Lessard J, Sauvageau G . Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 2003; 423: 255–260.
Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer 2006; 5: 67.
Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res 2006; 66: 6063–6071.
Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA 2007; 104: 973–978.
Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC . PI3 K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev 2008; 22: 436–448.
Phillips TM, McBride WH, Pajonk F . The response of CD24(-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst 2006; 98: 1777–1785.
Shi W, Alajez NM, Bastianutto C, Hui AB, Mocanu J, Ito E et al. Significance of PLK1 regulation by mir-100 in human nasopharyngeal cancer. Int J Cancer (submitted).
Shi W, Pataki I, MacMillan C, Pintilie M, Payne D, O'Sullivan B et al. Molecular pathology parameters in human nasopharyngeal carcinoma. Cancer 2002; 94: 1997–2006.
Makitie AA, MacMillan C, Ho J, Shi W, Lee A, O'Sullivan B et al. Loss of p16 expression has prognostic significance in human nasopharyngeal carcinoma. Clin Cancer Res 2003; 9: 2177–2184.
Yip KW, Shi W, Pintilie M, Martin JD, Mocanu JD, Wong D et al. Prognostic significance of the Epstein–Barr virus, p53, Bcl-2, and survivin in nasopharyngeal cancer. Clin Cancer Res 2006; 12: 5726–5732.
Cheung ST, Huang DP, Hui AB, Lo KW, Ko CW, Tsang YS et al. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein–Barr virus. Int J Cancer 1999; 83: 121–126.
Busson P, Ganem G, Flores P, Mugneret F, Clausse B, Caillou B et al. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinomas. Int J Cancer 1988; 42: 599–606.
Alajez NM, Mocanu JD, Shi W, Chia MC, Breitbach CJ, Hui AB et al. Efficacy of systemically administered mutant vesicular stomatitis virus (VSVDelta51) combined with radiation for nasopharyngeal carcinoma. Clin Cancer Res 2008; 14: 4891–4897.
Yip KW, Mao X, Au PY, Hedley DW, Chow S, Dalili S et al. Benzethonium chloride: a novel anticancer agent identified by using a cell-based small molecule screen. Clin Cancer Res 2006; 12: 5557–5569.
Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG . A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat Med 2007; 13: 1490–1495.
Li JH, Shi W, Chia M, Sanchez-Sweatman O, Siatskas C, Huang D et al. Efficacy of targeted FasL in nasopharyngeal carcinoma. Mol Ther 2003; 8: 964–973.
Riley T, Sontag E, Chen P, Levine A . Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008; 9: 402–412.
Acknowledgements
Dr. NM Alajez is a recipient of an Excellence in Radiation Research for the 21st Century (EIRR21) scholarship. This study was supported by a grant from the Canadian Institutes of Health Research (CIHR). We thank Kelly Stewart for assisting with the ROS experiments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by; M Oren
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Rights and permissions
About this article
Cite this article
Alajez, N., Shi, W., Hui, A. et al. Targeted depletion of BMI1 sensitizes tumor cells to P53-mediated apoptosis in response to radiation therapy. Cell Death Differ 16, 1469–1479 (2009). https://doi.org/10.1038/cdd.2009.85
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2009.85
Keywords
This article is cited by
-
The mechanisms and reversal strategies of tumor radioresistance in esophageal squamous cell carcinoma
Journal of Cancer Research and Clinical Oncology (2021)
-
Concurrent targeting of BMI1 and CDK4/6 abrogates tumor growth in vitro and in vivo
Scientific Reports (2019)
-
ROS homeostasis and metabolism: a dangerous liason in cancer cells
Cell Death & Disease (2016)
-
miR-200c Inhibits invasion, migration and proliferation of bladder cancer cells through down-regulation of BMI-1 and E2F3
Journal of Translational Medicine (2014)
-
Knockdown of RNF2 induces apoptosis by regulating MDM2 and p53 stability
Oncogene (2014)