Abstract
Metabolic enzymes have been shown to function as transcriptional regulators. p53, a tumor-suppressive transcription factor, was recently found to regulate energy metabolism. These combined facts raise the possibility that metabolic enzymes may directly regulate p53 function. Here, we discover that nucleocytoplasmic malate dehydrogenase-1 (MDH1) physically associates with p53. Upon glucose deprivation, MDH1 stabilizes and transactivates p53 by binding to p53-responsive elements in the promoter of downstream genes. Knockdown of MDH1 significantly reduces binding of acetylated-p53 and transcription-active histone codes to the promoter upon glucose depletion. MDH1 regulates p53-dependent cell-cycle arrest and apoptosis in response to glucose deprivation, suggesting that MDH1 functions as a transcriptional regulator for a p53-dependent metabolic checkpoint. Our findings provide insight into how metabolism is directly linked to gene expression for controlling cellular events in response to metabolic stress.
Similar content being viewed by others
Main
Two fundamental processes—energy metabolism and gene regulation—in living organisms are considered indirectly linked to each other due to their different functional locations. Recent findings that some metabolic enzymes function as transcriptional regulators have implicated direct coupling of energy metabolism with gene regulation.1, 2, 3 For instance, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) serves as a co-activator to regulate the expression of histone H2B.4 Nuclear GAPDH also activates p300/CBP and induces apoptotic genes.5 In addition, Arg5/6, a yeast metabolic enzyme involved in arginine biosynthesis, regulates the transcription of nuclear and mitochondrial target genes,6 and plant hexokinase 1 forms a nuclear glucose signaling complex core that directly modulates specific target gene transcription.7
Glucose is the central molecule for energy metabolism in glycolysis and in the tricarboxylic acid (TCA) cycle and mitochondrial respiration. Interestingly, cancer cells have long been known to have altered glucose metabolism by preferentially acquiring energy from glycolysis rather than from mitochondrial respiration.8 This metabolic shift to aerobic glycolysis in cancer cells is commonly referred to as the Warburg effect.9, 10 Recently, pyruvate kinase M2 isoform was found to be important for aerobic glycolysis and cancer metabolism.11 But the molecular mechanism underlying the Warburg effect is still unclear. Because tumor formation is basically caused by dysregulation of gene expression by a genetic or epigenetic alteration, the Warburg effect may be caused by dysregulated transcription.
p53 is the best-known transcription factor that controls cell-cycle arrest and cell death in response to a wide range of stresses.12, 13 However, the tumor-suppressive function of p53 has been mainly studied under conditions of DNA damage with γ-irradiation, UV and free radicals. Intriguingly, recent findings that p53 regulates glucose metabolism have invoked vigorous interest in the direct linkage between metabolism and p53-related cancer.14, 15, 16, 17 According to these findings, p53 in normal cells leads to upregulation of mitochondrial function and downregulation of glycolysis, suggesting that p53 mutation results in the metabolic switch to the Warburg effect found in cancer cells.
In addition, p53 has been suggested to be involved in metabolic diseases, such as aging, and diabetes.13, 18, 19, 20 Intriguingly, p53 downregulates the isoforms of glucose transporter (GLUT) genes.17, 21 Indeed, knockout of GLUT4, the insulin-sensitive glucose transporter, leads to retarded growth and decreased longevity in mice.22
On the basis of previous findings that p53 regulates energy metabolism and that metabolic enzymes serve as transcriptional regulators, we reasoned that some glucose metabolic enzymes may directly bind to p53 to reflect the metabolic state in gene transcription. In this study, we identify nucleocytoplasmic malate dehydrogenase-1 (MDH1) as a p53-binding metabolic enzyme by immunoprecipitation assay, and we demonstrate a mechanism whereby glucose depletion activates p53 transcription factor through MDH1. MDH1 serves as a transcriptional co-activator of p53 by moving to the nucleus and binding to the promoter of p53-downstream genes, and ultimately regulating p53-mediated cell-cycle arrest and cell death in response to glucose deprivation. Our results show that MDH1 signals glucose depletion by directly binding to p53 and activating p53-mediated cell-cycle arrest and apoptosis, and suggest that dysregulation of the metabolic enzyme MDH1 in p53 activation may lead to tumor formation and growth.
Results
MDH1 associates with p53
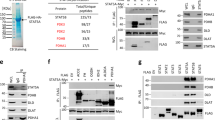
To address whether glycolysis and TCA cycle enzymes directly regulate the transcriptional activation of p53, we first cloned the human glucose metabolic enzymes into mammalian expression vectors (Supplementary Figure 1a, b; Supplementary Table 1) and screened for p53-binding metabolic enzymes through transient transfection of HA-p53 and Myc-tagged metabolic enzymes into human embryonic kidney 293 (HEK293) cells followed by immunoprecipitation assays. Interestingly, HA-p53 bound to Myc-tagged MDH1 but to malic enzyme-1 (ME1) (Figure 1a and b). A mammalian GST-fused p53 specifically bound to the C-terminal region of MDH1 (Figure 1c). To determine the binding region of MDH1 on p53, three different mutants of HA-p53 (ΔN, ΔDBD, ΔC) were transiently transfected into HEK293 cells followed by GST pull-down assay. GST-fused C-terminal domain of MDH1 could not associate with the DNA-binding domain (DBD)-deleted p53 mutant (ΔDBD; Figure 1d). When we tested whether recombinant GST-fused C-terminal domain of MDH1 bound to [35S]-labeled truncated mutants of p53 produced by in vitro translation, we found that purified recombinant GST-fused C-terminal domain of MDH1 could bind to [35S]-labeled in vitro-translated DBD-p53 (Figure 1e), indicating that the metabolic enzyme MDH1 specifically associates with p53.
