Abstract
Background:
The cellular autophagic response to radiation is complex. Various cells and tissues respond differentially to radiation, depending on both the dose of exposure and the time post irradiation. In the current study, we determined the autophagosomal and lysosomal response to radiation in lung cancer cell lines by evaluating the expression of the associated proteins, as well as the effect of relevant gene silencing in radio and chemosensitisation. Furthermore, tumour sensitisation was evaluated in in vivo autophagic gene silencing model after irradiation.
Methods:
A549 and H1299 cell lines were utilised as in vitro cancer models. Both cell lines were transfected with various small-interfering RNAs, silencing auto-lysosomal genes, and irradiated with 4 Gy. Cell growth response was evaluated with AlamarBlue assay. Western blot and confocal microscopy were utilised for the characterisation of the auto-lysosomal flux. Also, the H1299 cell line was stable transfected with small-hairpin RNA of the MAP1LC3A gene, and the tumour radiosensitisation in Athymic Nude-Foxn1nu was evaluated.
Results:
Following exposure to 4 Gy of radiation, A549 cells exhibited a significant induction of the autophagic flux, which was not supported by transcriptional activation of auto-lysosomal genes (LC3A, LC3B, p62, TFEB and LAMP2a), resulting in aggresome accumulation. Recovery of transcriptional activity and autophagy efficacy occurred 7 days post irradiation. Alternatively, H1299 cells, a relatively radio-resistant cell line, sharply responded with an early (at 2 days) transcriptional activation of auto-lysosomal genes that sustained an effective autophagosomal flux, resulting in adequate aggresome clearance. Subsequently, we tested the silencing of four genes (LC3A, LC3B, TFEB and LAMP2a), confirming a significant radiosensitisation and chemosensitisation to various chemotherapeutic agents, including cisplatin and taxanes. In mouse xenografts, exposure to radiation significantly reduced tumour growth (P<0.001), which was exacerbated among shLC3A-H1299 transfected tumours.
Conclusions:
The ability of lung cancer cells to survive after irradiation at 4 Gy depends on their ability to sustain a functional autophagic flux. Abrogation of such ability results in increased radiosensitivity and susceptibility to various chemotherapy agents. Selective inhibitors of cancer cell autophagic function may prove important for the eradication of lung cancer.
Similar content being viewed by others
Main
Lung cancer is the most common form of cancer worldwide, with an estimated 1.8 million new cases diagnosed in 2012, with high lethality even in operable patients (World Cancer Research Fund International, 2012). For non-small-cell lung cancer (NSCLC), radiotherapy improves survival in patients with incomplete resection, but the 5-year survival rate remains below 40% (Wang et al, 2015). The combination of radiotherapy with chemotherapy is the treatment of choice for locally advanced, inoperable disease (Bezjak et al, 2015). However, in-field control failure occurs in 50–70% of patients resulting in a 2-year survival rate <20% (Byhardt et al, 1998; Santana-Davila et al, 2015). It is evident that the development of novel agents targeting specific biological processes in the cancer cell are needed to improve the efficacy of radiotherapy and chemotherapy, with the ultimate goal of improving survival.
During the last decade, research efforts have focused on developing anti-angiogenic therapies and targeting the epidermal growth factor receptor pathway. However, clinical data from their combination with radiotherapy and chemotherapy are discouraging (Socinski et al, 2012a; Bradley et al, 2015). Tumour autophagic activity, though far less investigated, is emerging as an important area of clinical research due to its role in cancer cell resistance to chemotherapy and radiotherapy. Importantly, autophagic activity has been shown in clinicopathological studies to be elevated in patients with non-small-cell lung carcinoma and correlated with worse health outcomes (Karpathiou et al, 2011).
Autophagy accounts for the recycling of damaged cell proteins and organelles under stressful intratumoural conditions, such as hypoxia, acidity and nutrient deprivation, and is vital for cancer cell survival and growth (Parzych and Klionsky, 2014). As radiation damages proteins and cytoplasmic organelles, it is expected that an intact or intensified autophagic flux would be essential for cell recovery (Zois and Koukourakis, 2009). Indeed, blockage of LC3 autophagosomal membrane protein expression sensitised human endothelial cells to radiation (Kalamida et al, 2014). The interactions, however, of radiation with the autophagy machinery is quite complex and obscure, and only a limited number of published studies have focused on the effects of radiation on autophagic flux. Various cells and tissues respond differentially to radiation, depending on both the dose of exposure and the time post irradiation, resulting in contrasting publications in the literature (Ito et al, 2005; Moretti et al, 2007; Gorbunov and Kiang, 2009; Kim et al, 2010; Zois et al, 2011; Datta et al, 2014; Kalamida et al, 2014).
In the current study, we assessed the expression of the autophagosomal membrane-bound proteins light chain 3A (LC3A) and 3B (LC3B), in parallel with the expression of the p62/sequestosome protein, which is a carrier of waste material to autophagosomes, in human lung cancer cells post irradiation. Moreover, we determined the kinetics of transcription factor EB (TFEB) expression, which controls the biogenesis of various lysosomal and autophagy-relevant genes, lysosome-associated membrane protein 2a (LAMP2a) expression and Cathepsin-D intra-lysosomal enzyme expression. Evidence is provided that radioresistance is dependent on the inherent cancer cell’s ability to resist and overcome the radiation-induced blockage in autophagy.
Materials and Methods
The study has been approved by the Democritus University of Thrace Research and Ethics Committee.
