Abstract
The link between nucleocytoplasmic transport and apoptosis remains controversial. Nucleocytoplasmic exchange of molecules seems indeed essential for the initiation and execution of the apoptotic programme; but inhibition of nuclear transport factors may also represent a powerful apoptotic trigger. The GTPase Ran (together with its partners), first discovered to be essential in nucleocytoplasmic transport, has multiple key functions in cell biology, and particularly in spindle assembly, kinetochore function and nuclear envelope assembly. Among the Ran partners studied, NTF2 appears to be solely involved in nucleocytoplasmic transport. Here, we localised Ran and several of its partners, RanBP1, CAS and NTF2, at the nuclear membrane in the trypanosomatid Leishmania major. Remarkably, these proteins fused to GFP decorated a perinuclear ‘collar’ of about 15 dots, colocalising at nuclear pore complexes with the homologue of nucleoporin Sec13. In the other trypanosomatid Trypanosoma brucei, RNAi knockdown of the expression of the corresponding genes resulted in an apoptosis-like phenomenon. These phenotypes show that Ran and its partners have a key function in trypanosomatids like they have in mammals. Our data, notably those about TbNTF2 RNAi, support the idea that active nucleocytoplasmic transport is not essential for the initiation and execution of apoptosis, and, rather, the impairment of this transport constitutes an intrinsic signal for triggering PCD.
Similar content being viewed by others
Main
The link between nucleocytoplasmic transport and apoptosis remains controversial. On the one hand, nucleocytoplasmic exchange of molecules (reviewed in Ferrando-May et al.1) seems essential for the initiation and execution of the apoptotic programme. For example, inhibition of active nuclear import by wheat germ agglutinin or antibodies to importin-α2 or by mutations in RCC1 (regulator of chromosome condensation 1),3 prevents apoptosis. On the other hand, inhibition of nuclear transport factors has been reported as a powerful apoptotic trigger after a study on protein CC3. The latter impairs the function of several nuclear import receptors, such as transportin,4 and its overexpression predisposes tumour cells to apoptosis.5 Similarly, the reduction of importin-β and importin-α3 levels leads to an increase in the number of apoptotic cells.6 Besides, intracellular redistribution of nucleocytoplasmic transport factors is an early feature of apoptosis, which precedes caspase activation,1, 7 and which has been observed in response to cellular stresses including UV irradiation, oxidative stress and heat–shock8 and hyperosmotic stress.9 However, it is our opinion that all these reports do not allow inferring any firm conclusions on the link between the inhibition of the nucleocytoplasmic transport and apoptosis, in view of the diverse and essential functions of the proteins studied. In particular, the small, evolutionarily conserved, eukaryotic GTPase of the Ras family, Ran together with its partners, were first discovered to be essential in nucleocytoplasmic transport. But they were then reported as key factors during mitosis, being involved in mitotic spindle assembly, kinetochore function, centrosomal integrity/cohesion, and, at the late stage, nuclear envelope assembly. Remarkably, a common molecular mechanism, on the basis of the formation of a RanGTP gradient, seems to underlie the multiple functions of Ran,10 rendering it difficult to dissociate each of its functions. This RanGTP (and therefore RanGDP) gradient is created by means of the asymmetric localisation of regulators, either at the nuclear membrane or at the mitotic spindle or kinetochores or centrosomes. These regulators are, on the one hand, RCC1, a guanine nucleotide exchange factor (GEF), and, on the other hand, the GTPase-activating protein, RanGAP1, which requires the additional factor RanBP1/2 (Ran-binding proteins). These three proteins allow RanGTP levels being higher in defined regions of the nucleus. In general, transport factors (such as importin and exportin) sequester different proteins (which have to be transported through the nuclear membrane or are activators of cell processes) and inhibit their function until relieved by RanGTP in a spatial manner.10 To date, all partners of Ran have been described as involved in its multiple functions, with the exception of one: NTF2 (nuclear transport factor 2), that recycles RanGDP back to the nucleus, and therefore appears to be solely involved in nucleocytoplasmic transport.
In this study, we characterised the protein Ran (first described as rtb2, for ras homologue in Trypanosoma brucei),11 as well as several of its partners in parasitic protozoa of the trypanosomatid family (Leishmania major and T. brucei). Our aim was to understand their function in the cell biology and shed new light on the link between nucleocytoplasmic transport and programmed cell death (PCD). Leishmania and Trypanosoma are uniflagellated organisms responsible for a wide spectrum of human and animal diseases. Several features make them interesting models of the study of nucleocytoplasmic transport issues, hitherto essentially studied in mammals—their status of divergent eukaryotes, their nuclear membrane persisting throughout the cell cycle and the recent completion of their genome sequencing programmes.
Apoptosis is said to have evolved to regulate growth and development in multicellular organisms and is classically assumed to have appeared in phylogeny after the onset of multicellularity.12 However, apoptosis has also been found in a number of unicellular organisms, showing typical hallmarks of apoptosis. Furthermore, apoptosis in parasitic protozoa appears as a pivotal instrument of population density control to ensure a controlled and persistent parasitic load, and guarantee the survival of the host, at least until the transmission of the parasite to the next host is assured.13 Thus, apoptosis in protozoa has been described as ‘selfish altruism’,13 the organisms interacting as complex communities and not as individual entities.