MDH1 associates with p53. (a) p53 interacts specifically with MDH1. HEK293 cells were transfected with HA-p53 and metabolic enzymes, MDH1 and malic enzyme-1 (ME1). Lysates were subsequently immunoprecipitated using anti-Myc mAb and immunoblotted using anti-HA mAb. (b) Interaction of Myc-MDH1 with HA-p53 was detected from the lysate of HEK293 cells transfected with pcDNA3-HA-p53 and pcDNA3.1-MDH1-Myc. Anti-Myc immunoprecipitates were probed with anti-HA mAb. (c) MDH1 is composed of two domains, N-terminal domain (residues 1–155) and C-terminal domain (residues 156–334). p53 binds to the C-terminal domain of MDH1. HEK293 cells were transfected with various combinations of mGST-p53 and truncated MDH1 mutants. Cell lysates were precipitated with Glutathione Sepharose and probed with anti-Myc mAb. (d) The DNA-binding domain of p53 is critical for MDH1 binding. Recombinant GST-fused C-terminal MDH1 pulled down and bound truncated p53 mutant were detected with anti-HA mAb. (e) Bacterially purified recombinant C-terminal MDH1 binds to the [35S]-labeled in vitro-translated p53 DNA-binding domain. (f) MDH1-p53 interaction is enhanced by glucose starvation. Primary human lung fibroblast (L-132) cells (PD number 2) were cultured for 6 h in the presence or absence of glucose. Cell lysates were immunoprecipitated with anti-MDH1 pAb and probed with anti-p53 (DO.1) mAb. (g) HEK293 cells transfected with expression vectors for HA-p53 and Myc-MDH1 were cultured under normal or glucose-deprived conditions. Cell lysates were immunoprecipitated with anti-Myc mAb and subsequently probed with anti-HA mAb
We next investigated their endogenous interaction under glucose-deprived conditions. When we performed the immunoprecipitation from cell lysates of either glucose-starved or glucose-rich primary human lung fibroblast (L-132) cells, we found that immunoprecipitates of anti-MDH1 contained more p53 upon glucose withdrawal (Figure 1f), possibly indicating that MDH1 preferentially binds to p53 under glucose-deprived conditions. However, it may be because both p53 and MDH1 are more stabilized under glucose-starved conditions. To verify the preferential binding of p53 to MDH1, we transiently transfected HEK293 cells with HA-p53 and Myc-MDH1. After 24 h incubation, we depleted glucose from the media, harvested glucose-starved cells at the indicated time interval and tested the interaction between p53 and MDH1. Interestingly, Myc-MDH1 and HA-p53 associated more strongly under glucose-deprived conditions (Figure 1g).
Stabilization and nuclear localization of p53 by MDH1
We found that MDH1 binds to p53 and its protein amount is accumulated at approximately 1.5-fold higher by glucose deprivation (24 h) (Figure 1f; Supplementary Figure 2a). We thus examined that increased MDH1 affects p53 degradation by ubiquitin–proteasome pathway. To examine the effect of MDH1 on p53 stabilization, we infected primary human diploid fibroblast (HDF) cells with either adenoviral MDH1 (Ad-MDH1) or lentiviral shMDH1, and detected p53 ubiquitination after treatment with proteasome inhibitor, MG132. We first confirmed that infected MDH1 was increased at approximately 1.5-fold higher (Figure 2a; Supplementary Figure 2b). Overexpression of MDH1 led to a significant decrease in p53 ubiquitination (Figure 2a). In contrast, shRNA-mediated knockdown of MDH1 increased p53 ubiquitination (Figure 2b).
Stabilization and nuclear localization of p53 with MDH1. (a) Decrease of p53 ubiquitination by infection of Ad-MDH1 in primary HDF cells. (b) Increase of p53 ubiquitination by lentiviral shRNA-mediated knockdown of MDH1. In (a and b), primary HDF cells (PD number 10–13), infected with either adenoviral Myc-MDH1 (Ad-Myc-MDH1) or lentiviral shMDH1 (pLKO.1-shMDH1), were treated with MG132 (10 μM). Cell lysates were immunoprecipitated with anti-p53 (no. 9282) pAb and probed with anti-ubiquitin antibody. (c) MDH1 blocks the Mdm2-mediated nuclear export of p53. H1299 cells were transfected with different combinations of p53, Mdm2 and Myc-MDH1 expression vectors. Expressed MDH1 was stained with anti-Myc mAb and Rhodamine Red-X-conjugated anti-mouse antibody. p53 was stained with anti-p53(FL393) pAb and FITC-conjugated anti-rabbit antibody. (d) MDH1 moves to the nucleus in primary human lung fibroblast (L-132) cells (PD number 2) upon glucose withdrawal (6 h). MDH1 was stained with anti-MDH1 pAb/anti-rabbit Rhodamine Red-X-conjugated IgG. p53 was stained with anti-p53 (DO.1) mAb/anti-mouse FITC-conjugated IgG
We then examined the possibility that MDH1 blocks the nuclear export of p53 by Mdm2. When we transfected p53-deficient H1299 cells with p53, Mdm2 and/or Myc-MDH1, we observed that p53 was entirely located in the nucleus when expressed alone, but translocated into the cytoplasm when coexpressed with Mdm2 (Figure 2c). As expected, p53 redistributed to the nucleus when MDH1 was expressed with Mdm2, indicating that MDH1 blocks the Mdm2-mediated nuclear export of p53.
