Abstract
Background:
Alterations in protein glycosylation have been related to malignant transformation and tumour progression. We recently showed that low mRNA levels of Golgi alpha-mannosidase MAN1A1 correlate with poor prognosis in breast cancer patients.
Methods:
We analysed the role of MAN1A1 on a protein level using western blot analysis (n=105) and studied the impact of MAN1A1-related glycosylation on the prognostic relevance of adhesion molecules involved in breast cancer using microarray data (n=194). Functional consequences of mannosidase inhibition using the inhibitor kifunensine or MAN1A1 silencing were investigated in breast cancer cells in vitro.
Results:
Patients with low/moderate MAN1A1 expression in tumours showed significantly shorter disease-free intervals than those with high MAN1A1 levels (P=0.005). Moreover, low MAN1A1 expression correlated significantly with nodal status, grading and brain metastasis. At an mRNA level, membrane proteins ALCAM and CD24 were only significantly prognostic in tumours with high MAN1A1 expression. In vitro, reduced MAN1A1 expression or mannosidase inhibition led to a significantly increased adhesion of breast cancer cells to endothelial cells.
Conclusions:
Our study demonstrates the prognostic role of MAN1A1 in breast cancer by affecting the adhesive properties of tumour cells and the strong influence of this glycosylation enzyme on the prognostic impact of some adhesion proteins.
Similar content being viewed by others
Main
In the last years, altered protein glycosylation has been shown in various tumour types, where it often contributes to cancer progression and metastasis (Munkley and Elliott, 2016). At the cell surface, aberrant O- and N-glycans are found because of disturbed glycosylation of adhesion molecules and receptors in tumour cells.
N-glycosylation is a complex process that takes place during translation of target proteins by addition of glycan structures to the amino group of asparagine residues at the consensus motif asparagine-X-serine/threonine (NXS/T) in which X is any amino acid except proline. It starts with synthesis of a dolichol-bound oligosaccharide precursor in the endoplasmatic reticulum (ER), consisting of 14 sugar moieties, among them nine mannose residues. This oligosaccharide is then transferred to a suitable asparagine residue within the nascent polypeptide by the oligosaccharyltransferase (OST) protein complex. Now, the correct folding and secretion of the glycoprotein depends on trimming of the glycan precursor in the ER and Golgi (Figure 1). After removal of three glucose residues by glucosidases and one mannose by ER mannosidase MAN1B1, the glycoprotein is transferred to the Golgi with its N-glycans containing eight mannose residues, termed high-mannose glycans. Generally, few high-mannose glycans reach the cell surface, as additional mannose residues are then cleaved by Golgi mannosidases, which is the prerequisite for formation of complex or hybrid glycans (Moremen et al, 2012).
Principle steps of N-glycosylation and functional role of α 1,2-mannosidases. The transfer of an oligosaccharide precursor to a suitable asparagine residue within the nascent polypeptide is mediated by the oligosaccharyltransferase (OST) protein complex. Subsequent cleavage of three terminal glucose residues by DCS1 and GANAB and one mannose by the α-1,2-ER-mannosidase MAN1B1 leads to the formation of high-mannose glycan structures. Golgi α-1,2-mannosidases (MAN1A1, MAN1A2 and MAN1C1) cleave further α-1,2-bound mannose sugars from high-mannose glycans (Man8-9GlcNAc2), resulting in 5-mannose glycans (Man5GlcNAc2). This glycan structure is the substrate for various glycosyl transferases and mannosidases, which catalyse the formation of complex branched tri- or tetra-antennary N-glycans. ER=endoplasmatic reticulum; FuT=fucosyltransferases (i.e., FUT8); GalT=galactosyltransferases (i.e., B3GALT1); GnT=GlcNac transferases (i.e., MGAT family); ST=syalyltransferases (i.e., ST6GAL and ST3GAL).
The human 1,2-α mannosidases include seven related gene products that are members of CAZy glycosylhydrolase family 47 (GH47): the mannosyl-oligosaccharide 1,2-α-mannosidases MAN1A1, MAN1B1, MAN1A2, MAN1C1 and the ER degradation-enhancing α-mannosidase-like proteins EDEM1, EDEM2 and EDEM3 (Moremen and Nairn, 2014). In biochemical enzyme assays these mannosidases show overlapping substrate specificity, but also slight differences in enzyme activity and preference for specific mannose residues in high-mannose N-glycans (summarised by Moremen and Nairn (2014)). To date, there is only very little knowledge about the role of the different GH47 enzymes in human tissues and disease. MAN1A1, together with MAN1A2 and MAN1C1, belongs to the GH47 Golgi mannosidase I subfamily, which cleaves α-1,2-bound mannose sugars from high-mannose glycans (Man8-9GlcNAc2), resulting in 5-mannose glycans (Man5GlcNAc2). This glycan structure is the substrate for various glycosyl transferases that catalyse the formation of complex branched tri- or tetra-antennary N-glycans (Oliveira-Ferrer et al, 2017; Figure 1). α-mannosidase inhibitors have been previously described to increase surface high-mannose glycans and modulate cellular function, including cell adhesion (Scott et al, 2012).
In a prior study we investigated which glycosylation enzymes are associated with prognosis by analysis of microarray data in two breast cancer cohorts. Among the eight genes whose expression was significantly prognostic in both cohorts, three are involved in N-glycosylation, namely DPM1 (dolichyl-phosphate mannosyltransferase polypeptide 1), RPN1 (ribophorin) and the Golgi mannosidase MAN1A1 (mannosidase α-class 1A member 1). Whereas high mRNA levels of the first two genes correlated with poor prognosis, high MAN1A1 mRNA level was significantly associated with longer recurrence-free and overall survival, suggesting a tumour-suppressor function for this protein (Milde-Langosch et al, 2014).
To date, there is only little knowledge regarding the role of MAN1A1 in cancer progression. In a study on hepatocarcinoma cell lines, significant downregulation of MAN1A1 was observed in highly metastatic compared with non-metastatic cells (Liu et al, 2014), which also points to a tumour-suppressor role of this protein.
