Abstract
Bcl-2 family members regulate apoptosis in response to cytokine withdrawal and a broad range of cytotoxic stimuli. Pro-apoptotic Bcl-2 family members Bax and Bak are essential for apoptosis triggered by interleukin-3 (IL-3) withdrawal in myeloid cells. The BH3-only protein Puma is critical for initiation of IL-3 withdrawal-induced apoptosis, because IL-3-deprived Puma−/− cells show increased capacity to form colonies when IL-3 is restored. To investigate the mechanisms of Puma-induced apoptosis and the interactions between Puma and other Bcl-2 family members, we expressed Puma under an inducible promoter in cells lacking one or more Bcl-2 family members. Puma rapidly induced apoptosis in cells lacking the BH3-only proteins, Bid and Bim. Puma expression resulted in activation of Bax, but Puma killing was not dependent on Bax or Bak alone as Puma readily induced apoptosis in cells lacking either of these proteins, but could not kill cells deficient for both. Puma co-immunoprecipitated with the anti-apoptotic Bcl-2 family members Bcl-xL and Mcl-1 but not with Bax or Bak. These data indicate that Puma functions, in the context of induced overexpression or IL-3 deprivation, primarily by binding and inactivating anti-apoptotic Bcl-2 family members.
Similar content being viewed by others
Main
Proliferation and survival of myeloid cells depends on growth factors, such as interleukin-3 (IL-3). Loss of cytokine-signalling results in cessation of proliferation and cell death. Evading dependence on cytokine signals constitutes one step to malignancy.1 Apoptosis following cytokine deprivation is regulated by the Bcl-2 family of proteins. Overexpression of anti-apoptotic Bcl-2 family members, such as Bcl-2 or Bcl-xL2, 3 or the combined loss of the pro-apoptotic Bcl-2 family members Bax and Bak renders cells resistant to many apoptotic stimuli, including cytokine deprivation or overexpression of BH3-only proteins.4, 5, 6, 7
BH3-only proteins, a pro-apoptotic subgroup of the Bcl-2 family characterised by the presence of a single Bcl-2 Homology 3 (BH3) domain, are essential for initiation of apoptosis in conditions of cell stress or cytokine withdrawal.8 Analysis of knockout mice lacking one or more BH3-only proteins demonstrated death stimulus-specific and cell type-specific roles for these proteins. For example, lymphocytes from Bim−/− mice are resistant to cytokine deprivation and ionomycin9 whereas Bid, a BH3-only protein activated by caspases,10, 11 is required for Fas-mediated cell death in hepatocytes, but not in T lymphocytes.12, 13
Puma is a p53 inducible BH3-only protein.14, 15, 16 Analysis of Puma−/− mice demonstrated diminished apoptosis in response to p53-dependent stimuli, such as γ-irradiation and p53-independent death stimuli, such as cytokine deprivation, treatment with glucocorticoids or phorbol ester.17, 18, 19 Deletion of Puma protects primary and transformed myeloid progenitors from IL-3-withdrawal induced apoptosis.4, 18
Apoptosis induced by BH3-only proteins requires the neutralisation of anti-apoptotic Bcl-2 family members, including Bcl-2, Bcl-xL, Bcl-w, A1 and Mcl-1 and the activation of the pro-apoptotic, multi-domain Bcl-2 family members, Bax and Bak. The BH3-only proteins bind and repress the function of anti-apoptotic Bcl-2 family members with varying affinity. For example, Puma and Bim bind all Bcl-2-like pro-survival proteins whereas Bad binds Bcl-2, Bcl-xL and Bcl-w. Noxa binds Mcl-1 and A1.20, 21 BH3-only protein binding to Bax and Bak is restricted primarily to Bid and Bim (particularly BimS), and this binding activates Bax and Bak.22, 23 It is not clear whether Puma functions by repressing anti-apoptotic Bcl-2 family members, by directly activating Bax or by doing both. It is also unclear whether Puma requires the Bax-activating activity of Bid or Bim.
We have examined how Puma induces apoptosis by expressing Puma under an inducible promoter in IL-3-dependent HoxB8-immortalised myeloid progenitor cells (referred to as FDM cells for factor-dependent myeloid cells) and in murine embryonic fibroblasts (MEFs) derived from mice lacking the BH3-only proteins Bid and Bim, and cells lacking Bax, Bak or both. Induced Puma expression caused apoptosis of wt and Bid−/−;Bim−/− cells but not Bax−/−;Bak−/− cells, demonstrating that neither Bim nor Bid are required for Puma to induce apoptosis. Deletion of Bax or Bak alone did not inhibit Puma-dependent apoptosis. Puma immunoprecipitated Bcl-xL and Mcl-1, but not Bax or Bak. Our data are consistent with a model in which Puma functions to directly repress anti-apoptotic Bcl-2 family members to promote Bax activation but does not itself directly activate Bax.