As Mdm2 is a well-known ubiquitin ligase of p53, we examined whether MDH1 physically interrupts the p53-Mdm2 interaction (Supplementary Figure 3). To test the direct effect of MDH1 on the p53-Mdm2 interaction, we transiently transfected each of plasmid (HA-p53, Mdm2, Myc-MDH1) into HEK293 cells, respectively. Each of cell lysate was mixed with different combinations as indicated, immunoprecipitated with anti-HA antibody recognizing p53 and then immunoblotted with anti-Mdm2 antibody. An increasing amount of MDH1 did not interrupt the p53-Mdm2 interaction (Supplementary Figure 3a). In fact, we showed that MDH1 binds to the DBD of p53 (Figure 1d), whereas Mdm2 is known to bind to N-terminal region of p53. It is known that histone acetyltransferases (p300/CBP) compete with Mdm2 for binding to N-terminal region,23 we investigated the interaction of MDH1 with p300 by transient transfection (Supplementary Figure 3b). As expected, endogenous MDH1 could bind to p300, suggesting the possibility that MDH1 indirectly interrupt the p53-Mdm2 interaction. Meantime, as the interaction between MDH1 and p53 is strengthened under glucose-deprived conditions (Figure 1f and g), we investigated the localization of both p53 and MDH1 upon glucose withdrawal. MDH1 was dispersed in both cytoplasm and nucleus in glucose-rich media. However, MDH1 translocated to the nucleus and colocalized with p53 in glucose-free media (Figure 2d). Meantime we investigated whether translocation of MDH1 is caused by the enhancement of binding with p53. MDH1 in p53-deficient H1299 cells could translocate into nucleus under glucose-deprived conditions (Supplementary Figure 4), indicating that translocation of MDH1 into nucleus is not caused by binding with p53.
Transactivation of p53 by MDH1
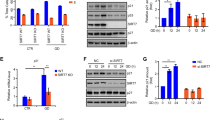
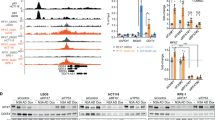
When H1299 cells were transfected with Myc-MDH1, HA-p53 and pG13-luciferase reporter gene vectors, increasing amounts of MDH1 upregulated the p53-driven luciferase activities (Supplementary Figure 5). We then investigated MDH1-mediated p53 transactivation using three p53-downstream luciferase reporter genes (Bax-, p21-, Noxa-luciferase) (Figure 3a). MDH1 increased the luciferase activities in p53-downstream luciferase reporter genes, but not in p53-responsive element (RE)-mutated and -deleted forms of the Noxa-luciferase reporter gene (Figure 3a). To examine whether MDH1 binds to the p53-RE in the promoters of downstream genes, we performed chromatin immunoprecipitation (ChIP) assays (Figure 3b). Endogenous binding of p53 to its promoters (p21, Bax) was further increased in glucose-deprived media. Intriguingly, endogenous MDH1 significantly bound to the p53-RE in the promoters of both Bax and p21 upon glucose deprivation (Figure 3b), but there was little or no difference in p53 knockout mouse embryonic fibroblasts (MEF) (p53−/−) (Figure 3b). To further investigate whether p53 mediates MDH1 binding to the promoters of p53-downstream genes, we performed the ChIP assay using anti-MDH1 antibody with either wild-type MEF (p53+/+) or p53 knockout MEF (p53−/−) under glucose-deprived conditions (Figure 3c). The binding of MDH1 to p53-downstream gene's promoters was significantly reduced in MEF (p53−/−) cells, indicating that MDH1 binds to the p53-RE in the promoters through its association with p53. We next examined the effect of MDH1 knockdown on the change of epigenetic codes around p53-RE in HDF cells under glucose-deprived conditions (Figure 3d). Intriguingly, p53 still bound to the Bax promoter, but acetylated-p53 was not detected in MDH1-knockdown HDF cells, even upon glucose removal. Moreover, both H3-AcK9 and H3-TriMK4 in MDH1-knockdown HDF cells were barely detected, but H3-TriMK9 was highly detected under glucose-deprived conditions (Figure 3d).
Transactivation of p53 by MDH1. (a) MDH1 increases p53 transactivation. H1299 cells were transfected with different combinations of pcDNA3-HA-p53 and pcDNA3.1-MDH1-Myc along with three different p53-downstream luciferase reporter genes. Luciferase activities were measured and normalized to protein content. Means±standard deviations for n=3 are shown. (b) ChIP analysis shows that MDH1 binds to p53-RE in the p53-downstream gene promoters. In MEF (p53+/+), but not in MEF (p53−/−) cells, MDH1 binding is increased by glucose deprivation. Input and immunoprecipitated DNAs from MEF cells were amplified by PCR using primer pairs covering the p53-RE in Bax and p21 promoters. (c) ChIP analysis shows that MDH1-binding to p53-RE is significantly reduced in MEF (p53−/−) cells under glucose-deprived conditions. After both MEF (p53+/+) and MEF (p53−/−) were starved (1 mM glucose) for 16 h, ChIP analysis was performed under the same conditions as in (b). (d) Knockdown of MDH1 in primary HDF cells changes the histone codes in p53-RE of the Bax promoter upon glucose depletion. Antibodies for histone codes used in ChIP analysis are described in Materials and methods section. (e) Real-time PCR analysis shows that mRNA increases of p53-downstream genes (Bax, p21, Noxa) are retarded in siMDH1-treated primary HDF cells under glucose-deprived conditions. MDH1 was knocked down by treatment of two sets of small interfering RNA corresponding to MDH1 gene as described in Materials and Methods section. Means±standard deviations for n=3 are shown
We next investigated the effect of MDH1 knockdown on the mRNA levels of p53-downstream genes by real-time PCR. The treatment of siMDH1 in HDF cells was first confirmed to reduce the mRNA level of MDH1 (Supplementary Figure 6). When the mRNA levels of p53-downstream genes (p21, Bax, Noxa) were measured in MDH1-knockdown HDF cells in glucose-free media, the upregulation of their mRNA levels in MDH1-knockdown cells was retarded in comparison with that in normal cells (Figure 3e). Taken together, MDH1 participates in p53 transactivation during glucose deprivation as a transcriptional co-activator.