Although apoptosis in protozoa presents numerous features similar to those seen in higher eukaryotes, it also exhibits specific characteristics. Many triggering events have been identified (see Duszenko et al.13 for a review), but apoptosis mechanisms remain largely unknown in these organisms. For example, caspases, the central executioners of metazoan apoptosis, have not been detected to date in these divergent organisms. Although metacaspase genes have been identified in the genome of all trypanosomatids, their involvement in apoptosis is still debated.14, 15, 16, 17 Moreover, no homologue for members of the Bcl-2 protein family has been identified in T. brucei. Finally, two genes whose transcription is upregulated late during the identification of concanavalinA-induced PCD in the procyclic form of T. brucei rhodesiense, a homologue of prohibitin and a protein called TRACK, homologous to the RACK family (receptor for activated kinase C).18 Owing to these peculiarities, here we preferred to use the term apoptosis-like or PCD for these parasitic pathogens.
In this study, using trypanosomatid organisms as models, we have focussed on Ran and several of its partners, including NTF2, and visualised NPCs as a low number of discrete perinuclear dots. Our results show that nucleocytoplasmic transport is not necessary for the entry of cells into PCD and suggest that, in the absence of proapoptotic external stimuli, the impairment of the nucleocytoplasmic RanGTP gradient is an intrinsic signal for PCD triggering.
Results
Identification of putative homologues of the GTPase Ran/Gsp1p, of Sec13 and of partners of Ran in trypanosomatid genomes
BLAST search for the homologue of the GTPase Ran/Gsp1p of Saccharomyces cerevisiae (accession number NP_013396) recognised the protein encoded by LmjF25.1420, a putative GTP-binding protein in the L. major genome, and Tb927.3.1120, in the T. brucei genome, with identity rates of 65.9 and 69.5%, and similarity rates of 77.7 and 78.6%, respectively. Sequence alignment using Align (http://www.ebi.ac.uk/emboss/align/) revealed the highly conserved active sites of the Ras family, Pfam, domain (Supplementary Figure S1). PSORT predicted a nuclear localisation (P=0.3) and a conserved monopartite nuclear localisation sequence (NLS) HRKK for these two proteins (Supplementary Figure S1 for the L. major protein).
Results of BLAST searches for the nucleoporin Sec13 and the partners of Ran/Gsp1p studied here are reviewed in Table 1 —RanGAP and RanBP1, which hydrolyse RanGTP in RanGDP; the RanGEF RCC1, CAS, which permits the recycling of importin-α; and NTF2, which recycles RanGDP back into the nucleus. It is noteworthy that, with the exception of Ran, which showed a high similarity rate with its yeast homologue, the other protein sequences exhibited medium (NTF2 and CRM1) or even weak (notably the four RCC1a – d proteins) similarity rates with their S. cerevisiae counterparts.
Localisation of the GTPase LmjRan and its partners at the nuclear membrane
The gene encoding each one of the proteins analysed here was introduced into expression vectors, which, after transfection into L. major cells, are maintained episomally and yield a constitutive expression of the recombinant protein; each protein was fused to the green fluorescent protein (GFP), either at the N- or the C-terminal end. The protein, LmjRan, was, in addition, expressed fused to the c-Myc tag at the N- or the C-terminal ends, thus, in total, under the form of four different recombinant proteins. It should be stressed that, in this study, all constructs for a given gene yielded identical results. The transcription of all recombinant genes was verified by northern blot.
The LmjRan protein, whether fused to the GFP or to the less bulky c-Myc tag, could be visualised at the nuclear membrane, remarkably as a succession of clearly individualised perinuclear dots (Figure 1a and b). This ‘beads collar’ image persisted throughout the cell cycle in this organism where the nuclear envelope is maintained during mitosis. To clarify the localisation of LmjRan, we wished to determine the subcellular localisation of the nucleoporin Sec13 homologue (encoded by LmjF32.0050). In agreement with it being constituent of the NPCs, and with its function as described in yeast19 and human,20 LmjSec13 localised in L. major at the nuclear membrane, as well as between the endoplasmic reticulum and the Golgi apparatus (Figure 1c). The perfect colocalisation of LmjRan with LmjSec13 at the nuclear membrane (Figure 1c–e) strongly suggests that the dots decorated by the LmjRan recombinant proteins are NPCs. Our conclusion was that LmjRan is mainly found at the nuclear pores in this organism, at any stage of the cell cycle, in contrast with its relocalisation at kinetochores during nuclear membrane disassembly in higher eukaryotes.
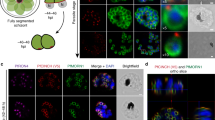
Ran is located at the nuclear periphery, at the NPCs and close to the chromosomal telomeric ends. (a and b) Subcellular localisation of LmjRan fused to the GFP (green) at the N-terminal end (LmjRan-GFPN) in non-dividing (a) and dividing (b) L. major cells. The nucleus and the single intensely staining mitochondrial DNA (kinetoplast) are DAPI stained (red). Inserts: phase contrast microscopical image of the same cell; bar=5 μm. (c – e) Colocalisation of the GFPN-fused LmjSec13 (c) and the c-MycC-fused LmjRan (d) at the nuclear periphery in a dividing cell with two nuclei (E=merged). (f – h) Subnuclear localisation of the GFPN-fused LmjRan (f) and the telomeric sequences as observed by FISH (g) (H=merged)
It is important to note that the number of dots in this ‘beads collar’ image (Figure 1a and b) was remarkably low and relatively constant: 15±3 per cell. Furthermore, this distribution could be paralleled to that of chromosomal ends at the nuclear periphery. Indeed, FISH experiments carried out in Leishmania revealed a significant clustering of telomeric extremities (Figure 1g), as also seen in some other protozoa, such as Plasmodium.21 As shown in Figure 1f–h, the 2 × 36 telomeres appeared clustered in about 15 discrete subnuclear compartments at the periphery of the nucleus and close to the NPCs.