Cell culture
A549 (human alveolar basal epithelial adenocarcinoma cells) and NCI-H1299 (human non-small-cell lung carcinoma cells) were purchased from ATCC (Wesel, Germany) (ATCC CCL-185 and ATCC CRL-5803, respectively) and all experiments were performed within 6 months. Details on the cell lines can be found at https://www.atcc.org/Products/All/CCL-185.aspx?slp=1#generalinformation and at http://www.atcc.org/Products/All/CRL-5803.aspx#characteristics and authentication of cell lines has been performed by ATCC and the details and methods of characterisation are available at http://www.atcc.org/Products/Cells_and_Microorganisms/Testing_and_Characterization/STR_Profiling_Analysis.aspx.
Cells were cultured under aseptic conditions using DMEM basal medium (31885-023, Gibco, Paisley, Scotland, UK) supplemented with 10% fetal bovine serum (FB-1000/500, Biosera, Boussens, France), 100 units per ml penicillin and 100 μg ml−1 streptomycin (15140-122, Gibco), and 2 mM L-glutamine (25030, Gibco). Cells were maintained at standard conditions (37 °C, 5% CO2) in humidified atmosphere and were used upon reaching 70–90% confluency.
Small-interfering RNAs
Small-interfering RNAs (siRNAs) targeting LC3A, LC3B, TFEB and LAMP2a were custom synthesised from Shanghai GenePharma Co., Ltd (Shanghai, China). Microtubule-associated proteins 1A/1B LC3A (MAP1LC3A) siRNAs were pooled as (5′-GCGAGUUGGUCAAGAUCAUTT-3′), (5′-GCUUCCUCUAUAUGGUCUATT-3′), (5′-CCUGCUGUGUGGUUCAUCUTT-3′), (5′-GCUGUAAGGAGGUACAGCATT-3′), microtubule-associated proteins 1A/1B LC3B (MAP1LC3B) siRNAs were pooled as (5′-GCCCUCUACUGAUUGUUAATT-3′), (5′-CUCCCUAAGAGGAUCUUUATT-3′), (5′-GCCUGUGUUGUUACGGAAATT-3′), TFEB siRNAs were pooled as (5′-CAGGCUGUCAUGCAUUACATT-3′), (5′-GACGAAGGUUCAACAUCAATT-3′), (5′-GCGGCAGAAGAAAGACAAUTT-3′), (5′-CCGAGACCUAUGGGAACAATT-3′) and LAMP2a siRNAs were pooled as (5′-GCAGUGCAGAUGACGACAATT-3′), (5′-GCACCAUCAUGCUGGAUAUTT-3′), (5′-CCCAGUGUCAUUAGAUAAUTT-3′) and (5′-GGCUACAACAGAACUUAAATT-3′).
Cells were seeded in six-well plates at a density of 1 × 105 and were transfected with a 100 nM concentration of each siRNA utilising Metafectene Pro (Biontex Laboratories GmbH, Munich, Germany) according to the manufacturer’s protocol. Transfection was performed for 24 h and the cells were used for evaluation 24 h later. The silencing efficiency of siRNAs was confirmed by western blot analysis.
Characterisation of cell viability and evaluation of radiosensitivity
The viability of A549 and H1299 cells was initially characterised following exposure to multi-dose irradiation (2–9 Gy). This range of doses starts from the dose of 2 Gy applied clinically in standard fractionation radiotherapy schemes used in NSCLC, increases to 4 Gy used in hypofractionated schemes and goes up to 9 Gy, as higher doses were 100% lethal for the two tested cell lines. Radiosensitisation of both cell types was then evaluated following transfection with siRNAs targeting LC3A, LC3B, TFEB and LAMP2a. Cells were seeded in a 96-well plate at a density of 250 cells per well. Following the protocol of multi-dose irradiation of the well columns within the same 96-well plate (Abatzoglou et al, 2013), exposure of cells to radiation was performed using a 6 MV beam of a Linear Accelerator (PRECISE, ELEKTA, Stockholm, Sweden) endowed with a MultiLeaf collimator (Stockholm, Sweden).
Cell viability was evaluated 7 days post IR using the AlamarBlue Cell Viability Reagent (DAL1100, Invitrogen, Carlsbad, CA, USA) assay, as previously described (Zachari et al, 2014). This is the time point of the nadir of cell counts, as re-growth (when feasible) starts thereafter (Zachari et al, 2014). Briefly, 10% v/v AlamarBlue was added to each well and after a 7 h incubation, fluorescence was measured (excitation: 530–560 nm and emission: 590 nm) by utilising a FLUOstar Omega filter-based multi-mode microplate reader (BMG Labtech, Germany). For each dose, six wells with irradiated cells were monitored and the mean value was used to plot the dose-response curves.
Bafilomycin A and chloroquine, chemical inhibitors of the autophagy flux, were also tested to evaluate radiosensitisation of A549 and H1299 cells. Two hundred and fifty cells cells were seeded per well in triplicates, followed by 24 h incubation with 100 nM of bafilomycin A or 5 μ M of chloroquine. The aforementioned technique was applied for experiments.