Enzymatic activity of MDH1 is not required for apoptosis upon glucose deprivation
To test whether MDH1 catalytic activity affects p53-mediated cellular apoptosis, we generated catalytically inactive mutant MDH1 (H187Y) by site-directed mutagenesis based on previous reports that the histidine residue of His-Asp pair is the critical base/acid catalyst in the active site of 2-hydroxy acid dehydrogenases.24 However, as there is little information about mutagenesis of human MDH1, we first measured the enzymatic activities of Flag-MDH1 (WT) and Flag-MDH1 (H187Y), which were purified from transfected HEK293 cells using an anti-Flag-M2-Agarose affinity column (Figure 4a). We confirmed that MDH1 (H187Y) lost almost all of its enzymatic activity (Figure 4b). Intriguingly, MDH1 (H187Y) bound to p53 (Figure 4c) and their interaction was more strengthened under glucose-deprived conditions (Figure 4d). In addition, MDH1 (H187Y) activated p53-driven luciferase reporter gene (Figure 4e). Moreover, exogenous MDH1 (H187Y) also triggered the nuclear localization of p53 (Figure 4f), suggesting that the catalytic activity of MDH1 is not required for the transactivation of p53.
The enzymatic activity of MDH1 is not necessary for p53 transactivation. (a) Silver staining of Flag-MDH1 (WT) and MDH1 (H187Y) purified from HEK293 cells. Purified proteins were confirmed by western blotting with anti-Flag M2 mAb. (b) MDH1 activities were measured at 340 nm using NADH and oxaloacetate using substrates. Means±standard deviations for n=3 are shown. (c) MDH1 (H187Y) binds to p53. HEK293 cells were transfected with HA-p53 and Myc-MDH1 (H187Y). Immunoprecipitates with anti-Myc were detected with anti-HA antibody. (d) p53-MDH1 (H187Y) binding is enhanced by glucose deprivation. HEK293 cells transfected with expression vectors for HA-p53 and Myc-MDH1 (H187Y) were cultured under normal or glucose-deprived conditions. Immunoprecipitates with anti-Myc were detected with anti-HA antibody. (e) MDH1 (H187Y) transactivates p53. H1299 cells were transfected with either MDH1 (WT) or MDH1 (H187Y) along with p53 and pG13-luciferase reporter genes. Luciferase activities were measured and normalized with protein content. Means±standard deviations for n=3 are shown. (f) MDH1 (H187Y) blocks the Mdm2-mediated nuclear export of p53. H1299 cells were transfected with different combinations of GFP-p53, Mdm2 and MDH1 (H187Y) expression vectors. MDH1 (H187Y) was stained with anti-Myc mAb and Rhodamine Red-X-conjugated anti-mouse antibody
To examine whether HDF cells with MDH1 (H187Y) exhibited increased cell-cycle arrest or apoptosis, we infected HDF cells with Ad-MDH1 (H187Y). As expected, MDH1 (H187Y) infection also induced G1-cell-cycle arrest (Figure 5a and b). Similar to MDH1 (WT), infection with Ad-MDH1 (H187Y) ultimately led to rapid cell death under glucose-deprived conditions (Figure 5c).
Ectopic expression of MDH1 affects cell-cycle arrest and apoptosis upon glucose deprivation. (a) Western blot analysis for the expression of MDH1 (WT), MDH1 (H187Y) and MDH2 in HDF cells after infection with Ad-Myc-MDH1 (WT), Ad-Myc-MDH1 (H187Y) and Ad-Myc-MDH2. Cell lysates were probed with anti-Myc mAb. (b) Infection of primary HDF cells with Ad-MDH1 and Ad-MDH1 (H187Y) triggers G1-cell-cycle arrest in glucose-rich media. (c) Both Ad-MDH1 (WT) and Ad-MDH1 (H187Y) induce more sensitivity to apoptosis upon glucose deprivation. Apoptotic HDF cells were analyzed by measuring the population of sub-G1 cells in PI-stained cells harvested at the indicated time. Means±standard deviations for n=3 are shown
MDH1 affects cell cycle and apoptosis upon glucose deprivation
Because p53 regulates cell cycle and apoptosis, we investigated whether MDH1 affects cell-cycle arrest and apoptosis through the activation of p53. First, we observed that infection with Ad-MDH1 triggered both phosphorylation (S15) and acetylation (K373/382) of p53 (Supplementary Figure 7a). In accordance with p53 modifications, p21 was upregulated in Ad-MDH1-infected cells (Supplementary Figure 7a). In addition, exogenous MDH1 bound to the p53-RE in the p21 promoter when cells were infected with Ad-MDH1 (Supplementary Figure 7b). Consistent with these data, infection with either Ad-MDH1(WT) or Ad-MDH1 (H187Y) triggered G1-cell-cycle arrest in primary HDF cells (Figure 5a and b). Meanwhile, mitochondrial MDH2 (Ad-MDH2) did not affect G1-cell-cycle arrest.
To further examine the function of MDH1 in apoptosis in response to glucose depletion, we measured the sub-G1 population in HDF cells infected with Ad-MDH1 (WT), Ad-MDH1 (H187Y), Ad-MDH2 and Ad-GFP. When glucose was removed from the media, the apoptotic rate in Ad-MDH1-infected HDF cells was increased more rapidly than that in Ad-GFP- or Ad-MDH2-infected cells (Figure 5c). Similar to MDH1 (WT), infection with Ad-MDH1 (H187Y) ultimately led to rapid cell death under glucose-deprived conditions (Figure 5c). Conversely, knockdown of MDH1 in both HDF and HCT116 cells retarded apoptosis in glucose-free media (Supplementary Figure 8).