Results
Enforced Puma expression induces apoptosis in Bid−/−;Bim−/− FDM cells
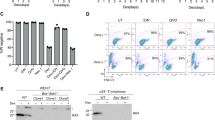
To investigate the mechanisms by which Puma kills cells, we cloned N terminally Flag-tagged murine Puma (iF-Puma), or a control protein (inducible enhanced green fluorescent protein, ieGFP), into a lentiviral expression vector in which gene expression can be controlled by addition of 4-Hydroxytamoxifen (4-OHT). We infected this lentivirus into multiple independent FDM lines derived from gene-deleted mice.24 We confirmed the inducible Puma expression by western blotting and immunoprecipitation (Figure 1a). The viability of wt, Bid−/−, Bim−/−, Bid−/−;Bim−/−, Bax−/−, Bak−/− and Bax−/−;Bak−/− FDM lines expressing iF-Puma was determined following treatment with 4-OHT. Induced expression of iF-Puma killed wt, Bid−/−, Bim−/−, Bid−/−;Bim−/−, Bax−/− and Bak−/− lines equally (Figure 1b and c). In contrast, Bax−/−;Bak−/− cells were completely resistant to iF-Puma expression. Induced expression of ieGFP was equally well tolerated by all cell lines tested. These data indicate that neither Bid nor Bim are required for overexpressed Puma to cause apoptosis. In contrast to previous reports,25 the presence of either Bax or Bak was sufficient for Puma to induce apoptosis in these cells, indicating that Puma overexpression induced apoptosis does not absolutely require Bax.
Enforced Puma expression kills Bid−/−;Bim−/− cells but not Bax−/−;Bak−/− cells. (a) Bax−/−;Bak−/− IL-3 dependent HoxB8 immortalised myeloid progenitor lines or MEF cells were infected with a lentivirus containing an inducible expression cassette for Flag-tagged Puma (iF-Puma). Puma expression was induced by treatment with 1 μ M 4-hydroxy tamoxifen (4-OHT) for 0, 4 or 8 h. Whole cell lysates were prepared and iF-Puma was immunoprecipitated using anti-Flag mAb (M2)-coupled agarose affinity gel. Immunoprecipitations were resolved on SDS-PAGE and then immunoblotted using antibodies to Puma. Immunoprecipitates are two times as concentrated as input lanes. An asterisk denotes non-specific bands. (b) Representative dot blots of IL-3 dependent HoxB8 immortalised myeloid progenitor lines of the indicated genotypes infected with a lentivirus containing iF-Puma and treated with 4-OHT for 24 h. Cell viability was determined by flow cytometric analysis after staining with FITC-conjugated Annexin V plus propidium iodide. (c) Multiple independent clones (n) infected with iF-Puma lentivirus were generated as in (b). Cell viability was determined by flow cytometric analysis after staining with FITC-conjugated Annexin V plus propidium iodide (PI) 24 h after addition of 4-OHT. Results shown represent means±S.E.M. of four independent experiments. Asterisk denotes a P-value of 0.02 (compared with 4-OHT-treated wt cells) using Student's t-test. (d) Wt, Bid−/−;Bim−/− and Bax−/−;Bak−/− IL-3 dependent HoxB8 immortalised myeloid progenitor lines were infected with a lentivirus encoding an inducible enhanced green fluorescent protein (ieGFP). Cell viability was measured and data presented as described above. Results shown represent means±S.E.M. of four independent experiments
Puma overexpression results in loss of mitochondrial membrane potential and cytochrome c release
Apoptosis triggered by BH3-only proteins results in loss of mitochondrial membrane potential and release of cytochrome c from the mitochondrial inter-membrane space into the cytosol. To determine whether these events occurred in our system of Puma expression, we induced the expression of Puma in FDM lines and stained cells with carbocyanine dye DiOC6(3) fluorochrome, which fluoresces in cells that retain mitochondrial membrane potential.26 Six hours after induction of Puma expression, there was substantial reduction in mitochondrial membrane potential in wt, Bax−/−, Bid−/−;Bim−/− and to a lesser extent in Bak−/− cells (Figure 2b). The delayed mitochondrial depolarisation of Bak−/− cells was consistent with increased viability of Bak−/− cells compared with wt cells 24 h after Puma expression (Figure 1). Although statistically significant, the physiological significance of this is questionable as the vast majority of Bak−/− cells died after Puma expression. Bax−/−;Bak−/− cells showed no reduction in DiOC6(3) fluorescence staining. Subcellular fractionation and immunoblotting detected cytoplasmic cytochrome c following Puma expression in wt and Bid−/−;Bim−/− cells but not in Bax−/−;Bak−/− cells (Figure 2c). Collectively these results indicate Puma expression triggered typical apoptotic changes, even in the absence of both Bid and Bim, or either of Bax or Bak.