Evaluation of chemosensitivity
Small-interfering RNA technology was employed in combination with five chemotherapuetic agents used widely in the treatment of lung cancer, in order to evaluate their effect when specific autophagolysosomic genes are silenced. siRNAs of LC3A, LC3B, TFEB or LAMP2a were transfected to A549 and H1299 cells, as described above, and cells were seeded in a 96-well plate at a density of 1000 cells per well. No combination of gene silencing was tested herein. Wild-type and silenced cells were, thereafter, exposed to the following treatment conditions: 100 nM albumin-bound paclitaxel (Abraxane), 2 nM docetaxel (Taxotere), 5 μ M liposomal doxorubicin (Caelyx), 30 μ M liposomal Cisplatin (Lipoplatin) or 10 μ M cisplatin (Platosin). We included the albumin-bound paclitaxel as this has been recently approved for the treatment of NSCLC (Socinski et al, 2012b) and, furthermore, we included the liposomal form of cisplatin that is under evaluation for apporval after major randomised studies performed (Stathopoulos et al, 2011). Liposomal doxorubicin has not been approved for NSCLC, although phase I/II studies have shown marked accumulation in lung tumours in humans and encouraging radiosensitisation properties (Koukourakis et al, 1999). Cell viability was evaluated after 24 h incubation with the chemotherapuetic agents, using the AlamarBlue assay, as described above. Drug efficacy was estimated in siRNA-transfected cells vs the control cells.
Immunoblotting
Western blot analysis was used for the characterisation of the autophagy markers for both A549 and H1299 cell lines following radiation exposure. Hence, cells irradiated with 4 Gy and cell lysates were collected at 2 and 7 days post irradiation. This dose was chosen as it allows ∼80% cell survival (3.7 Gy for A549 and 4.4 Gy for H1299 allow 80% cell survival), so we chose a mildly toxic for the cells radiation dose to study the effects of radiation on autophagy flux. Cells were irradiated utilising a Co60 unit (Theratron Elite 100, DBA MDS Nordion, Ottawa, ON, Canada). Cells were washed with PBS twice and lysed in a sucrose-based lysis buffer (0.25 M sucrose, 25 mM Tris-HCl, pH 7.4) containing protease inhibitors (complete mini protease inhibitor cocktail, Roche Diagnostics GmbH, Mannheim, Germany) and phosphatase inhibitors (phosphatase inhibitor cocktail, Cell Signaling Technology, Danvers, MA, USA). A differential centrifugation of the whole-cell lysates led to supernatant (cytoplasmic water soluble proteins) and pellet (membrane proteins) fractions. Total protein quantification was performed in the pellet fraction using the BCA Protein Assay Kit (#23225, Thermo Scientific, Pierce, Rockford, IL, USA) utilising FLUOstar Omega filter-based multi-mode microplate reader (BMG Labtech).
A total of 40 μg of each protein lysates were resolved by discontinuous SDS gels using 15 (LC3A and LC3B), 12 (p62, Cathepsin-D and TFEB) and 8% (LAMP2a) separation and 5% stacking gels, and they were transferred to the appropriate PVDF membrane. All different experimental condition lysates were loaded on the same gel and transferred on the same membrane. Following blocking with TBS (pH 7.6) containing 0.1% (v/v) Tween 20 and 5% (w/v) non-fat dried milk for 1 h at room temperature, membranes were hybridised at 4 °C overnight with primary anti-MAP1LC3A (1 : 1000, ab62720, Abcam, UK), anti-MAP1LC3B (1 : 500, 0231-100/LC3-5F10, Nanotools, Teningen, Germany), anti-SQSTM1/p62 (1 : 1000, ab56416, Abcam), anti-Cathepsin-D [CTD-19] (1 : 1000, ab6313, Abcam), anti-TFEB (1 : 500, A303-673A, Bethyl, Montgomery, TX, USA) and anti-LAMP2a (1/500, ab18528, Abcam). Then, the membranes were hybridised with the appropriate secondary antibodies (1 : 2000, Goat Anti-Rabbit IgG (H+L)-HRP Conjugate, #170-6515, Bio-Rad, Hercules, CA, USA and 1 : 2000 bovine anti-mouse IgG-HRP, P0447, DAKO, Glostrup, Denmark) and developed in Amersham ECL western blotting detection reagents and analysis system (RPN2209, GE Healthcare, Dornstadt, Germany). Each of these blots was then stripped, and the membranes were re-probed with a primary anti-beta actin antibody (1 : 5000, ab75186, Abcam). The images of the blots were captured utilising Chemidoc MP imaging system (Bio-Rad) and band densitometry analysis was performed using the Chemidoc-logismic (Hercules, CA, USA).
RNA isolation, cDNA synthesis and RT–qPCR
Total RNA was isolated from A549 and H1299 cells at 2 and 7 days post exposure to 4 Gy radiation using a Nucleospin RNA plus (Macherey-Nagel GmbH, Düren, Germany) kit. The quality (260/280 and 260/230 nm ratios) as well as the quantity of the RNA were evaluated and performed using a NanoDrop 2000 (Thermo Scientific). One microgram of total RNA was used to synthesise cDNA using a Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacturer’s instructions. The first step involved heating of the template–primer mixture at 65 °C for 10 min to ensure denaturation of RNA secondary structures if present. The second step involved the addition of the remaining components (Transcriptor RT reaction buffer, Protector RNase inhibitor, dNTPs and Reverse Trascriptase), the synthesis step at 55 °C for 30 min and the inactivation step at 85 °C for 5 min. All steps were performed in a thermal block cycler with a heated lid (Mastercycler pro, vapo.protect and eppendorf, Foster City, CA, USA).