GFP-fused homologues of Ran partners, the proteins RanBP1, CAS and one of both putative homologues of NTF2 (here termed LmjNTF2b) also localised at the nuclear membrane, like LmjRan, and with a similar arrangement (Figure 2a–c). On the contrary, the GFP-fused proteins that we identified by bioinformatics as homologues of RanGAP, RCC1a and NTF2a localised in the cytoplasm (not shown). We verified the correct expression of one of these proteins, RCC1a, of which the localisation was most unexpected, using a western blot with an anti-GFP primary antibody (not shown).
The Ran partners RanBP1, NTF2b and CAS are located like Ran at the nuclear periphery. Subcellular localisation of LmjRanBP1 fused to the GFP at the N-terminal end (RanBP1-GFPN) (a) and of LmjNTF2b (b) and LmjCAS (c) fused to GFP (green) at the C-terminal end. All three proteins localise at the nuclear periphery, clustered in 15±3 fluorescing dots. DAPI-staining of the nucleus and the kinetoplast is represented in red. Inserts as in Figure 1
Neither morphological modification nor cell growth curve differences were observed following the overexpression of any of these recombinant proteins (Supplementary Figure S2 for LmjRan; data not shown for the others).
RNAi of the GTPase Ran induces apoptosis
RNA interference (RNAi) is not functional in L. major but is efficient in T. brucei. The high degree of synteny and sequence homology between these genomes allows straightforward identification of most orthologous genes between both organisms. Therefore, to clarify the function of Ran in trypanosomatids, we inhibited the expression of Tb927.3.1120, the orthologue of LmjRan in T. brucei, an RNAi vector. This expression knockdown, attested by northern blot analysis, induced a significant decrease in cell growth from day 2 (Figure 3a). Clear modifications of the cell structure could be noted in tetracyclin-induced cells, in particular cell shrinkage, an ovoïd cell shape and a nucleus broken into multiple vesicles, reminiscent of an apoptotic phenotype (Figure 3b and Supplementary Figure S3a-c). To rule out off-target effects, we performed a second RNAi experiment using a non-overlapping sequence on the same gene, and obtained the same growth curve and phenotype (not shown). We then attempted to better define this apoptotic-like phenotype.
RNA interference of the GTPase Ran induces PCD. (a) Growth curves of non-induced (NI; closed losanges) and tetracyclin-induced (I; open squares) cells (means±S.D.; n=6). Cell growth decreased from day 1 after tetracyclin induction of TbRan RNAi in T. brucei procyclic forms. Insert: TbRan silencing was confirmed by northern blot, with the gene GPI8, constituvely expressed in procyclic trypanosomes, as a control (left lane: NI; right lane: I). (b) Microscopical observation of T. brucei NI and I cells on day 3; top, phase contrast; bottom, DAPI staining of the nuclei and kinetoplasts. I cells exhibited a morphological apoptotic phenotype with abnormal cellular and nuclear morphology. (c) FACS analysis: examples of raw histograms of NI and TbRan RNAi-I cells, stained with propidium iodide (PI) after permeabilization, on days 1, 2 and 3 postinduction. A peak of degraded DNA appearred as early as day 1 at the left of the G1 and G2 peaks. In NI cells, G1 and G2 peaks represent 66 and 33% of the population, respectively. (d) Histograms of TbRan RNAi NI (grey) and I (white) cells that possess degraded DNA (hypoploid cells; means±S.D.; n=3). Cells were harvested at different time points, permeabilised, stained with PI and analysed with an FACSCalibur flow cytometer to determine the rate of DNA degradation. I cells exhibited degraded DNA suggestive of apoptosis. (e) Histograms of TbRan RNAi NI (grey) and I (white) cells that were PI positive without ethanol permeabilisation, hence in necrosis and late apoptosis (means±S.D.; n=3); 98.5% of ethanol-permeabilised control cells were PI positive (not shown). This shows that the I cells were essentially not dying from necrosis. (f) Measure of phosphatidylserine exposure for NI and TbRan RNAi-I cells on days 1, 2 and 3: raw data for one experiment. This was repeated in quadriplicate and the mean percentages are shown in each quadrant. (g) Measure of phosphatidylserine exposure through the Annexin V assay (analysis of the data presented in panel f). TbRan RNAi NI cells are represented in grey colour and TbRan RNAi-I cells are in white (means±S.D.; n=4). This shows that the I cells were dying from PCD. (h) Measure of DNA double-strand nicks with the TUNEL assay, for TbRan RNAi NI (grey) and I (white) cells (means±S.D.; n=3). The data confirm the apoptotic cell death of TbRan RNAi-I cells. (i) Transfected trypanosomes were cultivated in the presence of different concentrations of the broad caspase inhibitor zVAD-fmk either without (NI; grey) or with (I; white) tetracycline, and cell density was monitored using a haemocytometer on day 2. The addition of zVAD-fmk had no effect on cell concentration and notably on the apoptotic-like phenotype in the I cells, showing that the apoptotic-like phenotype is caspase independent (means±S.D.; n=3)
Flow cytometer analysis of permeabilized T. brucei cells stained with propidium iodide (PI) showed a peak of sub-G1 cells, that is, cells with degraded DNA (hypoploid cells; Figures 3c and d), suggesting apoptosis.22 Moreover, PI staining of non-permeabilized-induced cells, which identifies late apoptotic and necrotic cells, remained below 6.5% during the 3 days of observation (Figure 3e), as opposed to 98.5% of control ethanol permeabilized cells (not shown). This shows that a large majority of cells did not die from necrosis.