Enforced Puma expression causes loss of mitochondrial membrane potential and mitochondrial release of cytochrome c. (a) IL-3 dependent HoxB8 immortalised myeloid progenitor lines of the indicated genotypes transduced with iF-Puma lentivirus were treated with 4-OHT for 6 h and then incubated with DiOC6(3) and analysed by flow cytometry. Representative histograms are shown. (b) Multiple independent IL-3-dependent HoxB8 immortalised myeloid progenitor lines (n) of the indicated genotypes transduced with iF-Puma lentivirus were treated with 4-OHT for 6 h and then incubated with DiOC6(3) and analysed by flow cytometry. The results show the mean±S.E.M. of three independent experiments. Asterisk denotes P-value of 0.002 (compared to 4-OHT treated wt cells) using Student's t-test. (c) Cells of the indicated genotypes transduced with iF-Puma lentivirus were treated with 4-OHT for 4 h and then fractionated into cytosolic and membrane fractions using digitonin. Lysates were resolved on SDS-PAGE and immunoblotted with antibodies to cytochrome c, VDAC and HSP70. Less cytochrome c was observed in the cytosolic fraction in Bid−/−;Bim−/− cells compared with wt cells which is probably accounted for by loading differences as proportionally the same amount of cytochrome c was released from wt cells as Bid−/−;Bim−/− cells. Loading variation also most likely accounts for variations in the levels of the mitochondrial channel protein VDAC. The mitochondrial channel protein VDAC was only detected in the membrane fractions, demonstrating the purity of the fractionation
Combined loss of Bid and Bim does not enhance clonogenic survival in myeloid progenitors after IL-3 deprivation
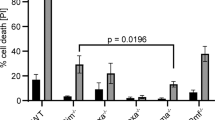
Puma is critical for IL-3 withdrawal-induced apoptosis of FDM cells, primary myeloid progenitors and mast cells.4, 18, 27 We used this model to further explore the role of Bim and Bid in Puma-mediated apoptosis. We generated multiple FDM lines from mice lacking Bid, Bim or both Bid and Bim. Bim−/− or Bid−/− cells were as susceptible to IL-3 withdrawal as those from wt mice (Figure 3a), as reported earlier.4 Bid−/−;Bim−/− cells died after IL-3 deprivation (Figure 3a) whereas Puma−/− cells were significantly protected from IL-3 withdrawal-induced apoptosis, and Bax−/−;Bak−/− cells were completely protected over the course of these experiments. FDM cells lacking both Bim and Bid or Puma treated with the BH3 mimetic drug ABT-737 also underwent apoptosis28 (Supplementary Figure S1) consistent with other reported results.29
Loss of Bid and/or Bim does not confer a clonogenic survival advantage to IL-3 dependent HoxB8 immortalised myeloid progenitor cells after cytokine deprivation. (a) Multiple independent IL-3-dependent HoxB8 immortalised myeloid progenitor lines (numbers indicated as ‘n’) of the indicated genotypes were deprived of IL-3 over a 3-day time course. Cell viability was determined by flow cytometry analysis after staining with FITC-conjugated Annexin V plus propidium iodide (PI) uptake. The results show means±S.E.M. of four independent experiments. Asterisks denote P-value <0.05 (compared with IL-3 deprived wt cells) at day 3 using Student's t-test. (b) Cells from (a) were plated in soft agar in the presence of IL-3 after the indicated times of culture in the absence of IL-3. Colonies were counted after 21 days and their relative clonogenicity determined. The results show means±S.E.M. of four independent experiments. Differences between wt and Puma−/− cells had P-value of 0.005 at day 3 using Student's t-test
Since survival following IL-3 withdrawal (as determined by Annexin V/PI staining) does not necessarily correlate with clonogenic survival,4, 24 we determined whether loss of Bid and/or Bim promoted long-term cell survival and retention of proliferative capacity. Cells cultured in the absence of IL-3 over 3 days were plated in soft agar containing abundant IL-3. When compared to Puma−/− cell lines, cells lacking both Bid and Bim had a 10-fold lower clonogenic survival and the clonogenic survival of Bid−/−;Bim−/− cells was not significantly different from wt cells (P=0.16, student's t-test). The mean relative clonogenicity after IL-3 withdrawal of Bid−/− lines was 1.8% and for Bid−/−;Bim−/− lines 2.5% compared to 0.2 and 0.5% for wt or Bim−/− lines (Figure 3b). These data demonstrate that most Bid−/−;Bim−/− FDM cells readily undergo apoptosis after IL-3 withdrawal and cannot form colonies when IL-3 is restored. Thus, in the context of physiological Puma expression following IL-3 deprivation, Bim and Bid are not required for apoptosis or clonogenic survival of FDM cells.
Puma causes Bax activation but does not require Bax for subcellular localisation and pro-apoptotic activity
Puma localisation to mitochondria is dependent on both its BH3 domain and certain C-terminal hydrophobic residues.30 To determine whether Puma localisation required Bax or Bak, eGFP-tagged Puma (ieGFP-Puma) cloned into the inducible lentiviral system was infected into wt, Bid−/−;Bim−/−, Bax−/−;Bak−/− and Apaf-1−/− MEFs, which were co-stained with a mitochondrial dye (MitoFluor Red) (Figure 4a). ieGFP-Puma was fully functional as it rapidly induced apoptosis in wt and Bid−/−;Bim−/− cells (Supplementary Figure S2), even before substantial ieGFP-Puma fluorescence was detected. Expressing ieGFP-Puma in Apaf-1−/− cells (which lack apoptosome formation) allowed ieGFP-Puma fluorescence to be more easily detected as a result of delayed or inhibited progression of apoptotic morphology.24 ieGFP-Puma localised to mitochondria in all genotypes tested, including Bax−/−;Bak−/− cells (Figure 4a) indicating Puma localisation does not require either Bax or Bak.
Puma localises to the mitochondria independently of Bax or Bak and results in the activation of Bax. (a) MEFs of the indicated genotypes were infected with a lentivirus encoding an expression cassette for an inducible eGFP-tagged Puma (ieGFP-Puma). Cells were induced to express ieGFP-Puma by the addition of 4-OHT for 4 h. Cells were counter-stained with the mitochondrial dye MitoFluor Red and the nucleic acid dye Hoechst. Live cells were imaged by confocal microscopy. (b) Cells of the indicated genotypes, which were induced to express ieGFP-Puma for 4 h, were fixed and stained with an antibody specific to the active conformation of Bax (6A7). Cells were counter-stained with the nucleic acid dye DAPI and imaged by confocal microscopy
To detect Bax activation by ieGFP-Puma, we co-stained cells with an antibody which recognises activated Bax. Expression of ieGFP-Puma in wt, Bid−/−;Bim−/− and Apaf-1−/− cells resulted in Bax activation (Figure 4b). The ieGFP-Puma signal co-localised with Bax in its active conformation. However, because the distribution of ieGFP-Puma is similar in Bax−/−;Bak−/− cells stained with MitoFluor Red, we could not conclude that co-localisation of Puma and activated Bax indicated direct interaction. Together, these results show that correct subcellular localisation of Puma did not require either Bax or Bak and that Bax activation in the presence of overexpressed Puma did not imply direct interaction between Puma and Bax. Furthermore, in our experimental systems, Bax activation by overexpressed Puma did not require Bid or Bim.