The fact that only MAN1A1, but no other GH47 enzymes act as a prognostic factor in our prior mRNA study is unexpected because of the functional overlap among these enzymes. Therefore, the present study aimed to further analyse the role of MAN1A1 on a protein level using tumour tissue samples from breast cancer patients. In addition, the functional consequences of mannosidase inhibition or reduced MAN1A1 expression were examined in breast cancer cell lines in vitro.
Materials and methods
Patient cohorts
MAN1A1 protein expression was analysed in a subcohort of 105 tumours from patients included in the microarray cohort (see below). The clinical and histological characteristics of these cases are shown in Table 1. Patients included in this retrospective study were treated between 1991 and 2002 and selected on the basis of tissue availability. Informed consent for the scientific use of tissue materials, which was approved by the local ethics committee (Ethik-Kommission der Ärztekammer Hamburg, #OB/V/03), was obtained from all patients. The study was performed in accordance to the principles of the Declaration of Helsinki and REMARK criteria (McShane et al, 2006). No radiotherapy, neoadjuvant chemotherapy or endocrine therapy had been administered before surgery. Follow-up data were available for 104 patients. The median overall survival was 141 months (mean 128 months, range 9–251 months), with a median recurrence-free interval of 120 months (mean 116 months, range 0–251 months).
MAN1A1, ALCAM and CD24 mRNA expression was analysed using microarray data (Affymetrix, Santa Clara, CA, USA) from 194 breast cancer samples from our clinic (Milde-Langosch et al, 2014). The data sets supporting the conclusions of this article are available in the Gene Expression Omnibus under the accession numbers GSE26971, GSE31519 and GSE46184 as described (Milde-Langosch et al, 2014).
Western blot analysis
Protein extraction from tumour samples and polyacrylamide gel electrophoresis were performed as described before (Milde-Langosch et al, 2015). MAN1A1 was detected by incubation with the rabbit anti-α-1,2-mannosidase IA antibody (Abcam, Cambridge, UK; ab140613), diluted 1 : 1000 in 1.5% milk powder and incubated overnight at 4 °C.
As a positive control, proteins from the breast cancer cell line MDA-MB231 were loaded in each gel. MAN1A1 expression was quantified using densitometry, and the band volume of each sample was calculated in relation to the MDA-MB231 control, which was set as 100%. As a loading control, the membranes were re-probed with GAPDH antibody (Santa Cruz Biotechnology, Heidelberg, Germany; sc-25778; 1 : 5000; 1 h at room temperature). To study the effect of mannosidase on N-glycosylated proteins, MDA-MB-231 and T47D cells were stimulated with the inhibitor kifunensine for 48 h and the following antibodies were used for western blot analysis: ALCAM mouse monoclonal antibody (diluted 1 : 400 in 1.5% milk powder in TBS-T buffer; NCL-CD166, Novocastra, Newcastle, UK), ICAM-1 mouse monoclonal antibody (1 : 5000 in 1.5% milk powder in TBS-T buffer; sc-8439, Santa Cruz Biotechnology) and B-CAM rabbit monoclonal antibody (diluted 1 : 5000 in 10% Blocking Reagent in TBS-T; clone EPR4165, LS-C138475, LifeSpan Biosciences, Seattle, WA, USA) incubated overnight at 4 °C. Secondary antibodies used (goat anti-mouse IgG-HRP, sc-2055; goat anti-rabbit IgG-HRP, sc-2054) were purchased from Santa Cruz Biotechnology and membranes were incubated for 1 h at room temperature. β-Actin (diluted in 1.5% milk powder in TBS-T buffer, sc-47778, Santa Cruz Biotechnology) served as the loading control.
Quantitative reverse transcriptase polymerase chain reaction
RNA was isolated with RNeasy Mini Kit (Qiagen, Hilden, Germany) and was reverse transcribed using Maxima First Strand cDNA Synthesis Kit (ThermoFisher Scientific; Pinneberg, Germany). Quantitative reverse transcriptase polymerase chain reaction (qRT–PCR) was performed with SYBR Premix Ex Taq (Takara, Kusatsu, Japan) using the Light Cycler (Roche, Mannheim, Germany). Primer sequences for MAN1A1, MAN1A2, MAN1B1 and MAN1B1 are listed in the Supplementary Figure S1C. The data were analysed based on the ΔΔCt method as described (Oliveira-Ferrer et al, 2014).
Breast cancer cell lines, treatments and stable transfections
The human breast cancer cell lines SKBR3, AU565 and HCC1954 were kindly provided by Wirtz (Stratifyer GmbH, Köln, Germany). The cell lines T47D and MDA-MB-231, MDA-MB-231SA and MDA-MB-231-Brain were a generous gift from Professor Harriet Wikman–Kocher (Institute of Tumour Biology, University Medical Center Hamburg, Eppendorf, Hamburg) and Dr Takara (University of Texas), respectively. All cell lines were cultured under standard conditions in a water-saturated atmosphere containing 5% CO2 at 37 °C.
To generate cells with reduced MAN1A1 expression, MDA-MB-231 cells were transfected with five expression vectors containing different shRNA sequences targeting human MAN1A1 and a scramble negative control (shRNA nc; Origene, Herford, Germany) using Lipofectamin (Invitrogen GIBCO/Life Technologies, Carlsbad, CA, USA) as previously described (Oliveira-Ferrer et al, 2014). After puromycin (1 μg ml−1, PAA Laboratories GmbH, Pasching, Austria) selection, the level of MAN1A1 protein and mRNA was determined with western blot analysis and RT–qPCR, and two cultures (MAN1A1 shRNA #2 and MAN1A1 shRNA #3) were chosen for further analysis.
Treatment with kifunensine
The cell lines MDA-MB-231 and T47D were seeded in culture flasks (Greiner Bio-One, Frickenhausen, Germany) and cultured overnight for adherence. Cells were incubated with the mannosidase inhibitor kifunensine (dissolved in DMSO, Sigma-Aldrich Chemie GmbH, Hamburg, Germany) in three different concentrations (1, 10 and 50 μ M) or DMSO for 48 h in serum-reduced medium (5% FBS), and functional assays were subsequently performed. In addition, protein extractions of kifunensine-treated cells (10 μ M for 48 h) were performed using the Qproteome Cell Compartment Kit (Qiagen) according to the manufacturer’s instructions. The respective cytosol, membrane and nucleus fractions were examined for the proteins ALCAM, ICAM-1 and BCAM using western blot analysis.