Samples were analysed in triplicate by quantitative real time PCR (RT–qPCR) using KAPA SYBR FAST qPCR Kit-Optimized for LightCycler 480 (KR0392—v7.14, KAPABIOSYSTEMS, Wilmington, MA, USA) on the LightCycler 480 Instrument II (Roche) in conjunction with gene sequence-specific forward and reverse primers. Oligonucleotide primers were designed to amplify hu;man MAP1LC3A: forward, 5′-CATGAGCGAGTTGTTCAAGA-3′; reverse, 5′-CCATGCTGTGCTGGTTCA-3′, human MAP1LC3B: forward, 5′-CGCACCTTCGAACAAAGAG-3′; reverse, 5′-CTCACCCTTGTATCGTTCTATTATCA-3′, human LAMP2a: forward, 5′-GCTCGTTCTGGTCTGCCTA-3′; reverse, 5′-TGTCAAATTAAGTTCCAATGCATAA-3′, human SQSTM1: forward, 5′-AGCTGCCTTGTACCCACATC-3; reverse, 5′-CAGAGAAGCCCATGGACAG-3′, human Transcription Factor EB (TFEB): forward, 5′-AACATGACAGCAAGCTCAGG-3′; reverse, 5′-CTGCATGCGCAACCCTAT-3′ and Human β-actin: forward, 5′-CCAACCGCGAGAAGATGA-3′; reverse, 5′-CCAGAGGCGTACAGGGATAG-3′. All sets of primers were purchased from Roche.
Quantitative analysis was performed using LightCycler software (LightCycler 480 SW 1.5.0 SP4 v1.5.0.39; Mannheim, Germany). The cycle threshold values were determined for the genes of interest and were normalised to those of β-actin using the ΔΔCt method. The experiments were performed in triplicates and repeated three times.
Stable transfected shLC3A cell lines
Human lung carcinoma H129 cell line was stably transfected with shLC3A plasmids (Genepharma Co., Ltd), carrying the geneticin resistance gene, with the following sequences:
5′-CACCGCGAGTTGGTCAAGATCATCCTTCAAGAGAGGATGATCTTGACCAACTCGCTTTTTTG-3′
5′-CACCGCTGTAAGGAGGTACAGCAGATTCAAGAGATCTGCTGTACCTCCTTACAGCTTTTTTG-3′
5′-CACCGGTGATCATCGAGCGCTACAATTCAAGAGATTGTAGCGCTCGATGATCACCTTTTTTG-3′
5′-CACCGCTTCCTCTATATGGTCTACGTTCAAGAGACGTAGACCATATAGAGGAAGCTTTTTTG-3′
The cells were plated in six-well plates, incubated for 24 h, and then transfected with 4 μg of the relevant small-hairpin RNA plasmids in DMEM medium without any antibiotics or FBS, using Metafectene Pro (Biontex Laboratories, GmbH) according to the manufacturer’s instructions. The medium was then replaced with fresh DMEM 6 h after the transfection, in order to prevent extensive cytotoxicity and cell stress. The medium was replaced with 0.5 mg μl−1 Geneticin (Abcam) containing DMEM 24 h following transfection. The cells were cultured in this medium for 15 days until the selection process was completed for the cells carrying the shLC3A plasmids providing geneticin resistance, as confirmed by the death of control/untransfected cells, which were not resistant to geneticin.
Confocal immunofluorescence
For immunofluorescence staining, human lung carcinoma A549 and H129 cell lines were grown on No. 1.5 glass coverslips and were fixed in 3.7% formaldehyde/PBS pH 7.4 for 20 min at 37 °C. Cells were then permeabilised in PBS/0.1% v/v Triton X-100 pH 7.4 for 5 min at room temperature. Cells were blocked in PBS/5% w/v BSA pH 7.4 and stained with various combinations of the following for 1 h at room temperature: anti-MAP1LC3A rabbit polyclonal antibody (1 : 500, ab62720, Abcam), anti-LC3B mouse monoclonal antibody (1 : 100, 0231-100/LC3-5F10, Nanotools), anti-p62 mouse monoclonal antibody (1 : 100, ab56416, Abcam), anti-LAMP2A rabbit polyclonal antibody (1 : 500, ab18528, Abcam), anti-Cathepsin-D mouse monoclonal antibody (1 : 200, ab6313, Abcam), anti-TFEB antibody (1 : 500, A303-673A, Bethyl) and anti- gamma H2AX rabbit polyclonal antibody (1 : 500, ab11174, Abcam). Cells were washed in PBS pH 7.4, incubated with appropriate CF 488 and 564 secondary antibodies (1 : 500. Biotium, Hayward, CA, USA) at room temperature, and the DNA was counterstained with Hoechst 33342 (1 μg ml−1; Sigma-Aldrich, St Louis, MO, USA). After final washes, the coverslips were mounted in a homemade Mowiol mounting medium. Imaging was performed on a customised Andor Revolution Spinning Disk Confocal System built around a stand (IX81, Olympus Corporation, Shinjuku, Tokyo, Japan) with a × 100 and a × 60 lens and a digital camera (Andor Ixon+885, Andor Technology Ltd., Belfast, UK) (CIBIT Facility, MBG-DUTH). Image acquisition was performed in Andor IQ 2 software (Andor Technology Ltd.). Optical sections were recorded every 0.3 μm. All confocal microscopic imagesare presented in two-dimensional (2D) maximum intensity projections of z-stack images (ImageJ 1.47v, National Institute of Health, Bethesda, MD, USA).