Puma binds endogenous Bcl-xL and Mcl-1 but does not co-immunoprecipitate Bax or Bak
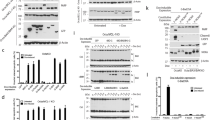
It has been previously reported that Puma can bind and activate Bax.31 Whilst we showed that Puma localised normally and induced apoptosis in cells lacking Bax, this did not exclude direct interaction between Puma and Bax. To test this iF-Puma was immunoprecipitated from wt, Apaf-1−/− and Bax−/−;Bak−/− MEFs 4 h after induction with 4-OHT (Figure 5). Apaf-1−/− cells permitted higher levels of Puma expression whilst maintaining normal expression of Bax and Bak. Puma was readily detected in Apaf-1−/− and Bax−/−;Bak−/− MEFs treated with 4-OHT with some background expression in uninduced cells (Figure 5a). Puma−/− cell lysates were an essential control to detect Puma, as the antibody also detects other proteins (Supplementary Figure S3). Endogenous Mcl-1 and Bcl-xL co-immunoprecipitated with Puma, which, due to ‘leakiness’ of iF-Puma expression, was also detectable in uninduced cells, but we could not detect an interaction between Puma and Bak or Bax (Figure 5a). We obtained similar results when iF-Puma was immunoprecipitated from FDM cells (Supplementary Figure S4). To control for specificity of these interactions in MEFs, we also expressed a loss-of-function iF-Puma 4E mutant (a mutated BH3 region based of published loss-of-function mutations in Bim20) (Figure 5b). The Puma 4E mutant, which ran at a higher molecular weight than wt Puma, did not co-immunoprecipitate Bcl-xL or Mcl-1. These data demonstrate that Puma interacted primarily with anti-apoptotic Bcl-2 family members. Together with results in Figure 4, this suggests that Bax activation induced by enforced Puma expression does not result from Puma binding directly to Bax or Bak, but results from Puma binding and inactivating Bcl-xL and Mcl-1.
iF-Puma co-immunoprecipitates with endogenous Bcl-xL and Mcl-1 but not with Bax or Bak. (a) iF-Puma was induced for 4 h with 4-OHT in MEFs of the indicated genotypes. iF-Puma was immunoprecipitated using anti-Flag mAb (M2)-coupled agarose affinity gel. Immunoprecipitations were resolved on SDS-PAGE and then immunoblotted using antibodies to Puma, Bcl-xL, Mcl-1, Bax and Bak. Input lanes show an equivalent input of proteins (lysed according to cell number) for the IP reactions. Immunoprecipitations were eluted in one-fifth of the volume of the lysate used for immunoprecipitation. The gap in the autoradiograph indicates where irrelevant lanes have been removed. All samples were run on the same gels (b) iF-Puma or a Puma 4E (loss of function) mutant expression was induced for 24 h with 4-OHT in Bax−/−;Bak−/− MEF lines. Wt iF-Puma or 4E mutant Puma was immunoprecipitated using anti-Flag mAb (M2)-coupled agarose affinity gel. Immunoprecipitated proteins were resolved on SDS-PAGE and immunoblots probed using antibodies to Puma, Bcl-xL and Mcl-1
We also immunoprecipitated Bax from lysates from cells induced to express iF-Puma (Figure 6). Endogenous Bax readily co-immunoprecipitated Bak and Bcl-xL. The ability of Bax to co-immunoprecipitate with Bak was unaffected by Puma expression whereas the interaction between Bax and Bcl-xL diminished following Puma induction. An apparent preservation of the association between Bax and Bcl-xL in Apaf-1−/− cells was explained by a more efficient immunoprecipitation of Bax from Apaf-1−/− compared with wt cells. The input levels of Bax were comparable between wt and Apaf-1−/− cells. Bid did no co-immunoprecipitate with Bax in the absence or presence of Puma expression. We immunoprecipitated endogenous Bcl-xL or Bax from SV40 transformed MEFs and FDM cells expressing inducible Puma to determine if the expression of Puma could disrupt a p53-Bcl-xL complex (Supplementary Figure S6). We could not detect any association between Bcl-xL and p53 in either cell types. In MEFs (Supplementary Figure S6A) this may be accounted for by SV40 inactivation p53 and because the association between Bcl-xL and p53 occurred in the context of death stimuli other than Puma overexpression.32, 33, 34 In FDM cells, p53 was below the limits of detection in the absence or presence of Puma (Supplementary Figure S6B) and detectable only after treatment with DNA-damaging agents (data not shown). These results, together with those in Figure 5, demonstrate that Bax and Bak dissociate from Bcl-xL when Puma binds to Bcl-xL, but do not support a direct interaction between Puma and either Bax or Bak. Moreover, enforced Puma expression did not result in an observable interaction between Bax and Bid.