p53 is involved in MDH1-mediated apoptosis under glucose-deprived conditions
To examine the function of p53 in MDH1-mediated apoptosis upon glucose deprivation, we infected p53-deficient H1299 cells with adenoviral forms of either p53 (Ad-p53) or Ad-LacZ, and measured the apoptotic cells in the sub-G1 population under glucose-deprived conditions. Interestingly, Ad-p53-infected H1299 cells were more sensitive to apoptosis upon glucose withdrawal (Figure 6a; Supplementary Figure 9). We next knocked down p53 by infecting HDF cells with the lentiviral form of shp53 and then observed the apoptotic cells under glucose-deprived conditions. As expected, p53 knockdown retarded the apoptotic rate in HDF cells upon glucose deprivation (Figure 6b; Supplementary Figure 10). The data indicated that p53 functions as a transcriptional regulator for apoptosis in response to glucose depletion. In addition, when p53-knockdown HDF cells were infected with the adenoviral form of Ad-MDH1, mRNA levels of p53-downstream genes (p21, Bax) did not increase further, even in glucose-free media (Figure 6c). To further confirm whether MDH1-mediated apoptosis occurs through p53, we infected MEF (p53+/+) and MEF (p53−/−) cells with Ad-MDH1 (Figure 6d). In Ad-MDH1-MEF (p53+/+) cells, the number of apoptotic cells gradually increased upon glucose withdrawal (Figure 6e). Meanwhile, Ad-MDH1-MEF (p53−/−) cells were relatively insensitive to apoptosis in glucose-free media (Figure 6e).
p53 is involved in MDH1-mediated apoptosis upon glucose deprivation. (a) Infection of H1299 with Ad-p53 accelerates apoptosis under glucose-deprived conditions. (b) shRNA-mediated knockdown of p53 in primary HDF cells retards apoptosis under glucose-deprived conditions. (c) Real-time PCR shows that infection with Ad-MDH1 cannot further increase mRNA levels of p53-downstream genes in p53-knockdown HDF cells even under glucose-deprived conditions. Means±standard deviations for n=3 are shown. (d) Western blot analysis of the expression of infected MDH1 and endogenous p53 in both MEF (p53+/+) and MEF (p53−/−) cells. (e) p53 is important for MDH1-mediated apoptosis under glucose-deprived conditions. MEF (p53+/+) and MEF (p53−/−) cells were infected with Ad-MDH1. Apoptotic cells were measured as in Figure 5c. Means±standard deviations for n=3 are shown. (f) Colony formation assays show that ectopic expression of MDH1 triggers apoptosis in a p53-dependent manner. H1299 or U2OS cells were transfected with either pcDNA 3.1-MDH1-Myc or empty vector (EV) and selected with G418 for 2 weeks. Colonies were counted following 1% crystal violet staining. Means±standard deviations for n=3 are shown. The figure shown is representative of three different experiments
We next performed colony formation assays to determine whether MDH1 was able to induce apoptosis in a p53-dependent manner. H1299 and U2OS cells were transfected with mammalian expression vectors for MDH1 or p53, and these transfected cells were selected with G418. Overexpression of MDH1 did not trigger apoptosis in p53-deficient H1299 cells, whereas overexpression of p53 resulted in significant apoptosis in H1299 cells (Figure 6f; Supplementary Figure 11). Conversely, overexpression of MDH1 reduced the colony formation in p53-wild-type U2OS cells, suggesting that MDH1 triggered p53-dependent apoptosis.
Discussion
p53 is a well-documented transcription factor that controls cell-cycle arrest, DNA repair and cell death and that ultimately contributes to the maintenance of genomic stability. These functions of p53 are thus defined as ‘the guardian of the genome’.12, 13 Recently, a novel function of p53 was discovered that regulates the ATP-generating pathways, that is, glycolysis and mitochondrial respiration, in glucose metabolism;25, 26, 27 this function was referred to as ‘the guardian of metabolism’. p53 downregulates glycolysis by inducing TIGAR, which causes a decline in Fru-2,6-P2 levels.15 p53 regulates glucose metabolism through an IKK-NF-κB pathway.17 In addition, p53 loss leads to increased PGM expression and enhanced glycolysis.14 Meanwhile, p53 guards mitochondrial respiration by maintaining the expression of SCOII, a component of the mitochondrial cytochrome oxidase complex.16 Consequently, in cancer cells, dysregulated p53 leads to the metabolic shift to aerobic glycolysis.
In this study, we demonstrate a mechanism whereby the metabolic enzyme MDH1 enhances the stabilization and transactivation of p53 in response to glucose depletion. The significance of this finding lies in the newly defined function of MDH1 in the activation of p53 as a transcriptional co-activator.
MDH1 belongs to a family of 2-hydroxy acid dehydrogenases.28 Interestingly, the 2-hydroxy acid dehydrogenases participate in gene regulation. Lactate dehydrogenase (LDH) has DNA-binding activity in the nucleus.29 LDH together with GAPDH is also part of the OCA-S complex for S-phase specific H2B transcription.4 C-terminal binding protein (CtBP), a reminiscent form of 2-hydroxy acid dehydrogenase, is a transcriptional corepressor that recruits histone deacetylases and demethylases.30 However, its catalytic ability is not required for transcriptional repression.31, 32 Like CtBP, the inactive mutant of MDH1 is able to trigger apoptosis in response to glucose depletion, indicating that the catalytic activity of MDH1 is not involved in connecting with transcription during glucose depletion. Nonetheless, we will not exclude any possibility that metabolic changes by overexpression of MDH1 or its mutant may affect cell cycle and apoptosis in response to metabolic stress.
MDH1 (NAD+-dependent malate dehydrogenase, EC 1.1.1.37) catalyzes the reversible conversion of malate to oxaloacetate in cytoplasm. The primary functions of MDH1 in metabolism are to transport NADH equivalents and metabolites across the mitochondrial membrane and to control the size of the TCA cycle pool.33, 34 Due to the importance of these functions, MDH1 appears to be a critical checkpoint for energy balance between mitochondria and cytoplasm. However, its catalytic activity, namely, the ability of conversion of malate to oxaloacetate, is unlikely to be involved in p53 activation under glucose-starved conditions. Nonetheless, we cannot exclude other possibilities: kinases (AMPK and Akt/PKB) that regulate the mode of p53 upon glucose withdrawal35, 36, 37 may induce a specific interaction between MDH1 and p53, or conformational changes in MDH1 caused by a change in the ratio of [NAD+]/[NADH] may induce the MDH1-p53 interaction, similar to CtBP regulation of p300-mediated transcriptional activation by monomer–dimer transition in an NADH-dependent manner.32 In addition, PARP-1 modifies the poly(ADP-ribosyl)ation at the p53-DBD using NAD+ as substrate, which leads to accumulation of p53 in nucleus.38 Because MDH1 binds to the p53-DBD, it will be interesting to investigate whether poly(ADP-ribosyl)ation affects the association between MDH1 and p53 upon glucose depletion.