Aggresome detection
Aggresome accumulation was determined in irradiated (4 Gy) A549 and H1299 cells at 2 and 7 days post irradiation. Aggresome visualisation was performed using the ProteoStat Aggresome detection kit for fluorescence microscopy (Enzo Life Sciences Ltd, Exeter, UK) according to the manufacturer’s instructions. Aggresome accumulation is directly linked to the inhibition of autophagy, specifically the ‘autophagic flux’. Nuclei were counterstained using Hoechst 33342. MG132, a proteasome inhibitor facilitating aggresome accumulation was used as a positive control. Stained cells were imaged by confocal microscopy with × 100 and × 60 magnification using a standard red filter (561 nm). A549 and H1299 lung cancer cell lines have been irradiated with 4 Gy, aggresomes have been detected 2 days and 7 days following radiation respectively, whereas non-irradiated cells have been used as control.
Evaluation of DNA damage
A549 and H1299 cells were grown on No. 1.5 glass coverslips to 90–100% confluency and exposed to 4 Gy irradiation. Cells were stained with anti-gamma H2AX antibody detecting DNA double strand breaks as nuclear foci at 30 min, 4 and 24 h post irradiation. DNA damage was evaluated using immunofluorescence and the mean value and standard deviation was plotted and statistically analysed.
Animal care and handling
Animal care and handling was carried out according to the guidelines set by Directive 86/609/EEC. All experimental procedures have been approved by the Veterinary Authorities for Animal Research in the Department of Experimental Surgery at the Democritus University of Thrace. Athymic nude mice were housed in the Easy Flow IVC Air Handling Unit (Tecniplast, West Chester, PA, USA), which consists of individually ventilated cages. Caging included sterile conditions of bedding, diet and water provided ad libitum, and the animals were maintained at an ambient temperature of 23 °C and at a photoperiod of 12 h light:12 h dark cycle.
Animal cancer cell line xenografts (CCL-xenografts)
The study has been approved by the Committee of Evaluation of Experimental Animal Research Protocols and by the local Veterinary authorities. Athymic nude mice - Hsd:Athymic Nude-Foxn1nu were purchased from Harlan Laboratories (San Pietro al Natisone (UD), Italy). Animals were used upon reaching 8 weeks of age. The animals were randomly divided into two groups (n=5 mice per group). Mice in both groups were injected subcutaneously with 1 × 107 H1299 cells (wild type or transfected with shLC3A). Subcutaneous implantations of the CCL-xenografts were made heterotopically in the top area of the right rear limb.
Cell preparations for subcutaneous injection were prepared by pelleting the cells via centrifugation, recording the volume of the dispersed cell pellet (<200 μl) and adding an equal volume of CultrexBME, type 3 (3632-005-02, Pathclear, Trevigen, Gaithersburg, MD, USA) to facilitate adherence of the cells in the subcutaneous injection space, as well as to consolidate tumour formation (Fridman et al, 2012).
Animals were checked daily after the implantation of CCL-xenografts and were irradiated with 6 Gy per day (using a 6MV LINAC) for 2 consecutive days, when tumours were ∼10 mm × 10 mm in size. Irradiation was directed towards the caudal region of the mouse, where the tumours were growing, using a customised apparatus and three-dimensional radiotherapy planning after CT-scanning of mice (Karagounis et al, 2016). Tumours were monitored for their growth for 20 consecutive days post irradiation.
Statistical analysis
Statistical analysis and graphical presentation was performed using GraphPad Prism Version 5.01a statistical package (GraphPad Software Inc., La Jolla, CA, USA). Paired and unpaired two-tailed t-tests were used for comparisons as appropriate. A P-value of <0.05 was used for determining the statistical significance.
Results
The effect of irradiation on LC3A autophagic flux
Western blot analysis of proteins was performed using the pelleted fraction of cell lysates, containing autophagosomes. LC3A protein was detected mainly in the membrane-bound LC3A-II (16 kDa) form, in both the cell lines.
In A549 cells, 4 Gy of irradiation significantly reduced levels of membrane-bound LC3A-II that remained low for at least 7 days post irradiation (relative band densities 1/0.3/0.4 at 0/2/7 days, respectively) (Figure 1A). Levels of p62 protein were reduced at 2 days post irradiation, but returned to normal after 7 days (relative band densities 1/0.5/0.8 at 0/2/7 days, respectively) (Figure 1A and B). Confocal microscopy revealed an increased autophagic flux as an early (2 days) response of A549 cells to radiation, as shown by intense LC3A/LAMP2a co-localisation (Figure 1C). However, this level of autophagosomal consumption was insufficient to induce the elimination of the accumulated damaged material, as shown by the presence of aggresomes in the cytoplasm (Figure 1D). This inadequate autophagic response tended to normalise after 7 days post irradiation. Levels of LC3A and p62 mRNA were initially unchanged post irradiaton, but were increased at 7 days (Figure 1E).