To confirm the apoptotic-like state, phosphatidylserine (PS) exposure was assayed through Annexin V staining (Figure 3f). At day 1 postinduction, no differences could be observed. At day 2, 13% of induced cells were Annexin V positive, against 1.4% in control non-induced cells, a difference that further increased with time (Figure 3g). The TUNEL technique also confirmed the entry into PCD after Ran RNAi (Figure 3h).
On the whole, our data clearly show that PCD is induced by Ran RNAi. This phenomenon appeared caspase-independent, as the addition of various concentrations of the broad caspase inhibitor zVAD-fmk had no effect on the RNAi phenotype (Figure 3i). Finally, no upregulation of the transcription of the orthologues of prohibitin and TRACK could be observed, as analysed by northern blot (data not shown).
Apoptosis in the absence of active nuclear transport
As Ran is involved in nucleocytoplasmic transport, and in view of the controversial link between the latter and PCD, we decided to further explore this link in trypanosomatids. Each of the potential partners of Ran was subjected to RNAi in T. brucei. The depletion of each protein identified above as associated to the NPC (NTF2b, RanBP1, CAS and CRM1) triggered PCD. The corresponding cell lines died (Figure 4a) presenting a typical apoptotic phenotype, as confirmed by Annexin V flow cytometer assay (Figure 4b). For the exportin CRM1 homologue, downregulation was performed by adding leptomycin B to the cell cultures, with the same results as those obtained by RNAi (data not shown). It is noteworthy that RanBP1, CAS and CRM1 participate in the diverse functions of Ran in the cell, whereas NTF2 constitutes a Ran partner solely involved in nucleocytoplasmic transport.23, 24 All in all, these data show that, in trypanosomatids, PCD can be directly triggered by the inhibition of NTF2 expression, therefore by the inhibition of the sole nucleocytoplasmic transport.
RNA interference of the Ran partners NTF2b, CAS, RanBP1 and CRM1 induces an apoptotic-like phenotype. (a) Growth curves for non-induced (NI; closed losanges) and tetracyclin-induced (I; open squares) T. brucei cells after transfection with the TbNTF2, TbCAS, TbRanBP1 and TbCRM1 RNAi vectors (means±S.D.; n=3). Cell growth decreased after depletion of each of the four Ran partners studied. Inserts: northern blots hybridised with the corresponding gene probe, plus a control with the GPI8 gene probe (left lane: NI; right lane: I). (b) Measure of phosphatidylserine exposure through the Annexin V assay. The induction of the TbNTF2b, TbCAS, TbRanBP1 and TbCRM1 RNAi triggered the PCD
On the other hand, RNAi knockdown of the proteins identified as putative homologues of RanGAP, RCC1 and NTF2a, but that localised in the cytoplasm and not at the nuclear membrane, had no effect on cell growth (Supplementary Figure S4). However, although the independent RNAi of each the four RCC1 homologues identified in T. brucei had no effect, RNAi directed simultaneously against the four RCC1 genes (termed a – d) induced PCD (Supplementary Figure S5); this suggests a redundancy between these homologues.
Discussion
The involvement of the small GTPase Ran in apoptotic cell death is subject to controversial debate.1, 25 Indeed, its role in the nucleocytoplasmic transport of apoptotic factors is necessary to apoptosis triggering (reviewed in Ferrando-May et al.1 and Fahrenkrog25), as shown, for example, by the expression of a dominant negative Ran GTPase protein.26 Conversely, in response to external stimuli triggering apoptosis, a change in the RanGTP/RanGDP gradient is observed, leading to the inhibition of active nuclear transport.7, 9 In this study, using trypanosomatid protozoa, we have (i) identified Ran and several of its main partners in this divergent cell model, (ii) specified their subcellular localisation at the NPCs and (iii) characterized the apoptotic phenotypes induced by the depletion of their expression.
Identification of Ran and its protein partners in trypanosomatids
Bioinformatic identification of Ran in the L. major database was straightforward, with an amino-acid similarity rate of 78% with the GTPase Ran/Gsp1p of S. cerevisiae. Colocalisation of the recombinant LmjRan, fused either to GFP or to c-Myc, with LmjSec13 at the nuclear pores confirmed this identification. As for putative homologues of the Ran partners, such as RanBP1, CAS and NTF2, homology rates were much less satisfactory (20–30%, Table 1). The validity of their identification essentially relies upon two kinds of data: (i) the same localisation as Ran, as about 15 dots in the nuclear membrane and (ii) the fact that the inhibition of their expression yields an apoptotic-like phenotype, as is the case for Ran. Both results strongly suggest that these proteins are associated to Ran's functionality. By contrast, although BLAST analysis of RanGAP identified one candidate protein with a reasonable similarity rate (34%) between the yeast and L. major genomes, no evidence, either from localisation or from RNAi knockdown, suggested any partnership of this protein with Ran. Similarly, when tested by RNAi, none of the four putative homologues of RCC1 could induce any phenotype. This result may be explained by two hypotheses: either there is a complementation event due to redundancy between these homologues during RNAi experiments, or none of these proteins is the actual homologue of RCC1. The fact that RNAi directed against the four RCC1 homologues induced PCD suggests that there is indeed a complementation, but does not allow to distinguish between both hypotheses. More work is needed to better characterize this potential nuclear transport actor.