Endogenous Bax binds to Bcl-xL in the absence of Puma. Endogenous Bax was immunoprecipitated from lysates of MEFs, which had been induced to express iF-Puma for 4 h with 4-OHT, using a Bax-specific antibody and protein G sepharose beads. Immunoprecipitated proteins were resolved on SDS-PAGE and immunoblots probed with antibodies to Bax, Bak, Puma, Bcl-xL, Mcl-1 and Bid. Immunoprecipitations are five times more concentrated than input lanes. An asterisk denotes non-specific bands. Arrows indicate bands corresponding to the indicated protein
Discussion
Apoptosis occurring by the intrinsic pathway is regulated by the Bcl-2 family of apoptosis regulators and requires either Bax or Bak. Cells lacking both these proteins are profoundly resistant to a broad range of apoptotic stimuli, including growth factor deprivation and treatment with cytotoxic drugs.35 Anti-apoptotic Bcl-2 family members, such as Bcl-xL and Mcl-1, inhibit the pro-apoptotic activity of Bax and Bak in healthy cells, and one mechanism by which the pro-apoptotic BH3-only proteins (such as Puma or Bim) promote activation of Bax and Bak is by direct binding and sequestering the pro-survival Bcl-2-like proteins. Consequently, loss of certain BH3-only proteins or overexpression of Bcl-2-like proteins protects cells against a broad range of apoptotic stimuli.2, 3, 4, 7, 36
Although Puma is a p53-inducible BH3-only protein,14, 16 the biological roles of Puma are not restricted to p53-dependent cell death stimuli.17, 18 For example, Puma is critical for glucocorticoid and phorbol ester-induced apoptosis in thymocytes, which occur normally in p53−/− cells.17 Puma binds all anti-apoptotic Bcl-2 family members with high affinity.21, 29 However, the question as to whether Puma also directly binds and activates Bax is a matter of conjecture.31, 25, 29
Enforced Puma expression induces apoptosis in several cell types.14, 15, 16, 25 In human glioblastoma multiforme (GBM) cell lines transfected with Bax antisense oligonucleotides, human colon carcinoma cells (HCT116) in which Bax was deleted by homologous recombination and cell free systems, Puma-induced apoptosis is Bax-dependent and characterised by a direct interaction between Puma and Bax.31, 25 Our data provide several compelling lines of evidence that Puma can also cause apoptosis independently of Bax. First, FDM cells lacking either Bax or Bak still underwent apoptosis after Puma expression, whereas cells lacking both these proteins were completely resistant (Figure 1a). Second, although Bax was activated in Puma-induced cell death in transformed MEFs (Figure 4b), Bax activation was not essential provided Bak was present. Third, Bax activation induced by Puma overexpression occurred in the absence of a detectable interaction between Puma and Bax (Figures 5 and 6). These data do not absolutely exclude a potential interaction between Puma and Bax (or Bak) as such an interaction may be below the limits of detection or disrupted during the process of immunoprecipitation. We could, however, readily co-immunoprecipitate Puma or Bax with other known interacting proteins, such as Bcl-xL, demonstrating the efficiency of our immunoprecipitation. The co-localisation of Puma with activated Bax by confocal microscopy did not constitute ‘prima facie’ evidence of an interaction between these two proteins because Puma localisation was unaffected by the deletion of Bax (Figure 4a). Therefore, what can be stated with certainty is that in our systems, enforced Puma expression is associated with Bax activation, but that Puma has no requirement for Bax to properly localise or induce apoptosis. We therefore conclude that there is no requirement for Puma to directly bind to Bax to cause apoptosis in our systems. A comparison of the binding affinities of Puma for the different anti-apoptotic Bcl-2 family members (IC50s in the low nanomolar range) with the affinity of Bax for Puma (IC50 of greater than 10 μM29) further argues that under most physiological conditions Puma does not interact directly with Bax or Bak.
How is Bax activated by Puma overexpression if not directly by Puma itself? The BH3-only proteins Bim and Bid may be required for Puma to induce apoptosis as these proteins are critical activators of Bax/Bak in some systems.21, 22 Bid and Bim are bound and inhibited by anti-apoptotic Bcl-2 family members in healthy cells until released when ‘sensitiser’ BH3-only proteins, such as Puma, bind to the anti-apoptotic Bcl-2 family members.22 Our data suggest that, in our cells at least, this is not the case. Overexpressed Puma readily triggered apoptosis in cells lacking both Bid and Bim. Enforced expression of Puma did not result in detectable amounts of Bid binding to Bax, and Bax assumed an active conformation in response to enforced Puma expression in the absence of Bim and Bid. Although we cannot exclude the possibility that Bax and Bak are activated by another protein following Puma expression, or that some Bax is already in an active conformation so when released from Bcl-xL is sufficient to amplify the apoptotic process, our data demonstrate that direct activation of Bax by Bim or Bid is not required for Puma overexpression to cause apoptosis. Puma expression is important for apoptosis induced by IL-3 deprivation in FDM cells.4 We found that FDM cells lacking both Bid and Bim remained normally susceptible to apoptosis following IL-3 deprivation (Figure 3). Bim expression in FDM cells is below the limits of detection under normal culture conditions but readily detectable following treatment with the histone deacetylase inhibitor, SAHA37 (Supplementary Figure S5). This indicates FDM cells have the potential to express Bim, but this does not occur to detectable levels following IL-3 deprivation. The genetic deletion of Bim removes the potential for even undetectable levels of Bim expression, so demonstrating that Bim is not required for IL-3 withdrawal-induced apoptosis in FDM cells.