Western blot images of the LC3A, LC3B and p62 proteins, assessed in the pellet fraction at 0, 2 and 7 days after exposure of A549 and H1299 lung cancer cells to 4 Gy of radiation. (A) Confocal microscopy images of A549 and H1299 cells after immunofluorescence for p62 (red) protein at 0, 2 and 7 days after exposure to 4 Gy of radiation. (B) Confocal microscopy images of A549 and H1299 cells after double immunofluorescence for LC3A (green) and LAMP2a (red) proteins, showing co-localisation (yellow) patterns, as a result of auto-lysosomal fusion at 0, 2 and 7 days after exposure to 4 Gy of radiation. (C) Confocal microscopy images of A549 and H1299 cells after immunofluorescence for aggresome (red) detection at 0, 2 and 7 days after exposure to 4 Gy of radiation. (D) Relative quantitative analysis of mRNA levels of the LC3A, LC3B and p62 mRNA levels at 0, 2 and 7 days after exposure of A549 lung cancer cells (1B) or H1299 cells to 4 Gy of radiation. (E) Confocal microscopy images of A549 and H1299 cells after double immunofluorescence for LC3B (red) and Cathepsin-D (green) proteins, showing co-localisation (yellow) patterns as a result of auto-lysosomal fusion at 0, 2 and 7 days after exposure to 4 Gy of radiation. (F) Western blot images of the TFEB, LAMP2a and Cathepsin-D proteins assessed in the pellet fraction, 0, 2 and 7 days after exposure of A549 and (G) H1299 lung cancer cells to 4 Gy of radiation. A full colour version of this figure is available at the British Journal of Cancer journal online.
In H1299 cells, the membrane-bound LC3A-II form was unchanged at 2 days post irradiation, whereas a significant reduction was noted at 7 days (relative band densities 1/0.9/0.6 at 0/2/7 days, respectively) (Figure 1A). Confocal microscopy confirmed that initially the autophagic flux remained unchanged, which was followed by an intense induction at 7 days (LC3A/LAMP2a co-localisation; Figure 1C). Levels of p62 protein showed an increase at 2 days, but returned to baseline values at 7 days (relative band densities 0.6/1/0.6 at 0/2/7 days, respectively) (Figure 1A and B). Unlike A549 cells, aggresome assessment of H1299 cells showed lack of accumulation in the cytoplasm (Figure 1D). Levels of LC3A and p62 mRNA were elevated at 2 days after irradiation and remained at high levels thereafter (Figure 1E).
The effect of irradiation on LC3B autophagic flux
Membrane-bound LC3B-II was detected in both cell lines in the pelleted fraction. The pattern of membrane-bound LC3B-II response to radiation was similar to LC3A in both cell lines (Figure 1A). In A549 cells, levels of LC3B were reduced 2 days post irradiation and returned to lower than normal values after 7 days (relative band densities 1/0.1/0.6 at 0/2/7 days, respectively). In H1299 cells, LC3B protein levels were reduced after 2 days, and were further reduced after 7 days (relative band densities 1/0.7/0.2 at 0/2/7 days, respectively). Co-localisation of LC3B with lysosomal Cathepsin-D was intensified on day 2 after irradiation in A549 cells and on day 7 in H1299 cells (Figure 1F). Levels of LC3B mRNA, however, showed a small initial increase that became intense at 7 days in A549 cells (Figure 1E), whereas LC3B mRNA rapidly increased in H1299 cells after 2 days and remained elevated 7 days after irradiation (Figure 1E).
The effect of irradiation on lysosomal markers
Lysosomal markers, TFEB, LAPM2a and Cathepsin-D, were assessed in the pelleted fraction using western blot. TFEB expression was low in A549 cells, but was sharply increased after 7 days (relative band densities 0.3/0.2/1 at 0/2/7 days, respectively) (Figure 1G). RT–qPCR showed similar levels of mRNA expression, decreasing on day 3 and increasing on day 7 post irradiation. In H1299 cells, levels of TFEB mRNA were significantly elevated on day 2 (Figure 1E). Lysosomal protein LAMP2a increased after irradiation in the A549 cell, whereas the Cathepsin-D protein showed an initial drop at 2 days that was restored at 7 days. The lack of transcriptional activity at 2 days that increased at 7 days for these genes may in part explain the findings. In the H1299 cell line, despite the robust autophagic activity, LAMP2a remained stable and Cathepsin-D further increased 7 days after irradiation, indicative of increased lysosomal genesis to support an enhanced autophagic flux, as suggested by the increased TFEB and LAMP2a mRNA levels that started early on day 2.
The effect of irradiation on γH2Ax kinetics
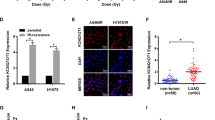
The mean number of γH2Ax foci formed at 30 min, 4 and 24 h post irradiation was assessed with confocal microscopy (Figure 2A–C). H1299 cells showed a significantly higher number of γH2Ax foci at 30 min post irradiation (P<0.01) and a slower regression at 4 h (P<0.001), as compared with A549 cells.
Detection of γ H2A? protein kinetics (mean fluorescent nuclear red foci detected with confocal microscopy), comparatively in A549 (white boxes) and H1299 (grey boxes) lung cancer cell lines after exposure to 4 Gy of radiation (at 0, 30 min, 4 and 24 h time points). (A) Typical confocal microscopy images of γH2Ax nuclear foci, induced at 30 min after 4 Gy of radiation, in (B) A549 and (C) H1299 cell lines. (D) Gene silencing efficacy assessed with western blot analysis and (E) with confocal microscopy. A full colour version of this figure is available at the British Journal of Cancer journal online.
Gene silencing and radiosensitivity
Silencing of four genes involved in autophagosome membrane formation (LC3A and LC3B), lysosomal membrane formation (LAMP2a) or lysosomal biogenesis (TFEB) was performed in both A549 and H1299 cells (Figure 2D and E).