Inhibition of the nucleocytoplasmic transport is an intrinsic triggering factor of PCD
Several studies have shown that external stimuli-triggering apoptosis in human cells, like oxidative7 or osmotic9 stress, induce a redistribution of Ran and its protein partners between the nuclear and cytoplasmic compartments, and therefore changes in the functionalities of these proteins. Yet, the authors brought no evidence that these nucleocytoplasmic redistributions were the direct cause, rather than the consequence, of apoptosis. Here, the expression inhibition of Ran and several of its partners constituted an apoptosis-like triggering factor in the absence of any external stimulus. This suggests that the impairment of Ran, such as that observed when triggering PCD by external stimuli, is not merely a consequence of the initiation of PCD, but can directly trigger it.
A major issue in interpreting our results comes from the multiplicity of Ran's functions in the cell (an issue often not mentioned in such studies). Indeed, Ran is involved not only in nucleocytoplasmic transport but also in the function of the kinetochore and centrosomes and, in metazoa, in the reconstitution of the nuclear membrane at the end of mitosis. The phenotypes induced by Ran RNAi can therefore be attributed to one or several of these functions (with the exception of the reconstitution of the nuclear membrane that, here, persists throughout mitosis). Our RNAi results for CAS, RanBP1 and CRM1 do not allow settling this issue, as these proteins, like other partners of Ran (RCC1, RanGAP, importins-α and -β), are associated to the multiple functions of this GTPase. By contrast, NTF2 is solely involved in the nuclear import of RanGDP.23, 24 Therefore, NTF2 depletion acts primarily on the nucleocytoplasmic gradient of RanGTP/RanGDP and it causes PCD. This shows that the disruption of this gradient is an intrinsic factor sufficient for triggering PCD. Besides, it should be noted that, in mammals, the redistribution of RanGTP/RanGDP and partners following proapoptotic external stimuli actually precedes caspase activation and nucleoporin cleavage, and is independent of these events.1 As a consequence, our data demonstrate that the inhibition of the nucleocytoplasmic transport may have a precursor role in the initiation and execution of PCD.
In total, two distinct mechanisms might involve the nucleocytoplasmic transport in this highly complex process that is apoptosis: (i) the transport through the nuclear membrane of apoptotic factors, which is necessary to the initiation and execution of apoptosis and (ii) the inhibition of this transport that appears to be intrinsically an apoptosis-triggering factor. It is awkward to resolve this apparent paradox in the present state of knowledge. In higher eukaryotes, the nucleocytoplasmic transport is altered during apoptosis in two ways (reviewed in Ferrando-May et al.1): (i) during the early phase, the size exclusion limit of the nuclear envelope is increased by a yet unknown mechanism, leading to passive redistribution of molecules that are smaller than this limit between the nucleus and cytoplasm; (ii) during the executive phase, caspases attack the NPCs, resulting in the passive redistribution of all molecules, whatever be their size. In this model, inhibition of passive transport should impede apoptosis triggering, a hypothesis supported by the apoptosis block induced by the addition of WGA, which protects nucleoporins from caspases attack.2 On the contrary, inhibition of active transport resulting from the increase of the size exclusion limit and/or destruction of NPCs should trigger apoptosis through the passive entry/exit of pro-/antiapoptotic molecules. In our work, although this remains highly speculative, Ran inhibition might induce an increase of the size exclusion limit of the nuclear pores, or lead to the recruitment of molecules distinct from caspases that would attack NPCs. It is noteworthy that the conception of a single model for a functional link between nucleocytoplasmic transport and apoptosis may also presently be made difficult by the use of largely distant models, including immortalised (cancer) cell lines, Drosophila, yeast or trypanosomes.
Are caspases involved in PCD in trypanosomatids?
The PCD induced by the silencing of Ran or its partners here is independent of caspase activity, as the specific inhibitor zVAD-fmk has no effect on its progression. This correlates well with the fact that no caspase genes have been identified in the genome of trypanosomatids, and most authors have described PCD in these organisms as caspase-independent. Metacaspases, which are clan CD cysteine peptidases harbouring the predicted secondary structure described for caspases,27 have also been identified in trypanosomatids. However, their direct involvement in cell death remains controversial. Indeed, the L. major metacaspase has been able to replace the yeast metacaspase in PCD,14 and the overexpression of L. donovani metacaspase 1 increased the sensitivity of amastigotes to PCD induced by H2O2.17 Nevertheless, in this study, (a) metacaspase 1 has a trypsin-like and not a caspase-like activity and (b) the authors did not demonstrate that metacaspases were actually necessary for the initiation and execution of apoptosis. Furthermore, the L. major metacaspase seems to have an essential function in cell-cycle progression rather than in PCD.28 And, in T. brucei, metacaspases appear to be associated with RAB11-positive endosomes,15 and not to intervene in apoptosis. Still, intriguingly, some reports have described ‘caspase-like’ activities in these protozoa.16, 29, 30, 31, 32, 33 The issue, therefore, is yet unanswered. Even if apoptosis-like in these divergent organisms appears mainly caspase-independent, it remains probable that other proteases are involved, but they would be either simply different or too distant to be identified from sequence data.
A valuable model for studying nuclear organisation around nuclear pores
There is now strong evidence of an intimate link between the distribution of nuclear pores and the organisation of nuclear structures and processes.34, 35, 36 According to our data, the nuclear pores in Leishmania exhibit a striking punctuated distribution, defining a reduced and relatively constant number of ‘domains’ at the nuclear periphery. This contrasts, however, with previous serial sectioning EM data that allowed Rout and Field37 to identify 200 – 300 nuclear pores per nucleus in T. brucei. This discrepancy might be explained either by the lower definition of the fluorescence imaging system, or by a clustering of the NPCs. Both hypotheses are not exclusive, and it is possible that we observe only those of the NPCS, which are clustered, hence fluoresce more intensely.