Thus, we conclude that the interaction between Puma and anti-apoptotic Bcl-2 family members, such as Mcl-1 and Bcl-xL, is sufficient for apoptosis to proceed in our cell lines and models of Puma expression. Our data demonstrate that in these models, Puma binds to endogenous anti-apoptotic proteins Bcl-xL and Mcl-1, and that interaction between Bcl-xL and Bax is diminished when Puma is overexpressed. Furthermore, we observed no requirement for the BH3-only proteins Bid and Bim, for Puma to induce apoptosis and we observed no direct interaction between Puma and Bax or Bak by co-immunoprecipitation. Together, these data support a model of Puma function in which inhibition of anti-apoptotic Bcl-2 family members is sufficient for Bax and Bak to undergo the conformational changes leading to their activation and apoptosis. Although these data do not permit us to conclude that this model of Puma function is true in all instances, our data do support other published data proposing this as a more general model of BH3-only protein function.29 It is important to note however, that even though Puma overexpression can kill Bid−/−;Bim−/− cells, this does not exclude the possibility that other proteins function to directly activate Bax when Puma is overexpressed. The tumour suppressor p53 is reported to this function when in the cytosol.32, 33, 34 Our failure detect an association between Bcl-xL and p53 in SV40 transformed MEFs (Supplementary Figure S6A) may be as a result of p53 inactivation by SV40 large T antigen and in FDM cells, p53 was undetectable in the presence or absence of Puma expression (Supplementary Figure S6B) and only detectable after DNA damage (data not shown). The association between Bcl-xL and Puma was observed in E1A/H-ras transformed MEFs subjected to UV irradiation and may not occur in the context of enforced Puma expression. We have shown earlier that overexpression of Puma in p53−/− FDM cells or transformed MEFs is as lethal as in wt cells,38 implying that p53 is not essential in this context.
Although our results indicate that neither Bid nor Bim are required in FDM lines for IL-3 withdrawal-induced apoptosis, it is clear the contribution of BH3-only proteins to apoptosis provoked by growth factor deprivation varies from one cell type to another. For example, deletion of Bim potently protects lymphoid cells,9 granulocytes39 and mast cells,40 from cytokine deprivation. Puma is more critical for cytokine deprivation-induced apoptosis of untransformed bone marrow-derived progenitors and FDM cells.18, 24 That FDM cells lacking Bim are not protected from IL-3 deprivation indicates Bim is redundant for this apoptotic stimulus in these cells, but clearly does not exclude a role for Bim in growth-factor deprivation in other cell types. Cytokine withdrawal-induced apoptosis in several cell types from Bim−/−;Puma−/− double knockout mice showed that Bim and Puma have overlapping functions.41
Although it is possible that BH3-only proteins have different mechanisms of action dependent on cell type or the observed mechanisms of action may vary with the experimental model, our results show that direct activation of Bax or Bak by Bim, Bid or Puma is not required for apoptosis induced by enforced expression of Puma, and that deletion of Bid and Bim does not prevent cells from undergoing apoptosis when growth factor is removed. Instead, inactivation of Mcl-1 and Bcl-xL (and other anti-apoptotic Bcl-2 family members) by Puma is sufficient for Bax and Bak to activate and initiate apoptosis.
Materials and Methods
Generation of IL-3-dependent cell lines
Factor-dependent myeloid (FDM) cells were generated from E14 mice by co-culturing single cell suspensions from fetal liver with fibroblasts producing a HoxB8 expressing retrovirus in the presence of 0.25 ng/ml IL-3. Puma−/−, Bid−/−, Bim−/−, Bax−/−, Bak−/−, Bax−/−;Bak−/−, p53−/− and Bid−/−;Bim−/− mice have been described earlier.4, 9, 13, 17, 24, 29, 42 All mice were derived from C57BL/6 embryonic stem (ES) cells or from 129SV-derived ES cells and resulting mice back-crossed onto a C57BL/6 background for at least 10 generations. Cell lines were tested for IL-3 dependence by determining their ability to proliferate in the absence of IL-3 in soft agar. No cell lines were able to generate colonies under such conditions. All cells were cultured in DMEM (low glucose; Gibco) with 10% fetal calf serum (FCS; JRH Laboratories) supplemented with 0.25 ng/ml IL-3 (R&D systems). Murine embryonic fibroblasts (MEFs) were generated as described earlier.13, 20
Cell viability assays
IL-3 was removed by washing cells three times in PBS and then culturing them in DMEM with 10% FCS without IL-3. Cell viability was determined by staining cells with FITC-coupled Annexin V (Invitrogen) in a balanced salt solution including 5 mM CaCl2 plus propidium iodide (PI; 1 μg/ml; Sigma Aldrich) followed by flow cytometric analysis (LSR II; Becton Dickinson).
Clonogenic survival assays
Clonogenic survival assays were performed as described earlier.4, 24 Briefly, 1 × 104 cells per ml were cultured in DMEM, 10% FCS with or without IL-3. At various time points, predetermined numbers of cells were plated in 6-well plates in DMEM, 20% FCS, 0.5 ng/ml IL-3 and 0.3% soft agar. After 21 days the numbers of colonies were counted and expressed as a percentage relative to the number of colonies generated per 1000 cells that had never been deprived of IL-3. At least three independent clones of each genotype were used in each experiment.