Static cell adhesion assay
To estimate the adherence of tumour cells to endothelial cells, static cell adhesion assays were performed as described (Dippel et al, 2013). One day before the experiment started, tumour cells were labeled with CellTrackerTM Green CMFDA (Molecular Probes, Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol. Human pulmonary microvascular endothelial cells (HPMECs, PromoCell, Heidelberg, Germany) were seeded in black 96-well plates (Greiner Bio-One) and grown to confluence. HPMECs were stimulated with tumour necrosis factor α (TNFα; Sigma-Aldrich GmbH) 4 h before the addition of tumour cells. Tumour cells treated with kifunensine or those with reduced MAN1A1 expression (MAN1A1 shRNA #2, #3), as well as their respective control cells, were washed with DPBS and detached with AccuMax (eBioscience, San Diego, CA, USA). Then, culture medium of the HPMEC was removed and 80 000 tumour cells per well were added directly and incubated for 40 min at 37 °C. Afterwards, wells were washed three times with DPBS containing MgCl2 and CaCl2. Finally, fluorescence was measured with a fluorescence plate reader (Infinite 200 PRO multimode reader, Tecan, Männedorf, Switzerland).
Dynamic cell flow adhesion assay
Cell adhesion under flow conditions was analysed with ibiTreat slides (IBIDI, Martinsried, Germany) as described (Dippel et al, 2013; Oliveira-Ferrer et al, 2014). The channels of these slides were coated with HPMECs (80 000 cells per channel) and cultivated until confluence for ∼48 h. Then, cells were stimulated with TNFα (10 ng ml−1) for 4 h, which has previously been shown to be the optimal activation period. MDA-MB-231 cells were adjusted to 105 cells per ml in serum-free medium. The perfusion pump created a constant flow of the cell suspension through the flow chamber of 8.5 ml h−1, corresponding to a shear stress of 0.25 dyn cm−2. Cell adhesion was digitally recorded for 2 min with a video camera mounted on the microscope. Adherent cells were counted using CapImage software (Dr. Zeintl, Heidelberg, Germany) and given as a percentage of adherent control cells per minute.
Proliferation assay
To analyse the proliferation of MDA-MB-231 MAN1A1 shRNA clones, 5 × 103 cells of MAN1A1 shRNA #2, MAN1A1 shRNA #3 or nc shRNA clones in culture medium with either 1% or 10% FBS were seeded in 96-well plates and cell proliferation was analysed using the Cell Proliferation Kit (MTT, Roche) after 24, 48 and 72 h as previously described (Oliveira-Ferrer et al, 2014).
Cytotoxicity assay
Transfected MDA-MB-231 cells were seeded (1 × 104 cells per well) in 96-well plates, incubated overnight and treated with camptothecin in seven different concentrations (1–0.01 μg ml−1) for 24 h. Cell viability was determined using the MTT kit (Roche).
Migration and motility assays
To study cell migration, cells were seeded with 2 × 105 cells per well in six-well plates (Greiner Bio-One). After 48 h, a wound-healing assay was performed as described previously (Oliveira-Ferrer et al, 2014). Subsequently, the migration potential of the cells was determined by analysing the gap closure of four different areas in duplicates after four different time points (10, 24, 30 and 48 h) with the ImageJ Wound Healing Tool (Wayne Rasband, National Institute of Health).
Statistics
Statistical analysis was conducted using SPSS software Version 24 (IBM SPSS Statistics, Armonk, NY, USA). Correlations between the MAN1A1 mRNA and protein expression values were examined using two-sided Pearson tests. For further statistical analysis, all tumour cases were divided into four groups of similar size (quartiles) representing low, moderate/low, moderate/strong and strong MAN1A1 expression.
χ2-tests were used to examine the correlations between MAN1A1 expression and clinicopathological factors. For prognostic parameters, the following groups were compared: histological grading (G1/2 vs G3); breast cancer stage (I/II vs II vs IV); nodal status (positive vs negative); ER and PR status (positive vs negative); presence of bone, lung, visceral or brain metastasis (positive vs negative); and molecular subtype (luminal vs HER2-enriched vs triple-negative). Survival curves were plotted by Kaplan–Meier analysis. Differences between survival curves were evaluated by log-rank tests. Probability values less than 0.05 were regarded as statistically significant.
Results
MAN1A1 protein expression and correlation with mRNA data
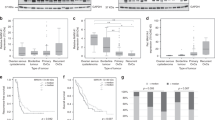
Using western blot analysis in 105 breast cancer samples, an at least minimal MAN1A1 protein expression was detected in all tumours. Yet, in contrast to MDA-MB-231 and other cell lines that showed the expected band at ∼70 kDa, one or more additional bands at ∼60 kDa were detected in most tissue samples (Figure 2A). As the function of these smaller proteins is not clear, we quantified both the 70 kDa- and the combined 60-kDa bands separately using densitometry.
MAN1A1 protein expression in clinical tumour tissue samples. (A) Representative western blot analysis showing MAN1A1 expression (ca. 70 and 60 kDa) in two breast cancer cell lines and 15 clinical tumour tissue samples. As a loading control, GAPDH expression is shown. (B, C) Kaplan–Meier analysis showing correlations of high MAN1A1 protein expression with longer disease-free survival (B) and overall survival (C). P-values after log-rank tests comparing four groups (quartiles Q1–Q4) or two groups (quartiles Q1–Q3 vs Q4) are shown. (D–I) Correlation of MAN1A1 protein expression with clinical and histological tumour parameters. P-values after χ2-tests are given. Significant associations of high MAN1A1 expression with negative nodal status (D) and high to moderate differentiation (G1/2; E), as well as a weak association with a luminal molecular subtype, are shown (F). In addition, low MAN1A1 levels correlated significantly with the appearance of brain metastases (G), whereas a trend was observed for bone and lung metastases (H, I).