Besides, our FISH experiments showed that the telomeric ends of the 36 chromosomes also congregate in a reduced number of clusters similar to that of the NPCs. Considering the key importance of nucleocytoplasmic transport in cell biology, and the role of Ran and its partners in fundamental processes, such as kinetochore and centrosome function, we postulate that this original distribution of nuclear pores here should powerfully structure the functional architecture of the nucleus.
The characterisation of apoptosis-like mechanisms in protozoa is only starting to be unravelled, and their study might bring surprises. Moreover, increasing knowledge of the molecular mechanisms involved in PCD in these organisms should not only shed light upon the evolutionary aspects of this phenomenon, but also open novel avenues in drug development for the fight against the corresponding parasitic diseases.
Materials and Methods
Parasites
Leishmania major ‘Friedlin’ promastigotes (MHOM/IL/81/Friedlin) were grown as described previously.38 Procyclic forms of the 29-13 line of Trypanosoma brucei for the RNAi experiments were grown at 27°C in SDM-79 (Sigma®) supplemented with 10% FCS, 7 μg/ml hemin, 30 μg/ml of hygromycin and 10 μg/ml of geneticin.
Construction of the Leishmania GFP- and c-Myc-fused protein expression vectors pTH6nGFPc and pTH6cGFPn
The different genes were PCR-amplified from genomic DNA using the oligonucleotide primers listed in Supplementary Table S1, containing MfeI (CAATTG) and HpaI (GTTAAC) restriction sites. The PCR products were cloned into pGEM-Teasy (Promega) and then into both vectors pTH6cGFPn and pTH6nGFPc.39 In the expression vector c-Myc-Ran, the GFP gene was replaced by the c-Myc tag. The conservation of the reading frame of the fusion proteins was systematically confirmed by nucleotide sequence analysis.
Leishmania major transfection
A total of 5.107 cells grown to mid-log phase were resuspended with 80 μg of plasmid DNA. Electroporation was performed in a Bio-Rad Gene pulser 2 electroporator using the following conditions: square wave protocol, 1500 V, 25 μF, 2 pulses of 0.5 ms and 10 s between the pulses. The day after, the selective antibiotic hygromycin B (Sigma®) was added at 30 μg/ml, and stable transfectants were obtained between 1 and 2 weeks after transfection.
Microscopy and fluorescence imaging
For intracellular localisation analysis of GFP fusion proteins and immunofluorescence, cells were treated essentially as described elsewhere.39 Briefly, for visualisation of GFP recombinant proteins, transfected cells were grown to mid-log phase, fixed in 4% paraformaldehyde and air-dried on microscope immunofluorescence slides. For immunofluorescence detection, cells were fixed and air-dried on microscope glass slides as described above. Slides were then incubated with 1 : 10 dilution of mouse antihuman c-Myc monoclonal antibody (Chemicon®) and revealed with rabbit antimouse IgG antibody conjugated with the Alexa Fluor 546 dye (Molecular Probes®). All slides were finally mounted with Mowiol (Calbiochem®) and 4,6-diamino-2-phenylindole (DAPI). Leishmania cells were viewed by phase contrast, and fluorescence was visualised using appropriate filters, on a Zeiss Axioplan 2 microscope with a × 100 objective. Digital images were captured using a Photometrics CoolSnap CDD camera (Roper Scientific®) and processed with MetaView (Universal Imaging®).
ImmunoFISH was conducted on promastigote forms of L. major fixed with 4% paraformaldehyde and 4% acetic acid. An immunofluorescence assay was first performed as described above. Slides were then hybridised with a heat-denatured telomeric probe (reported elsewhere40) under a sealed rubber frame at 94°C for 2 min and overnight at 37°C. The hybridisation solution contained 50% formamide, 10% dextran sulphate, 2 × SSPE, 250 μg/ml salmon sperm DNA and ≈100 ng of dsDNA probe labelled by using the fluorescein high-prime kit (Roche Applied Sciences®). After hybridisation, parasites were sequentially washed in 50% formamide/2 × SSC at 50°C, 2 × SSC at 37°C, 4 × SSC at room temperature and 100 mM Tris·HCl/150 mM NaCl/0.5% (vol/vol) Tween 20. Parasites were finally mounted in Vectashield (Vector Laboratories®) and imaged as described above.
Western blot
A total of 2 × 108 cells were centrifuged and washed with a protease inhibitor cocktail (Roche®). Then cells were lysed 10 min at 100°C in loading buffer (62.5 mM Tris-HCl pH6.8, 25% glycerol, 2% SDS, 0.01% bromophenol blue and 5% β-mercaptoethanol). Lysates were separated on a 10% SDS-PAGE gel and transferred onto a PVDF membrane. The membrane was saturated with PBS, 5% skimmed milk and 0.05% Tween20 for 1 h. Mouse anti-GFP primary antibody (Roche®), diluated at 1 : 1000, was detected with a goat antimouse antibody conjugated to the alkaline phosphatase (1 : 7500; Promega®). Finally, the membrane was revealed with BCIP (0.165 mg/ml) and NBT (0.33 mg/ml).
RNAi in T brucei
Different portions of the T. brucei genes identified in GeneDB were PCR-amplified from genomic DNA, using the oligonucleotide primers listed in Supplementary Table S2, with the HindIII (AAGCTT) and SacII (CCGCGG) restriction sites. The different PCR products were cloned into pGEM-Tesay (Promega®) and then into p2T7tiB/GFP. Transfection and RNAi induction were as described.41 Briefly, 10 μg of linearised plasmid DNAs were transfected into 3.107 cells of the 29–13 cell line. An exponential protocol was used with 1500 V and 25 μF as parameters. Transfectants were grown under selective pressure with 5 μg/ml of phleomycin during 15 – 20 days before induction by the addition of 1 μg/ml of tetracyclin.