Our findings in this study provide evidence that MDH1 directly regulates the p53-dependent apoptosis upon glucose deprivation and suggest that cross talk between energy metabolism and p53 transactivation is involved in maintaining cellular metabolic state and further determining cell death. Hence, further studies examining the importance of MDH1 in p53 transactivation will advance our understanding of metabolic enzymes as transcriptional regulators in p53-related diseases.
Materials and Methods
Cells
Primary HDF cells were isolated from the foreskin of a 7-year-old boy, obtained during an operation with parents’ consent at Seoul National University Hospital. HDF cells were regularly trypsinized and the number of population doublings (PD) was monitored for further experiments. These HDF cells were obtained from Dr. Jin Ho Chung (Seoul National University College of Medicine, Korea). MEF (p53+/+) and MEF (p53−/−) were obtained from Dr. Han-Woong Lee (Yonsei University, Korea). Primary human lung fibroblast (L-132) cells, HEK293, H1299, U2OS and HCT116 cells were purchased from ATCC. Cells were cultured in DMEM containing 10% (v/v) fetal bovine serum and 50 U/ml of streptomycin and penicillin.
DNA and transfection
Mammalian expression vectors for full-length or deletion mutants of p53 were described previously.39 The expression vectors for full-length or truncated MDH1 mutants were obtained by inserting PCR fragments into pcDNA3.1-Myc (Invitrogen) and pGEX-4T-1 (Amersham). p53-driven luciferase reporter genes were described previously.39 The point mutant used in this study was made using a QuickChange site-directed mutagenesis kit (Stratagene). More detailed information about the DNAs used in this study is available on request. DNA transfections were carried out using Lipofectamine reagent according to the manufacturer's instructions (Invitrogen). For adenoviral vectors for Myc-MDH1 and Myc-MDH2, PCR was carried out using pcDNA3.1-MDH1-Myc and pcDNA3.1-MDH2-Myc as templates, and PCR products were inserted into adenoviral vectors as described previously.40
Immunoprecipitation and immunoblotting
Cells were lysed with lysis buffer (20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% (v/v) NP-40, 1 × protease inhibitor cocktail (Roche)), and cell lysates were incubated with the appropriate antibody in the presence of protein A/G beads (Santa Cruz Biotechnology). Immunoprecipitates were boiled with SDS sample buffer, loaded into SDS-PAGE gels, transferred to nitrocellulose membranes and probed with suitable antibodies. The following antibodies were used in this study: anti-β-actin mAb and anti-Flag mAbs were purchased from Sigma; anti-HA and anti-Myc mAbs from Covance; anti-p53 (DO-1) mAb, anti-ubiquitin (P401), anti-p21 (sc-6246), anti-Bax (sc-493) and anti-GST (B-14) from Santa Cruz Biotechnology; anti-acetyl-p53 (K373/382, 06-758) from Upstate; anti-p53 pAb (no. 9282) and anti-phospho-p53 (S15, no. 9284) from Cell Signaling Technology. Affinity-purified rabbit anti-MDH1 polyclonal antibody was generated against human MDH1 (amino acids 321–334, TEEKESAFEFLSSA) (LabFrontier Co., Ltd, Seoul, Korea).
Real-time PCR
Total RNAs were extracted with TriZol® reagent (Invitrogen) and reverse transcribed (AMV Reverse Transcriptase XL, Takara, Japan). mRNA levels were quantified by real-time PCR with SYBR® Green qPCR Kit (Finnzymes, F-410L) in the iQ5 Real-Time PCR Detection System (Bio-Rad). Results were normalized to total input DNA using the 2−ΔΔCT calculation method.41 The primers for real-time PCR were as follows:
Human p21 (forward: 5′-GCAGACCAGCATGACAGATTT-3′, reverse: 5′-GGATTAGGGCTTCCTCTTGGA-3′), human Bax (forward: 5′-TCTACTTTGCCAGCAAACTGGTGC-3′, reverse: 5′-TGTCCAGCCCATGATGGTTCTGAT-3′), human Noxa (forward: 5′-ACTGTTCGTGTTCAGCTC-3′, reverse: 5′-GTAGCACACTCGACTTCC-3′), human β-actin (forward: 5′-ACGTTGCTATCCAGGCTGTGCTAT-3′, reverse: 5′-TTAATGTCACGCACGATTTCCCGC-3′).
RNA interference
A mixture of two sets of siMDH1s that was used to knockdown MDH1 (siMDH1-1: 5′-GAAAGGAAGUUGGUGUUUA-3′; siMDH1-2: 5′-GGAUCUUACUGCAAAGGAA-3′) was synthesized by Samchully Pharmaceuticals (Seoul, Korea). As a control, siGFP was used as previously described.39 Primary HDF cells were treated with 100 nM of siMDH1 using Oligofectamine reagent (Invitrogen). For viral-mediated RNA interference of MDH1, a mixture of lentiviral vectors containing MDH1-targeting sequences (pLKO.1-shMDH1 (RHS3979-9595927 and RHS3979-9595931)) was used. For knockdown of p53, pLKO.1-shp53 (RHS3979-9572439 and RHS3979-9572440) was used. As a control, pLKO.1-scrambled was also used. These lentiviral vectors were purchased from Open Biosystems.