Expression values of the 70-kDa bands, relative to MDA-MB-231, ranged from 16 to 684%, with a median expression of 163%. For the 60-kDa bands, expression relative to the same MDA-MB-231 band ranged from 0 to 637% (median 77%). Both bands correlated strongly with each other by Pearson tests (correlation coefficient 0.877). In addition, both the 70-kDa- and the 60-kDa bands showed significant correlations with MAN1A1 mRNA expression as obtained by microarray analysis as shown before (Milde-Langosch et al, 2014). Pearson coefficients for correlations with the Affymetrix 208116_s_at probeset for both protein bands were 0.493 and 0.491, respectively. For the second probeset 221716_at, these values were 0.429 and 0.441, respectively. These data point to an at least moderate extent of transcriptional regulation of the MAN1A1 expression in tumour tissue. In the following chapters, we confine ourselves to the MAN1A1 70-kDa band, yet similar results were obtained for the smaller band (Supplementary Table S1).
Correlation of MAN1A1 expression with clinicopathological variables
For correlations with clinical and histological prognostic tumour parameters, as well as the follow-up information, the cohort was divided into quartiles of similar size (25–27 samples) with low, moderate/low, moderate/strong and strong MAN1A1 expression. Using χ2-tests, we found significant correlations of low MAN1A1 protein expression with nodal involvement and high grading (P=0.002, P=0.047; Figures 2D and E). On the basis of the mRNA expression of ESR1, PGR and ERBB2, the molecular subtypes of the samples of this cohort had been previously defined (Milde-Langosch et al, 2013). When comparing these groups, differences in MAN1A1 protein levels were observed, with a lower MAN1A1 expression in triple-negative tumours compared with HER2-enriched or luminal carcinomas (P=0.057; Figure 2F). Regarding the immunohistochemically determined oestrogen receptor (ER) expression and the tumour stage, no significant correlations were found (Supplementary Table S1).
In most cases of tumour recurrence, the site(s) of metastasis is well documented for our cohort. Interestingly, tumours, which later developed metastases of the bone, lung or brain were characterised by lower MAN1A1 expression compared with tumours without these metastases (Figures 2G–I). For brain metastasis, this difference reached statistical significance (P=0.043; Figure 2G). No difference in MAN1A1 expression was observed for cases with and without visceral/hepatic metastases (Supplementary Table S1).
Using Kaplan–Meier analysis comparing the four quartiles, we found significant inverse correlations of MAN1A1 expression with survival (Figures 2B and C). In cases with high MAN1A1 protein levels (quartile 4), only few events of recurrence or death were observed. On the contrary, tumours with low/moderate MAN1A1 expression showed relatively short disease-free intervals (mean RFS of 103, 113, 152 and 210 months in quartiles 1–4) and overall survival (mean OAS of 146, 144, 177 and 218 months in quartiles 1–4; log-rank: P=0.005 and P=0.050, respectively). Accordantly, the 10-year survival rate for recurrence-free intervals and overall survival increased with MAN1A1 expression (RFS: 44, 46, 60 and 79% for quartile 1–4; OAS: 59, 53, 67 and 84% in quartiles 1–4). In a multivariate analysis, we could not confirm the prognostic role of MAN1A1 expression adjusted for classical prognostic parameters, such as nodal involvement and grading (Supplementary Table S2).
Influence of MAN1A1 expression on the prognostic influence of target proteins
Our analyses have shown a clear correlation of low MAN1A1 mRNA and protein levels – presumably resulting in large amounts of high-mannose glycans – with high tumour aggressiveness. This suggests that the nature of N-glycans (high-mannose vs complex types) might influence the biological properties of target proteins, which affect tumour progression and metastasis. On the basis of this hypothesis, we analysed the prognostic value of selected highly N-glycosylated proteins, comparing tumours with low vs high MAN1A1 expression. For this purpose, we used the mRNA microarray data of our previously described Hamburg breast cancer cohort (Milde-Langosch et al, 2014).
The role of the activated leukocyte cell adhesion molecule (ALCAM) in breast cancer has been studied by us and others, resulting in partly conflicting results. As we found that the prognostic role varies depending on the treatment (Ihnen et al, 2008), we only included chemotherapy-treated patients (n=100) in our stratified analysis. After dividing this cohort into quartiles based on ALCAM mRNA (Affymetrix no. 201992_at) levels, Kaplan–Meier analysis revealed the expected correlation of high ALCAM expression with longer RFS (P=0.049; mean RFS: 104, 129, 142 and 155 months in quartiles 1–4), whereas the association with overall survival did not reach statistical significance (not shown). In a stratified analysis we found that this correlation was highly dependent on MAN1A1 levels: only in tumours with relatively high MAN1A1 mRNA level (>median), ALCAM expression was significantly associated with longer DFS (Figures 3A and B). Ten-year DFS rates in that high-MAN1A1 group were 36% in cases with low ALCAM mRNA levels (Q1) vs 81% in tumours with higher ALCAM levels (Q2–4; P=0.001), with the mean DFS times of 89 and 165 months, respectively. Similar results were shown regarding overall survival (P=0.639 and 0.031 for cases with low vs high MAN1A1 expression (not shown)).
Influence of MAN1A1 expression on the prognostic role of ALCAM and CD24. Expression of MAN1A1 and ALCAM (A, B) or MAN1A1 and CD24 (C, D) were analysed in clinical tumour tissue samples, based on cDNA microarray data. Regarding the ALCAM or CD24 expression data, the cases were divided into four quartiles for Kaplan–Meier analysis and log-rank tests, stratified for tumours with low (<median) or higher (>median) MAN1A1 mRNA expression. High CD24 and low ALCAM expression correlated significantly with shorter overall survival only in cases with a higher mannosidase MAN1A1 expression (B, D).