DNA content
To determinate the DNA content, a PI staining method was used. For this purpose, cells were washed with PBS, resuspended in 500 μl of iced 70% ethanol, vortexed 1 min and incubated at 4 °C. After centrifugation, cells were resuspended in PBS and 10 mg/ml RNAse, placed 20 min at 37°C, centrifuged, incubated 10 – 30 min on ice with 2.5% PI and immediately analysed with a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA) with the BD CellQuest™ Pro software.
Phosphatidylserine exposure
Exposed PS was detected on the outer membrane of cells using the Annexin-V-FLUOS staining kit (Roche®). Cells were washed in PBS and incubated for 10–15 min at 4 °C with the incubation buffer of the kit. Fluorescence was measured using an FACS analysis.
TUNEL test
To detect DNA double-strand breaks, we applied the TUNEL test, using the in situ cell death detection kit, fluorescein, obtained from Roche. A total of 1.107 cells were fixed with paraformaldehyde 2%, laid on an immunoslide and permeabilized with a 0.1% triton and 0.1% sodium citrate solution. The reaction solution from the kit was then added, before observation with a fluorescence microscope.
Abbreviations
- CAS:
-
cellular apoptosis susceptibility protein
- DAPI:
-
4,6-diamidino-2-phenylindole
- FISH:
-
fluorescent in situ hybridisation
- GFP:
-
green fluorescent protein
- NES:
-
nuclear exclusion signal
- NLS:
-
nuclear localisation signal
- NPC:
-
nuclear pore complex
- NTF2:
-
nuclear transport factor 2
- PCD:
-
programmed cell death
- RanBP:
-
Ran-binding protein
- RanGAP:
-
Ran GTPase-activating protein
- RanGEF:
-
Ran guanine exchange factor
- RCC1:
-
regulator of chromosome condensation 1
- RNAi:
-
RNA interference
- TUNEL:
-
terminal uridine deoxynucleotidyl transferase dUTP nick-end labelling
References
Ferrando-May E, Cordes V, Biller-Ckovric I, Mirkovic J, Gorlich D, Nicotera P . Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis. Cell Death Differ 2001; 8: 495–505.
Yasuhara N, Eguchi Y, Tachibana T, Imamoto N, Yoneda Y, Tsujimoto Y . Essential role of active nuclear transport in apoptosis. Genes Cells 1997; 2: 55–64.
Shi WY, Skeath JB . The Drosophila RCC1 homolog, Bj1, regulates nucleocytoplasmic transport and neural differentiation during Drosophila development. Dev Biol 2004; 270: 106–121.
King FW, Shtivelman E . Inhibition of nuclear import by the proapoptotic protein CC3. Mol Cell Biol 2004; 24: 7091–7101.
Whitman S, Wang X, Shalaby R, Shtivelman E . Alternatively spliced products CC3 and TC3 have opposing effects on apoptosis. Mol Cell Biol 2000; 20: 583–593.
Quensel C, Friedrich B, Sommer T, Hartmann E, Kohler M . In vivo analysis of importin alpha proteins reveals cellular proliferation inhibition and substrate specificity. Mol Cell Biol 2004; 24: 10246–10255.
Kodiha M, Chu A, Matusiewicz N, Stochaj U . Multiple mechanisms promote the inhibition of classical nuclear import upon exposure to severe oxidative stress. Cell Death Differ 2004; 11: 862–874.
Miyamoto Y, Saiwaki T, Yamashita J, Yasuda Y, Kotera I, Shibata S et al. Cellular stresses induce the nuclear accumulation of importin alpha and cause a conventional nuclear import block. J Cell Biol 2004; 165: 617–623.
Kelley JB, Paschal BM . Hyperosmotic stress signaling to the nucleus disrupts the Ran gradient and the production of RanGTP. Mol Biol Cell 2007; 18: 4365–4376.
Joseph J . Ran at a glance. J Cell Sci 2006; 119: 3481–3484.
Field MC, Field H, Boothroyd JC . A homologue of the nuclear GTPase ran/TC4 from Trypanosoma brucei. Mol Biochem Parasitol 1995; 69: 131–134.
Vaux DL, Haecker G, Strasser A . An evolutionary perspective on apoptosis. Cell 1994; 76: 777–779.
Duszenko M, Figarella K, Macleod ET, Welburn SC . Death of a trypanosome: a selfish altruism. Trends Parasitol 2006; 22: 536–542.
Gonzalez IJ, Desponds C, Schaff C, Mottram JC, Fasel N . Leishmania major metacaspase can replace yeast metacaspase in programmed cell death and has arginine-specific cysteine peptidase activity. Int J Parasitol 2007; 37: 161–172.
Helms MJ, Ambit A, Appleton P, Tetley L, Coombs GH, Mottram JC . Bloodstream form Trypanosoma brucei depend upon multiple metacaspases associated with RAB11-positive endosomes. J Cell Sci 2006; 119: 1105–1117.
Kosec G, Alvarez VE, Aguero F, Sanchez D, Dolinar M, Turk B et al. Metacaspases of Trypanosoma cruzi: possible candidates for programmed cell death mediators. Mol Biochem Parasitol 2006; 145: 18–28.
Lee N, Gannavaram S, Selvapandiyan A, Debrabant A . Characterization of metacaspases with trypsin-like activity and their putative role in programmed cell death in the protozoan parasite Leishmania. Eukaryot Cell 2007; 6: 1745–1757.