Radiosensitivity
Gene silencing resulted in significant radiosensitisation, evident in both cell lines. In A549 cells, the radiation dose required to induce 50% growth inhibition 7 days after irradiation (day when the nadir of cell counts is noted; data not shown), was 6.2 Gy in control cells and was reduced to 1.9, 4.3, 3.2 and 2.5 Gy in cells with silenced LC3A, LC3B, TFEB and LAMP2a genes, respectively (Figure 3A). In relatively radioresistant H1299 cells, the radiation dose demanded to induce 50% growth inhibition was 8.6 Gy in control cells and was reduced to 4.2, 6.5, 4.5 and 3.4 Gy when the LC3A, LC3B, TFEB and LAMP2a genes were silenced, respectively (Figure 3A).
Radiation dose-response viability curves of A549 and H1299 cells, after silencing of the LC3A, LC3B, TFEB or LAMP2a gene expression. (A) Radiation dose-response viability curves of A549 and H1299 cells, after exposure to chloroquine or bafilomycin A. (B) Cell viability after exposure of A549 or H1299 cells to chemotherapeutic drugs, after silencing of the LC3A, LC3B, TFEB or (C) LAMP2a gene expression.
Incubation of the cell lines with the autophagy blockers chloroquine and bafilomycine also resulted in sensitisation to radiation (Figure 3B). The 50% growth inhibition in A549 cell line (7 days after irradiation) was obtained with 6.2 Gy in control cells and was reduced to 3.9 and 4.4 Gy, when cells were incubated with chloroquine and bafilomycine, respectively. For the H1299 cell line, the dose was reduced from 8.6 Gy to 5.8 and 3.5 Gy, when cells were incubated with chloroquine and bafilomycine, respectively.
Chemosensitivity
The effect of various chemotherapeutic agents (albumin-bound paclitaxel, liposomal doxorubicin, liposomal cisplatin, cisplatin and docetaxel) on cancer cell viability was assessed after 24 h incubation at selected mildly toxic concentrations, in control cells and cells with silenced LC3A, LC3B, TFEB and LAMP2a genes (Figure 3C). The drug concentration chosen was based on previous cell viability experiments with a wide range of doses (data not shown).
The proliferation ability of cells was examined for 4 consecutive days after gene silencing for LC3A, LC3B, TFEB or LAMP2a (data not shown). Silencing of LC3A was the only experiment that resulted in reduced cell growth in A549 cells, whereas no effect was noted for rest of the genes. No effect was noted in H1299 cells for the silencing of any of the genes investigated.
In addition to the suppressive effect of siLC3A on cell growth alone, silencing of the LC3A gene resulted in an additive, sensitising effect with chemotherapeutic agents in the A549 and H1299 cells. Silencing of the TFEB gene resulted in increased sensitisation in A549 cells for all chemotherapeutic agents examined, but only for liposomal doxorubicin-exposed H1299 cells. Silencing the LAMP2a gene sensitised H1299 cells to all the drugs examined except from paclitaxel, whereas this was confirmed only for the liposomal doxorubicin and docetaxel in A549 cells (Figure 3C).
Xenografts
As LC3A gene silencing showed the most potent radiosensitising effect in the radioresistant H1299 cell line, we developed stable shLC3A-transfected H1299 cells (shH1299; Figure 4A) for xenograft experiments. Following cell implantation, shH1299 cells exhibited a slightly slower growth rate compared with wild-type H1299 cells (Figure 4B). Having reached median 2D dimensions of around 1 cm2, tumours were irradiated with two consecutive fractions of 6 Gy. Wild-type H1299 cells continued their growth, entering an accelerated phase of growth 12 days later. The shH1299 tumours experienced a loss of growth up to 12 days post irradiation and then they restarted their growth (Figure 4B). The difference was statistically significant at all post irradiation time points (P<0.001).
Expression levels of the LC3A protein, in the wild-type H1299 cells and in the stably transfected with shLC3A plasmids H1299 cells (shH1299), as detected with western blot and confocal immunofluorescence microscopy. (A) Growth of the wild-type H1299 and the shH1299 tumours following subcutaneous implantation in nude mice, 20 days before and (B) 20 days after two fractions of 6 Gy of irradiation (* means P-value <0.001).
Discussion
Autophagy is an essential pathway regulating the recycling of intracellular waste material and organelles, and promoting cell survival and energy balance. Stress factors, such as fasting, may accelerate autophagy to allow energy acquisition. On the other hand, damaging agents, such as radiation and chemotherapy, force cells to intensify autophagic pathways to remove damaged organelles and membranes. Suppression of autophagy may, therefore, render cells vulnerable to various stresses and damaging agents. Overactivation of autophagy that is often inherent in a large percentage of human tumours including lung cancer, has been associated with poor post-operative overall survival (Karpathiou et al, 2011), presumably a result of increased cancer cell survival and metastatic ability (Giatromanolaki et al, 2015).