CD24 is another strongly N-glycosylated protein, which has been reported to has an important role in breast cancer progression (Kwon et al, 2015). In our cohort of breast cancer samples with cDNA microarray data, high CD24 expression (Affymetrix no. 266_s_at) was significantly associated with shorter DFS and OAS (P=0.013 and P=0.007; mean DFS rates: 189, 156, 137 and 112 months in quartiles 1–4; not shown). In a stratified analysis, this prognostic impact was only found in tumours with high MAN1A1 expression, whereas in cases with low MAN1A1 levels CD24 lost its significant role (Figures 3C and D). In the former group, DFS rates in quartiles 1–4 were 203, 165, 153 and 105 months, respectively (P=0.005). A similar difference was shown regarding the influence of CD24 on overall survival (log-rank test: P=0.399 and 0.001 for tumours with low and high MAN1A1 mRNA expression; not shown), and for the other CD24 probesets present on the used Affymetrix chips.
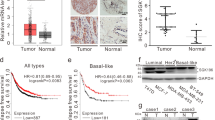
MAN1A1 expression in breast cancer cell lines
MAN1A1 expression was further analysed by western blot analysis in the following breast cancer cell lines: T47D and MCF7 (ER+ and PR+), SKBR3, AU565 and HCC1954 (HER2+) and MDA-MB-231, MDA-MB-231-Br and MDA-MB-231-SA (TNBC). In concordance with the results on patient samples, no significant correlation between MAN1A1 expression and the molecular subtype of the cell lines could be observed. Interestingly, we detected a remarkably decreased MAN1A1 expression in the bone-seeking (MDA-231-SA) and brain-seeking (MDA-231-BR) clones compared with their parental human breast cancer cell line MDA-MB-231 (Figure 4).
MAN1A1 protein expression in breast cancer cell lines. (A) Western blot analysis showing MAN1A1 expression in breast cancer cell lines: T47D and MCF7 (ER+ and PR+), SKBR3, AU565 and HCC1954 (HER2+) and MDA-MB-231, MDA-MB-231-BR and MDA-MB-231-SA (TNBC). As a loading control, GAPDH expression is shown. (B) MAN1A1 expression was quantified by densitometry, normalised to GAPDH expression and the relative expression of each sample was calculated in relation to MDA-MB231 cells, which was set as 1.
Influence of MAN1A1 on the adhesive properties of breast cancer cells
Once tumour cells have entered into the vascular system, adhesion to the EC bed represents a key step in order to successfully extravasate and form distant metastasis. As most cell adhesion molecules involved in this process are strongly glycosylated, we aimed to analyse the impact of mannosidase activity on this particular step of the metastatic cascade. Therefore, cell lines with high endogenous MAN1A1 expression were treated with the α-mannosidase I inhibitor kifunensine (Elbein et al, 1990), and subsequently tumour cell adhesion to human endothelial cells was evaluated. As the lung is a major metastatic site in breast cancer, HPMECs were chosen for these experiments. In order to study the impact of tumour cell glycosylation on adhesion, kifunensin has been applied exclusively to the tumour cell lines and not to the endothelial cells in the following experiments.
For the static adhesion assays, T47D and MDA-MD-231 cells were treated with 1, 10 and 50 μ M kifunensine for 48 h, labelled with CMFDA fluorescence dye and incubated on HPMEC for 30 min. Stimulation with TNFα, which has been described to upregulate diverse adhesion molecules on ECs, was performed before each adhesion assay. The quantification of adherent tumour cells showed a significant and dose-dependent increase in MDA-MB-231 cell adhesion to both untreated, as well as TNFα-stimulated ECs (Figures 5A and B). Kifunensine treatment of T47D cells, which endogenously express higher levels of MAN1A1 than MDA-MB-231 (Figure 4), also led to a significant increase in tumour cell adhesion on both TNFα-treated or untreated ECs, but only at the highest concentration of 50 μ M (Figures 5C and D).
Influence of MAN1A1 expression on the adhesion of breast cancer cells to endothelial cells. (A–F) After treatment with different concentrations of the mannosidase inhibitor kifunensine, tumour cell adhesion to activated (+TNFα) and not activated (−TNFα) endothelial cells was analysed under static (MDA-MB-231: A, B; T47D; C, D) and dynamic conditions (MDA-MB-231: (E, F). (G) Western blot analysis showing a mass shift in ALCAM, ICAM-1 and BCAM after treatment of T47D and MDA-MB-231 cells with 10 μ M kifunensine. (H) Western blot analysis (upper panel) and subsequent quantification (lower panel) of MAN1A1 knockdown in MDA-MB-231 cells, showing MAN1A1 expression in stably transfected MDA-MB-231 cells with scramble shRNA (nc) and two different MAN1A1 specific shRNA sequences (#2 and #3). (I–L) Tumour cell adhesion to activated (+TNFα) and control (−TNFα) endothelial cells in MAN1A1 knockdown MDA-MB-231 cells compared with the nc cells under static (I, J) and cell flow conditions (K, L). Values are means±s.d. (n=3). *P<0.05, **P<0.005.
Mimicking physiological conditions, we further performed dynamic adhesion assays with capillaries that were coated with EC cells. Here, MDA-MB-231 cells treated with kifunensine (average value from three experiments; n=2.5 cells per min) exhibited significantly increased adhesion to non-activated endothelial monolayers compared with the control (average value from three experiments; n=0.3 cells per min; Figure 5E), and the same trend was observed on TNFα-stimulated ECs. Here, the average amount of adherent control cells was 2, whereas 4.5 cells attached to the EC monolayer after kifunensine treatment (Figure 5F; Ø vs 10 μ M: P=0.065; Ø vs 50 μ M: P=0.1).
The mannosidase-inhibiting activity of kifunensine, which leads to a modification of N-glycan structures on the cell, could be corroborated by western blot analysis on kifunensine-treated vs untreated breast cancer cells. Here, the N-glycosylated adhesion molecules ALCAM, ICAM-1 and BCAM showed a molecular mass shift in both MDA-MB-231 and T47D cells after treatment with kifunensine (Figure 5G). Cell fractionation experiments corroborated the impact of kifunensine on the glycosylation pattern of those CAMs located at the cell surface (Supplementary Figure S2).