Detection of cell cycle and apoptosis
The cells were infected with viral particles and further incubated for 30 h (for adenoviral infection) or 72 h (for lentiviral infection). The infected cells were then cultured in DMEM with or without glucose (25 mM). For FACS analyses of cell cycle and apoptosis, cells were fixed in 50% (v/v) ethanol and stained with 1 ml of a solution containing RNase A (50 μg/ml) and propidium iodide (50 μg/ml). At least 10 000 cells were collected and analyzed with the Coulter Epics XL™ flow cytometer (Beckman-Coulter, USA). Percentages of cells in G1 or sub-G1 were calculated with Multicycle for Windows software (Beckman-Coulter).
Chromatin immunoprecipitation assay
Primary HDF or MEF cells grown in glucose-rich media (25 mM) or glucose-starved (1 or 0 mM) for 16 h were treated with formaldehyde. Lysates of nuclear chromatin (100–200 μg) were immunoprecipitated with anti-p53 (FL-393, Santa Cruz Biotechnology) pAb, anti-MDH1 pAb, anti-acetyl-p53 (K373/382, 06-758, Upstate) pAb, anti-H3 (ab1791, Abcam) pAb, anti-Ac-H3K9 (no. 9671, Cell Signaling) pAb, anti-Tri-MeH3K9 (ab8898, Abcam) pAb or anti-Tri-MeH3K4 (07-473, Upstate) pAb. As a control, anti-rabbit IgG (Santa Cruz Biotechnology) was used. The primer sequences used for PCR were as follows: Mouse p21 (forward: 5′-CCTTTCTATCAGCCCCAGAGGATA-3′, reverse: 5′-GGGACATCCTTAATTATCTGGGGT-3′), mouse Bax (forward: 5′-CTGTCCTTGAACTCAGAGAGATGG-3′, reverse: 5′-GGCTATCCTGGAACTCACTTTTGA-3′), human Bax (forward: 5′-TAATCCCAGCGCTTTGGAAG-3′, reverse: 5′-TGCAGAGACCTGGATCTAGCAA-3′).
Ubiquitination assay
HDF cells were infected with Ad-MDH1 (for 30 h) or lentiviral shMDH1 (for 72 h). Infected cells were treated with 10 μM of the proteasomal inhibitor MG132 (Calbiochem) overnight. Cell lysates were immunoprecipitated with anti-p53 polyclonal antibody (no. 9282, Cell Signaling) and probed with anti-ubiquitin mAb (P401, Santa Cruz Biotechnology).
Immunofluorescent staining
To detect the localization of MDH1 after 6-h glucose depletion, primary human lung fibroblast (L-132) cells (PD number 2) were fixed with 4% (w/v) paraformaldehyde, permeabilized with 0.5% Triton X-100 and blocked with 2% (w/v) bovine serum albumin in phosphate-buffered saline. The cells were then stained with anti-MDH1 polyclonal antibody and Rhodamine Red-X-conjugated anti-rabbit antibody (Jackson Immunoresearch) and with anti-p53 (DO.1) monoclonal antibody and FITC-conjugated anti-mouse antibody (Jackson Immunoresearch). Immunofluorescence was examined using a Zeiss LSM 510 laser scanning microscope.
Colony formation assay
p53-wild-type U2OS and p53-null H1299 were transfected with either empty vector (pcDNA3.1-Myc) or pcDNA3.1-MDH1-Myc expression vector. After 36-h incubation, cells were trypsinized and replated at 1 : 1000 dilution in 60 mm culture dishes. After incubation for 24 h, cells were selected with 1 mg/ml G418 (Calbiochem) for 2 weeks. The colonies were fixed with methanol and the number of colonies was counted following 1% crystal violet staining.
MDH1 purification
HEK293 cells were plated on 10 cm dishes and transfected with pcDNA3-Flag or pcDNA3-Flag-MDH1 (WT or H187Y mutant). After 48 h of incubation, cells were lysed with lysis buffer (25 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% (v/v) NP-40, 1 mM EDTA, 1 mM DTT, 1 × protease inhibitor cocktail, 1 mM PMSF), and soluble lysates (10 mg) were incubated with anti-Flag-M2-agarose (A-2220, Sigma) for 1 h. Immunoprecipitates were washed five times with lysis buffer and three times with 0.1 M potassium phosphate buffer (pH 7.4). Flag-MDH1s were eluted with 300 μl elution buffer (0.1 M potassium phosphate (pH 7.4), 0.1 μg/μl 3 × Flag peptide (F4799, Sigma)). The purity of these proteins was confirmed by silver staining.
Measurement of MDH1 activity
The MDH1 activity is determined by measuring the decrease in absorbance at 340 nm resulting from the oxidation of NADH. A 100 μl volume of enzyme was added to a 96-well plate containing 50 μl of reaction buffer (final concentration of 0.1 M potassium phosphate (pH 7.4), 6 mM oxaloacetic acid, 3.75 mM NADH). Activity was measured by following the change in spectrophotometer at 340 nm over a period for 10 min.
Abbreviations
- MDH1:
-
malate dehydrogenase-1
- GAPDH:
-
glyceraldehyde-3-phosphate dehydrogenase
- TCA:
-
tricarboxylic acid
- ChIP:
-
chromatin Immunoprecipitation
- GLUT:
-
glucose transporter
- HEK293:
-
human embryonic kidney 293
- p53-RE:
-
p53-responsive element
References
Shi Y, Shi Y . Metabolic enzymes and coenzymes in transcription-a direct link between metabolism and transcription? Trends Genet 2004; 20: 445–452.
Bhardwaj A, Wilkinson MF . A metabolic enzyme doing double duty as a transcription factor. Bioessays 2005; 27: 467–471.
Ladurner AG . Rheostat control of gene expression by metabolites. Mol Cell 2006; 24: 1–11.
Zheng L, Roeder RG, Luo YS . S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 2003; 114: 255–266.
Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol 2008; 10: 866–873.
Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M . Regulation of gene expression by a metabolic enzyme. Science 2004; 306: 482–484.
Cho YH, Yoo SD, Sheen J . Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 2006; 127: 579–589.
Warburg O . On respiratory impairment in cancer cells. Science 1956; 124: 269–270.
Gatenby RA, Gillies RJ . Why do cancers have high aerobic glycolysis? Nat Rev Cancer 2004; 4: 891–899.
Shaw RJ . Glucose metabolism and cancer. Curr Opin Cell Biol 2006; 18: 598–608.
Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumor growth. Nature 2008; 452: 230–234.
Vogelstein B, Kinzler KW . Cancer genes and the pathways they control. Nat Med 2004; 10: 789–799.
Levine AJ, Hu W, Feng Z . The P53 pathway: what questions remain to be explored? Cell Death Differ 2006; 13: 1027–1036.
Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G et al. Glycolytic enzymes can modulate cellular life span. Cancer Res 2005; 65: 177–185.
Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006; 126: 107–120.
Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O et al. p53 regulates mitochondrial respiration. Science 2006; 312: 1650–1653.
Kawauchi K, Araki K, Tobiume K, Tanaka N . p53 regulates glucose metabolism through an IKK-NF-κB pathway and inhibits cell transformation. Nat Cell Biol 2008; 10: 611–618.
Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 2002; 415: 45–53.
Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev 2004; 18: 306–319.
Campisi J . Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 2005; 120: 513–522.
Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E . The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res 2004; 64: 2627–2633.
Katz EB, Stenbit AE, Hatton K, DePinho R, Charron MJ . Cardiac and adipose tissue abnormalities but not diabetes in mice deficient in GLUT4. Nature 1995; 377: 151–155.
Wadgaonkar R, Collins T . Murine double minute (MDM2) blocks p53-coactivator interaction, a new mechanism for inhibition of p53-dependent gene expression. J Biol Chem 1999; 274: 13760–13767.
Birktoft JJ, Banaszak LJ . The presence of a histidine-aspartic acid pair in the active site of 2-hydroxyacid dehydrogenases. X-ray refinement of cytoplasmic malate dehydrogenase. J Biol Chem 1983; 258: 472–482.
Assaily W, Benchimol S . Differential utilization of two ATP-generating pathways in regulated by p53. Cancer Cell 2006; 10: 4–6.
Bensaad K, Vousden KH . p53: new roles in metabolism. Trends Cell Biol 2006; 17: 286–291.
Green DR, Chipuk JE . p53 and metabolism: inside the TIGAR. Cell 2006; 126: 30–32.
Birktoft JJ, Fernley RT, Bradshaw RA, Banaszak LJ . Amino acid sequence homology among the 2-hydroxy acid dehydrogenases: mitochondrial and cytoplasmic malate dehydrogenases form a homologous system with lactate dehydrogenase. Proc Natl Acad Sci USA 1982; 79: 6166–6170.
Ronai Z . Glycolytic enzymes as DNA binding proteins. Int J Biochem 1993; 25: 1073–1076.
Shi Y, Sawada J, Sui G, Affar EB, Whetstine JR, Lan F et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 2003; 422: 735–738.
Grooteclaes M, Deveraux Q, Hildebrand J, Zhang Q, Goodman RH, Frisch SM . C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc Natl Acad Sci USA 2003; 100: 4568–4573.
Kim JH, Cho EJ, Kim ST, Youn HD . CtBP represses p300-mediated transcriptional activation by direct association with its bromodomain. Nat Struct Mol Biol 2005; 12: 423–428.
Webb LE, Hill EJ, Banaszak LJ . Conformation of nicotinamide adenine dinucleotide bound to cytoplasmic malate dehydrogenase. Biochemistry 1973; 12: 5101–5109.
Grant PM, Roderick SL, Grant GA, Banaszak LJ, Strauss AW . Comparison of the precursor and mature forms of rat heart mitochondrial malate dehydrogenase. Biochemistry 1987; 26: 128–134.
Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 2005; 18: 283–293.
Thoreen CC, Sabatini DM . AMPK and p53 help cells through lean times. Cell Metab 2005; 1: 287–288.
Levine AJ, Feng Z, Mak TW, You H, Jin S . Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev 2006; 20: 267–275.
Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M et al. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol 2007; 9: 1175–1183.
Roe JS, Kim H, Lee SM, Kim ST, Cho EJ, Youn HD . p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol Cell 2006; 22: 395–405.
He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B . A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA 1998; 95: 2509–2514.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25: 402–408.
Acknowledgements
We thank Dr. H-W Lee and Dr. JH Chung for invaluable materials and JW Choi for initiating the cloning of metabolic enzymes. This work was supported by KOSEF grants from the National Research Laboratory (ROA-2007-000-20002-0), the Center for Aging and Apoptosis Research at Seoul National University (R11-2002-097-050050-0) and the Center for Functional Analysis for Human Genome (3344-20060070).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited KH Vousden
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, S., Kim, J., Cho, E. et al. A nucleocytoplasmic malate dehydrogenase regulates p53 transcriptional activity in response to metabolic stress. Cell Death Differ 16, 738–748 (2009). https://doi.org/10.1038/cdd.2009.5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2009.5
Keywords
This article is cited by
-
Permission to pass: on the role of p53 as a gatekeeper for aneuploidy
Chromosome Research (2023)
-
Low glucose metabolite 3-phosphoglycerate switches PHGDH from serine synthesis to p53 activation to control cell fate
Cell Research (2023)
-
Cardio-onco-metabolism: metabolic remodelling in cardiovascular disease and cancer
Nature Reviews Cardiology (2022)
-
Dysregulated metabolic pathways in age-related macular degeneration
Scientific Reports (2020)
-
Maintaining homeostasis by controlled alternatives for energy distribution in plant cells under changing conditions of supply and demand
Photosynthesis Research (2019)