Cloning and lentiviral expression
The coding sequence of mouse Puma was either amino terminally tagged with Flag or Enhanced Green Fluorescent Protein (eGFP) and cloned into pF5x UAS SV40 puromycin lentivirus system.38, 43 Cells were infected with GEV16 lentivirus and the pF5x UAS SV40 containing the gene of interest. Cells resistant to both hygromycin and puromycin were selected and tested for gene expression. Expression of Puma or eGFP was induced by treatment with 1 μM 4-Hydroxytamoxifen (4-OHT). For a more detailed description of the lentiviral system and cloning details, see Supplementary Information.
Immunoprecipitation and immunoblotting
Cells were either lysed in Onyx buffer (20 mM Tris-HCl pH 7.4, 135 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 10% Glycerol, 1% Triton X-100) or fractionated into the membrane and cytosolic compartments using digitonin (see Supplementary Information). Flag-tagged Puma was immunoprecipitated (IP) using anti-Flag mAb (M2)-coupled agarose affinity gel (Sigma Aldrich) and eluted using Flag peptide (Sigma Aldrich) according to the manufacturer's instructions. Bax IP was performed using antibody-coupled protein G sepharose beads and eluted with 0.1 M glycine pH 3 for 15 min. After removal of beads, eluates were neutralised in 0.05 M Tris-HCl pH 7.4, 0.15 M NaCl. All lysates and IP material were boiled in protein sample buffer for 5 min and resolved on either gradient (4–20%) or 12% SDS-PAGE (BioRad). The following antibodies were used for western blotting: rat monoclonal anti-Bax (gift of D Huang, The Walter and Eliza Hall Institute of Medical Research, unpublished), mouse monoclonal anti-Bax (Sigma Aldrich), rabbit polyclonal anti-Bak (Sigma Aldrich), rabbit polyclonal anti-Bak NT (UBI), rabbit polyclonal anti-Bcl-x (R&D Systems), rat monoclonal anti-Bid,13, 29 rabbit polyclonal anti-Puma NT (ProSci Integrated), mouse monoclonal anti-cytochrome c (BD Pharmingen), mouse monoclonal anti-p53 (BD Pharmingen), mouse monoclonal anti-VDAC (gift from D Thorburn; Murdoch Children's Research Institute, Melbourne, Australia), mouse anti-HSP70 (gift from R. Anderson, Peter McCallum Cancer Institute, Melbourne, Australia), rabbit polyclonal anti-Mcl-1 (Rockland), donkey polyclonal anti-rabbit IgG coupled to HRP (Amersham), goat polyclonal anti-mouse IgG coupled to HRP (Sigma Aldrich) and goat polyclonal anti-rat IgG coupled to HRP (Amersham).
Mitochondrial membrane potential
Loss of mitochondrial membrane potential was analysed as described earlier.4 Briefly, a stock solution of 10 mM 3,3′-dihexyloxacarbocyanine iodide (DiOC6(3); Molecular Probes) was diluted to 0.4 μM in DMEM plus 10% FCS. Cells were washed once in PBS and then resuspended in diluted DiOC6(3) for 30 min at 37 °C. Cells were then washed and resuspended in medium and analysed by flow cytometry in the FL-1 channel.
Immunofluorescence staining and confocal microscopy
To detect activated Bax, cells were fixed in 1% paraformaldehyde for 5 min at room temperature. Cells were then washed in PBS/2% FCS and probed with 5 μg/ml anti-Bax mAb (6A7; Becton Dickinson) in PBS/2% FCS and 0.3% saponin overnight at 4 °C. Cells were then washed and incubated with goat anti-mouse IgG antibody coupled to PE (1/200; Chemicon). Cells were mounted onto cover slips using ProLong Gold anti-fade with DAPI (Invitrogen) and imaged using a Leica TCS SP2 SE laser scanning confocal microscope using a 63 × 1.2 water immersion objective. Live cells imaging was performed after cells were grown in chamber slides. Cells were counter-stained with MitoTracker Red 580 (Invitrogen) and Hoechst 33342 (Calbiochem). Cells were imaged at 37 °C, 5% CO2 using a Leica TCS SP2 SE laser scanning confocal microscope using a 63 × 1.3 glycerol immersion objective. All images were analysed using Leica software.
Abbreviations
- 4-OHT:
-
4-Hydroxytamoxifen
- BH:
-
Bcl-2 homology
- FDM:
-
factor-dependent myeloid cells
- IL-3:
-
interleukin-3
- MEF:
-
murine embryonic fibroblasts
- PI:
-
propidium iodide
References
Hanahan D, Weinberg RA . The hallmarks of cancer. Cell 2000; 100: 57–70.
Vaux DL, Cory S, Adams JM . Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 1988; 335: 440–442.
Strasser A, Harris AW, Cory S . bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell 1991; 67: 889–899.
Ekert PG, Jabbour AM, Manoharan A, Heraud JE, Yu J, Pakusch M et al. Cell death provoked by loss of interleukin-3 signaling is independent of Bad, Bim, and PI3 kinase, but depends in part on Puma. Blood 2006; 108: 1461–1468.
Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell 2000; 6: 1389–1399.
Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell 2001; 8: 705–711.
Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 2001; 292: 727–730.
Huang DC, Strasser A . BH3-Only proteins-essential initiators of apoptotic cell death. Cell 2000; 103: 839–842.
Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 1999; 286: 1735–1738.
Luo X, Budihardjo I, Zou H, Slaughter C, Wang X . Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 1998; 94: 481–490.
Li H, Zhu H, Xu CJ, Yuan J . Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998; 94: 491–501.
Yin XM, Wang K, Gross A, Zhao Y, Zinkel S, Klocke B et al. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature 1999; 400: 886–891.