As kifunensine does not specifically inhibit MAN1A1 (Golgi class I mannosidase IA) but also other type I α-mannosidases, such as endoplasmatic reticulum α-1,2-mannosidase I (MAN1B1), Golgi class I mannosidases IB (MAN1A2) and IC (MAN1C1), we also generated stably transfected MDA-MB-231 cell lines with reduced MAN1A1 expression and analysed them in static and dynamic adhesion assays. Two different MAN1A1-specific shRNA sequences (#2 and #3) were used in order to produce respective sublines with 60% and 95% MAN1A1 knockdown, respectively (Figure 5H).
In static adhesion assays, we observed a significantly increased adhesion of MDA-MB-231 cells with reduced MAN1A1 expression (MAN1A1-shRNA cells #2 and #3) to activated endothelial cells compared with controls (scramble shRNA: nc; Figure 5J). This effect could not be reproduced with unstimulated ECs. Here, only MAN1A1-shRNA #3 cells, which were characterised by a more efficient MAN1A1 knockdown than MAN1A1-shRNA #2 (Figure 5H), showed a slightly enhanced adhesion to endothelial cells (Figure 5I).
Further, no significant differences in the adhesive behaviour of MAN1A1 knockdown cells compared with control ones were found in the dynamic assays, either on stimulated or unstimulated ECs (Figures 5K and L).
In order to rule out the possibility that the MAN1A1 silencing was neutralised by a compensatory upregulation of other α-mannosidases, qRT–PCR for MAN1A2, MAN1B1 and MAN1C1 was performed with the MAN1A1 knockdown cells and controls. MAN1A1 knockdown in MDA-MD-231 cells leads to a weak downregulation of MAN1A2 and MAN1C, whereas no consistent results in both clones were obtained for MAN1B1 (Supplementary Figure S1B).
Influence of MAN1A1 on proliferation, migration and apoptotic potential of breast cancer cells
The effect of reduced MAN1A1 expression on further cellular properties was analysed in vitro using stably transfected MDA-MB-231 cells. No influence of MAN1A1 knockdown on cell growth was observed in normal growth medium and under serum-reduced conditions (not shown). In addition, cell viability analysis after exposure to the cytotoxic agent camptothecin showed no significant differences in the amount of apoptotic cells in control MDA-MB-231 cells and those with reduced MAN1A1 expression (not shown). In contrast, in an in vitro scratch assay, we observed a significantly retarded wound closure of MDA-MB-231 cells with reduced MAN1A1 expression (MAN1A1 shRNA #2 and #3) compared with control ones. Here, a slight difference was already evident after 10 h, and this trend became more pronounced and statistically significant after 24, 30 and 48 h (Supplementary Figure S3).
Discussion
On the basis of our experimental data and investigations of clinical tumour tissue, we could clearly show that the mannosidase MAN1A1 has a metastasis-suppressor function in breast cancer. Weak MAN1A1 protein expression correlates with a higher probability of lymph node involvement, metastasis to the lung, bone or brain, and is associated with a significantly shorter disease-free and overall survival. Comparison of western blot results and mRNA expression data in cell lines and tumour tissue also indicates that MAN1A1 levels are subjected to transcriptional regulation.
Mannosidases have an important role in the maturation of N-glycans in the Golgi apparatus. In the absence of mannosidases, N-glycans stay in the high-mannose form, whereas in the presence of these enzymes mannose residues are cleaved and replaced by other sugars, leading to the formation of the highly variable complex N-glycans. MAN1A1 is not the only mannosidase involved in this process (Moremen and Nairn, 2014). Yet, although in vitro data point to a partial redundancy between MAN1A1 and other enzymes such as MAN1A2 or MAN1C1, MAN1A1 is the only member of this group whose expression correlates with clinical tumour characteristics, pointing to an outstanding role of this mannosidase. To date, we cannot explain the unexpected presence of a second MAN1A1 band of ∼60 kDa in addition to the expected band of ca. 70 kDa in western blot analysis of tumour tissues. As there are hardly any MAN1A1 western blot studies with clinical samples, it might have thus far been overlooked. Yet, regarding the strong correlation between expression values of both bands and the similar results of statistical analysis (Supplementary Table S1), we have no doubt that both are functional MAN1A1 protein variants.
Low mannosidase activity in the Golgi results in the accumulation of high-mannose glycans in the cells. In breast cancer tissue samples, N-glycan profiles of membrane proteins showed marked increases of high-mannose N-glycans, but also of triantennary complex glycans, compared with matched normal tissue (Liu et al, 2013). Similarly, in a breast tumour mouse model, N-glycan profiling of serum proteins showed a significant elevation of high-mannose glycans in the presence of breast cancer (de Leoz et al, 2011). This indicates that tumour progression might be accompanied to some extent by reduced mannosidase activity, which fits to our data, possibly leading to a higher abundance of high-mannose structures as compared with normal cells. Yet, regarding the redundancy of mannosidases and the activity of other glycosylation enzymes in tumour cells, we cannot equate MAN1A1 expression with a certain glycoform, and additional investigations including glycan-MS will be necessary to clarify the function of the specific glycans. In this context, we have observed an association between MAN1A1 mRNA levels and GNA (Galanthus nivalis) binding, a lectin with high affinity to α-1-3 and α-1-6-linked high mannose structures (Supplementary Figure S4).
To date, little is known about the role of the mannosidase MAN1A1 in human tumours. In a small study on transcriptional profiling of glycogenes, MAN1A1 was previously found to be downregulated in metastatic hepatocellular cancer (HCC) cell lines and orthotopic xenograft tumours, as compared with non-metastatic HCC controls (Liu et al, 2014). Our results on breast cancer samples point to a similar role of this enzyme in metastasis of this entity. In humans, more than 2000 proteins, including many adhesion proteins and membrane receptors, harbour amino-acid motifs suitable for N-glycosylation. Many of these proteins are involved in metastasis, and correct N-glycosylation might be crucial for their function. In our cohort, the membrane proteins ALCAM and CD24, which are known to have a role in breast cancer progression, were only significantly prognostic in tumour tissue samples if there was a relatively high MAN1A1 expression. We assume that tumour cells with normal or high levels of MAN1A1 are able to ‘correctly’ glycosylate ALCAM and CD24, such that these factors are consequently able to fully exert their function, showing a prognostic impact for breast cancer patients. In contrast, low levels of MAN1A1 would lead to an aberrant glycosylation, thereby impairing ALCAM and CD24 function and their influence on patient survival.