Welburn SC, Murphy NB . Prohibitin and RACK homologues are up-regulated in trypanosomes induced to undergo apoptosis and in naturally occurring terminally differentiated forms. Cell Death Differ 1998; 5: 615–622.
Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C et al. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 1996; 84: 265–275.
Enninga J, Levay A, Fontoura BM . Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol Cell Biol 2003; 23: 7271–7284.
Scherf A, Figueiredo LM, Freitas-Junior LH . Plasmodium telomeres: a pathogen's perspective. Curr Opin Microbiol 2001; 4: 409–414.
Krysko DV, Vanden Berghe T, D’Herde K, Vandenabeele P . Apoptosis and necrosis: detection, discrimination and phagocytosis. Methods 2008; 44: 205–221.
Ribbeck K, Lipowsky G, Kent HM, Stewart M, Gorlich D . NTF2 mediates nuclear import of Ran. EMBO J 1998; 17: 6587–6598.
Smith A, Brownawell A, Macara IG . Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol 1998; 8: 1403–1406.
Fahrenkrog B . The nuclear pore complex, nuclear transport, and apoptosis. Can J Physiol Pharmacol 2006; 84: 279–286.
Gil A, Andres-Pons A, Fernandez E, Valiente M, Torres J, Cervera J et al. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol Biol Cell 2006; 17: 4002–4013.
Uren AG, O’Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV et al. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell 2000; 6: 961–967.
Ambit A, Fasel N, Coombs GH, Mottram JC . An essential role for the Leishmania major metacaspase in cell cycle progression. Cell Death Differ 2008; 15: 113–122.
Das M, Mukherjee SB, Shaha C . Hydrogen peroxide induces apoptosis-like death in Leishmania donovani promastigotes. J Cell Sci 2001; 114: 2461–2469.
Lee N, Bertholet S, Debrabant A, Muller J, Duncan R, Nakhasi HL . Programmed cell death in the unicellular protozoan parasite Leishmania. Cell Death Differ 2002; 9: 53–64.
Sen N, Das BB, Ganguly A, Mukherjee T, Bandyopadhyay S, Majumder HK . Camptothecin-induced imbalance in intracellular cation homeostasis regulates programmed cell death in unicellular hemoflagellate Leishmania donovani. J Biol Chem 2004; 279: 52366–52375.
Sen N, Das BB, Ganguly A, Mukherjee T, Tripathi G, Bandyopadhyay S et al. Camptothecin induced mitochondrial dysfunction leading to programmed cell death in unicellular hemoflagellate Leishmania donovani. Cell Death Differ 2004; 11: 924–936.
Singh G, Jayanarayan KG, Dey CS . Novobiocin induces apoptosis-like cell death in topoisomerase II over-expressing arsenite resistant Leishmania donovani. Mol Biochem Parasitol 2005; 141: 57–69.
Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA . Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 2004; 117: 427–439.
Drummond SP, Rutherford SA, Sanderson HS, Allen TD . High resolution analysis of mammalian nuclear structure throughout the cell cycle: implications for nuclear pore complex assembly during interphase and mitosis. Can J Physiol Pharmacol 2006; 84: 423–430.
Maeshima K, Yahata K, Sasaki Y, Nakatomi R, Tachibana T, Hashikawa T et al. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J Cell Sci 2006; 119: 4442–4451.
Rout MP, Field MC . Isolation and characterization of subnuclear compartments from Trypanosoma brucei. Identification of a major repetitive nuclear lamina component. J Biol Chem 2001; 276: 38261–38271.
Dubessay P, Blaineau C, Bastien P, Pages M . Chromosome fragmentation in leishmania. Methods Mol Biol 2004; 270: 353–378.
Dubessay P, Blaineau C, Bastien P, Tasse L, Van Dijk J, Crobu L et al. Cell cycle-dependent expression regulation by the proteasome pathway and characterization of the nuclear targeting signal of a Leishmania major Kin-13 kinesin. Mol Microbiol 2006; 59: 1162–1174.
Dubessay P, Ravel C, Bastien P, Stuart K, Dedet JP, Blaineau C et al. Mitotic stability of a coding DNA sequence-free version of Leishmania major chromosome 1 generated by targeted chromosome fragmentation. Gene 2002; 289: 151–159.
Blaineau C, Tessier M, Dubessay P, Tasse L, Crobu L, Pages M et al. A novel microtubule-depolymerizing kinesin involved in length control of a eukaryotic flagellum. Curr Biol 2007; 17: 778–782.
Acknowledgements
We thank Pierre Travo (Platform RIO Imaging, Montpellier) for expert advice and assistance, as well as Professor Jacques Clot for allowing access to FACS facilities at the Laboratoire d’Immunologie (CHU of Montpellier, France) and François Renaud (UMR2724 CNRS-University Montpellier 1-IRD) for support and encouragement. We also thank Yvon Sterkers notably for the western blots and Laurence Berry-Sterkers for help in microscopy. MC was a recipient of a grant from the Ministère de l’Enseignement Supérieur et de la Recherche.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by JM Hardwick
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Casanova, M., Portalès, P., Blaineau, C. et al. Inhibition of active nuclear transport is an intrinsic trigger of programmed cell death in trypanosomatids. Cell Death Differ 15, 1910–1920 (2008). https://doi.org/10.1038/cdd.2008.132
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.132
Keywords
This article is cited by
-
RNA interference screen reveals a high proportion of mitochondrial proteins essential for correct cell cycle progress in Trypanosoma brucei
BMC Genomics (2015)
-
Comparative genomics of proteins involved in RNA nucleocytoplasmic export
BMC Evolutionary Biology (2011)
-
The trypanosome flagellar pocket
Nature Reviews Microbiology (2009)