Kaufmann T, Tai L, Ekert PG, Huang DC, Norris F, Lindemann RK et al. The BH3-only protein bid is dispensable for DNA damage- and replicative stress-induced apoptosis or cell-cycle arrest. Cell 2007; 129: 423–433.
Nakano K, Vousden KH . PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 2001; 7: 683–694.
Han J, Flemington C, Houghton AB, Gu Z, Zambetti GP, Lutz RJ et al. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc Natl Acad Sci USA 2001; 98: 11318–11323.
Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B . PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell 2001; 7: 673–682.
Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036–1038.
Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 2003; 4: 321–328.
Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L et al. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood 2005; 106: 4131–4138.
Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 2005; 17: 393–403.
Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell 2005; 17: 525–535.
Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ . Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2002; 2: 183–192.
Youle RJ . Cell biology. Cellular demolition and the rules of engagement. Science 2007; 315: 776–777.
Ekert PG, Read SH, Silke J, Marsden VS, Kaufmann H, Hawkins CJ et al. Apaf-1 and caspase-9 accelerate apoptosis, but do not determine whether factor-deprived or drug-treated cells die. J Cell Biol 2004; 165: 835–842.
Ming L, Wang P, Bank A, Yu J, Zhang L . PUMA Dissociates Bax and Bcl-X(L) to induce apoptosis in colon cancer cells. J Biol Chem 2006; 281: 16034–16042.
Korchak HM, Rich AM, Wilkenfeld C, Rutherford LE, Weissmann G . A carbocyanine dye, DiOC6(3), acts as a mitochondrial probe in human neutrophils. Biochem Biophys Res Commun 1982; 108: 1495–1501.
Ekoff M, Kaufmann T, Engstrom M, Motoyama N, Villunger A, Jonsson JI et al. The BH3-only protein Puma plays an essential role in cytokine deprivation induced apoptosis of mast cells. Blood 2007; 110: 3209–3217.
Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 2005; 435: 677–681.
Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 2007; 315: 856–859.
Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L . PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci USA 2003; 100: 1931–1936.
Cartron PF, Gallenne T, Bougras G, Gautier F, Manero F, Vusio P et al. The first alpha helix of Bax plays a necessary role in its ligand-induced activation by the BH3-only proteins Bid and PUMA. Mol Cell 2004; 16: 807–818.
Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell 2003; 11: 577–590.
Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR . PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 2005; 309: 1732–1735.
Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 2004; 303: 1010–1014.
Lindsten T, Thompson CB . Cell death in the absence of Bax and Bak. Cell Death Differ 2006; 13: 1272–1276.
Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005; 120: 237–248.
Lindemann RK, Newbold A, Whitecross KF, Cluse LA, Frew AJ, Ellis L et al. Analysis of the apoptotic and therapeutic activities of histone deacetylase inhibitors by using a mouse model of B cell lymphoma. Proc Natl Acad Sci USA 2007; 104: 8071–8076.
Callus BA, Ekert PG, Heraud JE, Jabbour AM, Kotevski A, Vince JE et al. Cytoplasmic p53 is not required for PUMA-induced apoptosis. Cell Death Differ 2007; 15: 213–215.
Villunger A, Scott C, Bouillet P, Strasser A . Essential role for the BH3-only protein Bim but redundant roles for Bax, Bcl-2, and Bcl-w in the control of granulocyte survival. Blood 2003; 101: 2393–2400.
Alfredsson J, Puthalakath H, Martin H, Strasser A, Nilsson G . Proapoptotic Bcl-2 family member Bim is involved in the control of mast cell survival and is induced together with Bcl-XL upon IgE-receptor activation. Cell Death Differ 2005; 12: 136–144.
Erlacher M, Labi V, Manzl C, Bock G, Tzankov A, Hacker G et al. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction. J Exp Med 2006; 203: 2939–2951.
Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev 2005; 19: 1294–1305.
Dunning CJ, McKenzie M, Sugiana C, Lazarou M, Silke J, Connelly A et al. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J 2007; 26: 3227–3237.
Acknowledgements
We thank Abbott Laboratories for providing ABT-737, David CS Huang, Philippe Bouillet and John Silke for valuable discussions and providing reagents, Mathew Burton for help with confocal microscopy, Ewa Michalak for providing us with Puma null mice and Peter Czabotar for supplying recombinant Bax. This work is supported by the National Health and Research Council (NHMRC) project grants 384404 and 436936 and NHMRC program grant 257502 (DLV and AS). PGE is supported by NHMRC Career Development Award and the Sylvia and Charles Viertel Senior Medical Fellowship. TK is supported by the Swiss National Science Foundation.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Edited by C Borner
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Jabbour, A., Heraud, J., Daunt, C. et al. Puma indirectly activates Bax to cause apoptosis in the absence of Bid or Bim. Cell Death Differ 16, 555–563 (2009). https://doi.org/10.1038/cdd.2008.179
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.179
Keywords
This article is cited by
-
Mito-priming as a method to engineer Bcl-2 addiction
Nature Communications (2016)
-
Homocysteine induces PUMA-mediated mitochondrial apoptosis in SH-SY5Y cells
Amino Acids (2016)
-
Mitochondrial targeted peptides for cancer therapy
Tumor Biology (2015)
-
Flavokawain B, a kava chalcone, inhibits growth of human osteosarcoma cells through G2/M cell cycle arrest and apoptosis
Molecular Cancer (2013)
-
Akt1 is the principal Akt isoform regulating apoptosis in limiting cytokine concentrations
Cell Death & Differentiation (2013)