In mouse models, CD24-positive cells display high tumourigenic properties and metastatic potential, partly because of CD24’s capacity to bind to P-selectin (Rostoker et al, 2015). Mannosidase activity is the prerequisite for formation of complex N-glycans potentially harbouring selectin-binding structures like Lewis antigens. This might be the background of the observation that, in tumours with low MAN1A1 expression, CD24 has no prognostic influence. Taken together, our data indicate that reduced MAN1A1 expression leading to aberrant N-glycosylation might have a superior prognostic role compared with adhesion proteins whose function is dependent on proper glycosylation.
Extensive research has been carried out on the role of protein glycosylation in leucocyte trafficking (Huo and Xia, 2009; Sperandio et al, 2009). Moreover, some studies have explored how modulation of N-glycans in endothelial cells serves, especially during inflammation, as a mechanism to control leucocyte–endothelial adhesion (Chacko et al, 2011; Scott et al, 2013). Here, the presence of high-mannose structures on cell adhesion molecules, such as ICAM-1, as a consequence of mannosidase inhibition leads to increased monocyte rolling and adhesion. In this context, it is reasonable to assume that tumour cells might use a similar mechanism to augment adhesion to vascular walls, thereby promoting extravasation and subsequent colonisation of distant tissue. Indeed, in the present study we show for the first time an increased adhesion of two breast cancer cell lines on endothelial cells after treatment with the mannosidase inhibitor kifunensine (under static and dynamic conditions). The observation that treatment of the endothelial cells with TNFα does not strongly increase this effect suggests that this adhesion is independent of selectins, as these are upregulated by TNFα stimulation.
Kifunensine is a potent inhibitor of mannosidase I enzymes, including endoplasmic reticulum α-1,2-mannosidase I (MAN1B1) and Golgi class I mannosidases IA (MAN1A1), IB (MAN1A2) and IC (MAN1C1) with Ki values of 130 and 23 nM, respectively (Elbein et al, 1990). It has been described to cause a shift in the structure of N-glycans from complex chains to high-mannose structures. This effect could be corroborated in our breast cancer cell lines, where lower molecular mass bands could be detected in western blots for ALCAM, ICAM-1 and BCAM after treatment with kifunensine. For ICAM-1, this lower MW isoform has been previously described to contain high-mannose N-glycans in endothelial cells (Scott et al, 2013).
Similar to mannosidase inhibition by kifunensine, specific knockdown of MAN1A1 in the cell line MDA-MB-231 led to enhanced adhesion to activated endothelial cells in static adhesion assays, whereas no impact on adhesion could be observed in experiments under flow conditions. This weaker effect of MAN1A1 knockdown compared with kifunensine inhibition might be attributed to some extent to the residual endogenous MAN1A1 enzyme activity present in the transfected cells, which could be sufficient to conduct at least partial N-glycan trimming, but certainly also to the presence of other active mannosidase class I family members, which can partially compensate the reduced MAN1A1 activity.
Nevertheless, a compensatory upregulation of other mannosidases in response to MAN1A1 deregulation can be excluded. We did not observe any significant correlation between MAN1A1, MAN1B1, MAN1A2, MAN1C1, EDEM1, EDEM2 and EDEM3 transcriptional levels in tumour tissue samples, indicating the lack of a compensatory regulation of other GH47 mannosidases in tumours with reduced MAN1A1 levels. Similarly, no compensatory upregulation of MAN1B1, MAN1A2 or MAN1C1 could be observed in MAN1A1 knockdown MDA-MB-231 cells (Supplementary Figure S1).
In contrast to cell proliferation and apoptotic potential, which were not influenced by MAN1A1 expression, we found a slowdown of cell migration after MAN1A1 knockdown in breast cancer cells, suggesting that an increase in high-mannose and a decrease in complex N-glycans might reduce cell motility. These results correspond well to those obtained in studies on the N-acetylglucosaminyl-transferase V (MGAT5), which is involved in the synthesis of complex branched N-glycans: knockdown of MGAT5 expression in fibrosarcoma cells resulted in decreased levels of complex glycans, increased cell–cell contacts due to altered glycosylation of cadherins and decreased migration and invasion. Conversely, MGAT5 overexpression in different cancer cells led to high cellular motility and invasion because of its influence on N-glycosylation of integrins or laminins (Oliveira-Ferrer et al, 2017). Yet, reduced cell motility in MAN1A1 knockdown cells does not correspond to the characteristics of low-MAN1A1 tumours in vivo, suggesting a minor impact of this feature on tumour cell aggressiveness.
In conclusion, the results of our present study corroborate a tumour-suppressive role of the Golgi mannosidase MAN1A1 in breast cancer, where MAN1A1 expression levels significantly affect recurrence-free and overall survival. MAN1A1-mediated glycosylation influences tumour cell adhesion of breast cancer cells to lung endothelial cells, and strongly affects the prognostic impact of well-characterised adhesion proteins such as ALCAM and CD24. Thus, the understanding of glycan alterations may provide new options for therapeutic intervention.
Change history
20 March 2018
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Chacko BK, Scott DW, Chandler RT, Patel RP (2011) Endothelial surface N-glycans mediate monocyte adhesion and are targets for anti-inflammatory effects of peroxisome proliferator-activated receptor gamma ligands. J Biol Chem 286: 38738–38747.
de Leoz ML, Young LJ, An HJ, Kronewitter SR, Kim J, Miyamoto S, Borowsky AD, Chew HK, Lebrilla CB (2011) High-mannose glycans are elevated during breast cancer progression. Mol Cell Proteomics 10, M110.002717-1-9.
Dippel V, Milde-Langosch K, Wicklein D, Schumacher U, Altevogt P, Oliveira-Ferrer L, Janicke F, Schroder C (2013) Influence of L1-CAM expression of breast cancer cells on adhesion to endothelial cells. J Cancer Res Clin Oncol 139: 107–121.
Elbein AD, Tropea JE, Mitchell M, Kaushal GP (1990) Kifunensine, a potent inhibitor of the glycoprotein processing mannosidase I. J Biol Chem 265: 15599–15605.
Huo Y, Xia L (2009) P-selectin glycoprotein ligand-1 plays a crucial role in the selective recruitment of leukocytes into the atherosclerotic arterial wall. Trends Cardiovasc Med 19: 140–145.
Ihnen M, Muller V, Wirtz RM, Schroder C, Krenkel S, Witzel I, Lisboa BW, Jaenicke F, Milde-Langosch K (2008) Predictive impact of activated leukocyte cell adhesion molecule (ALCAM/CD166) in breast cancer. Breast Cancer Res Treat 112: 419–427.
Kwon MJ, Han J, Seo JH, Song K, Jeong HM, Choi JS, Kim YJ, Lee SH, Choi YL, Shin YK (2015) CD24 overexpression is associated with poor prognosis in luminal A and triple-negative breast cancer. PLoS One 10: e0139112.
Liu T, Zhang S, Chen J, Jiang K, Zhang Q, Guo K, Liu Y (2014) The transcriptional profiling of glycogenes associated with hepatocellular carcinoma metastasis. PLoS One 9: e107941.
Liu X, Nie H, Zhang Y, Yao Y, Maitikabili A, Qu Y, Shi S, Chen C, Li Y (2013) Cell surface-specific N-glycan profiling in breast cancer. PLoS One 8: e72704.
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM (2006) REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat 100: 229–235.
Milde-Langosch K, Karn T, Muller V, Witzel I, Rody A, Schmidt M, Wirtz RM (2013) Validity of the proliferation markers Ki67, TOP2A, and RacGAP1 in molecular subgroups of breast cancer. Breast Cancer Res Treat 137: 57–67.
Milde-Langosch K, Karn T, Schmidt M, zu Eulenburg C, Oliveira-Ferrer L, Wirtz RM, Schumacher U, Witzel I, Schutze D, Muller V (2014) Prognostic relevance of glycosylation-associated genes in breast cancer. Breast Cancer Res Treat 145: 295–305.
Milde-Langosch K, Schutze D, Oliveira-Ferrer L, Wikman H, Muller V, Lebok P, Pantel K, Schroder C, Witzel I, Schumacher U (2015) Relevance of betaGal-betaGalNAc-containing glycans and the enzymes involved in their synthesis for invasion and survival in breast cancer patients. Breast Cancer Res Treat 151: 515–528.
Moremen KW, Nairn AV (2014) Mannosidase, Alpha, class 1 (MAN1A1 (Golgi alpha-mannnosidase IA), Man1A2 (Golgi alpha-mannosidase IB), MAN1B1 (ER alpha-mannosidase I), MAN1C1(Golgi alpha-mannosidase IC)). In Handbook of Glycosyltransferases and Related Genes, Taniguchi N, et al. (eds), pp 1297-1312. Springer: Japan.
Moremen KW, Tiemeyer M, Nairn AV (2012) Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol 13: 448–462.
Munkley J, Elliott DJ (2016) Hallmarks of glycosylation in cancer. Oncotarget 7: 35478–35489.
Oliveira-Ferrer L, Legler K, Milde-Langosch K (2017) Role of protein glycosylation in cancer metastasis. Semin Cancer Biol 44: 141–152.
Oliveira-Ferrer L, Rossler K, Haustein V, Schroder C, Wicklein D, Maltseva D, Khaustova N, Samatov T, Tonevitsky A, Mahner S, Janicke F, Schumacher U, Milde-Langosch K (2014) c-FOS suppresses ovarian cancer progression by changing adhesion. Br J Cancer 110: 753–763.
Rostoker R, Abelson S, Genkin I, Ben-Shmuel S, Sachidanandam R, Scheinman EJ, Bitton-Worms K, Orr ZS, Caspi A, Tzukerman M, LeRoith D (2015) CD24(+) cells fuel rapid tumor growth and display high metastatic capacity. Breast Cancer Res 17: 78.
Scott DW, Chen J, Chacko BK, Traylor JG Jr, Orr AW, Patel RP (2012) Role of endothelial N-glycan mannose residues in monocyte recruitment during atherogenesis. Arterioscler Thromb Vasc Biol 32: e51–e59.
Scott DW, Dunn TS, Ballestas ME, Litovsky SH, Patel RP (2013) Identification of a high-mannose ICAM-1 glycoform: effects of ICAM-1 hypoglycosylation on monocyte adhesion and outside in signaling. Am J Physiol Cell Physiol 305: C228–C237.
Sperandio M, Gleissner CA, Ley K (2009) Glycosylation in immune cell trafficking. Immunol Rev 230: 97–113.
Acknowledgements
We are grateful to the excellent technical assistance of Oliver Schnapauff. We also thank Sarah Bryan for her collaboration on language revision.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
RM Wirtz is an employee of a diagnostic company. The remaining authors declare no conflict of interests.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Legler, K., Rosprim, R., Karius, T. et al. Reduced mannosidase MAN1A1 expression leads to aberrant N-glycosylation and impaired survival in breast cancer. Br J Cancer 118, 847–856 (2018). https://doi.org/10.1038/bjc.2017.472
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.472
Keywords
This article is cited by
-
N-glycan profiling of tissue samples to aid breast cancer subtyping
Scientific Reports (2024)
-
Different glycosylation profiles of cystatin F alter the cytotoxic potential of natural killer cells
Cellular and Molecular Life Sciences (2024)
-
Transcriptomic comparison of bone marrow CD34 + cells and peripheral blood neutrophils from ET patients with JAK2 or CALR mutations
BMC Genomic Data (2023)
-
Debaryomyces hansenii supplementation in low fish meal diets promotes growth, modulates microbiota and enhances intestinal condition in juvenile marine fish
Journal of Animal Science and Biotechnology (2023)
-
Unconventional protein post-translational modifications: the helmsmen in breast cancer
Cell & Bioscience (2022)