The autophagic response of cancer cells to radiation remains unclear and the development of sensitising therapeutic agents targeting autophagy is an area of intense investigation. Conflicting and limiting evidence exists on whether radiation suppresses or activates autophagic flux in normal and cancer cells. Liu et al (2014) noticed an acceleration of autophagic flux following administration of five consecutive fractions of 2 Gy in A549 lung cancer cells. Of interest, Chen et al (2015) reported that very low doses of radiation, several cGy, induced autophagy in the same cell line, which renders cells resistant to subsequent higher doses of irradiation. Ito et al (2005) suggested that radiation induces autophagy in glioma cells, whereas Moretti et al (2007) suggested that radiation-induced autophagy occurs only in cells unable to undergo apoptosis. Datta et al (2014) found that exposure to sublethal irradiation suppresses autophagic activity in the intestinal tissue of mice, whereas Gorbunov and Kiang (2009) reported an intensified autophagy in the Paneth cells of the small intestine. We found that mouse lung irradiation induces a significant suppression of Beclin 1 and LC3A gene expression and accumulation of the LC3-II form and p62 in the cytoplasm, compatible with a suppressive effect of radiation on lung autophagy (Zois et al, 2011).
The effects of radiation on autophagy are very complex and the experimental procedures available provide data difficult to properly interpret. For example, LC3-II form protein accumulation recorded by western blot or confocal microscopy may represent blocked autophagosomal fusion with lysosomes, blocked auto-lysosomal lysis or may even suggest an increased autophagosomal flux in the context of an overexpression of the autophagy gene cascade. Accumulation of p62/sequestosome protein is considered a sign of blocked autophagy (Pankiv et al, 2007), as this carrier protein of waste molecules to autophagosomes is degraded after auto-lysosomal fusion. Nevertheless, intensification of p62 transcription may lead to overexpression in the context of an increased autophagy flux. TFEB is a transcriptional factor that resides on the lysosomal membranes and translocates to the nuclei to activate expression of a variety of genes involved in lysosomal biogenesis and autophagy activation (Settembre et al, 2011). Increased accumulation in the cytoplasm can be compatible with lysosomal accumulation, but could also occur in the context of TFEB gene overexpression. LC3, p62 or TFEB genes can be transcriptionally overactive in cancer cells either as a response to environmental conditions or as an inherent, presumably oncogene related, state.
In the current study, we showed that 4 Gy of radiation can induce early or late expression of the LC3A, LC3B and p62 mRNA in H1299 and A549 lung cancer cell lines, respectively. It seems, therefore, that the ability of cells to transcriptionally support an enhanced autophagic flux demanded post-radiation exposure due to cell damage, differs among cell lines. An early supportive reaction should be related to a better adaptation to the radiation stress and, indeed, H1299 cells are more radioresistant than the A549.
The early (at 2 days) noted reduction of the membrane-bound form of LC3-II in A549 cells should be interpreted as an attempt of the cells to accelerate the autophagic flux. This, however, stumbles onto an unparalleled transcriptional support, resulting in exhaustion of the available autophagosomes and to the accumulation of aggresomes, confirming that A549 cells fail to adequately digest the proteins and organelles damaged by radiation. In contrast, H299 cells sustained stable levels of LC3-II membrane-bound proteins and exhibited no aggresome accumulation, compatible with an adequate maintenance of the autophagic flux. This flux was transcriptionally supported by the early activation of the LC3, p62 and TFEB/LAMP2a lysosome-related genes. At 7 days post irradiation, surviving A549 cells exhibited an increased transcriptional activity of LC3s and p62, as well as of TFEB and LAMP2a genes. The LC3-II protein and aggresome content was reduced, suggesting a disentanglement of their autophagic flux. H1299 cells continued to transcriptionally support the autophagic machinery and no signs of blockage of autophagy flux could be noted at the protein level.
We, therefore, characterised two lung cancer cell lines with differential autophagic responses to 4 Gy of ionising radiation. One radiosensitive, A549, which exhibited impaired ability to sustain an adequate autophagic flux during the first days after irradiation, and one radioresistant, H1299, which suffered no evident autophagic blockage in the context of an early intensification of autophagy gene transcriptional activity. Although the hypothesis emerging from this observation demands thorough investigation in multiple cell lines, a similar correlation has been previously reported in two prostate cancer cell lines, the PC3 being impressively radioresistant and sustaining an intact autophagic flux after irradiation and the DU145 radiosensitive cell line that exhibits a severe blockage of the autophagic flux (Koukourakis et al, 2015). The γH2Ax nuclear foci formation at 30 min was higher in the resistant H1299 cell line and the regression of foci slower compared with the relatively more sensitive to radiation A549 cells. Slow DNA dsb DNA repair is certainly an indicator of reduced cell ability to overcome radiation-induced DNA damage, which however has not been verified in the current experiments using the γH2A? microscopy assay. This unexpected finding is however in accordance with a previous study by Toulany et al (2014), showing that different levels of radiosensitisation of lung cancer cells by cisplatin was not related to differences in DNA double strand break repair. It is therefore suggested that although DNA repair ability contributes to cell radiosensitivity, this is probably not the only factor, so that the observed increased radiosensitivity of A549 cells should be attributed to other contributors, such as the impaired post-irradiation autophagy restoration herein identified.
As the ability to sustain an adequate autophagic flux was postulated to be important in the survival of lung cancer cells post irradiation, we performed silencing of four genes involved in autophagosome formation (LC3A and LC3B), lysosomal formation (LAMP2a) or lysosomal biogenesis (TFEB), in both A549 and H1299 cell lines. We found that suppression of either of these genes resulted in significant radiosensitisation and also sensitised lung cancer cells, although not invariably, to chemotherapeutic agents used to treat lung cancer patients, such as cisplatin and taxanes. In xenografts, shLC3A-H1299 tumours suffered a significantly increased radiation-induced damage above wild type, leading to important growth delay. Forty years ago, Pazmiño and Yuhas (1974) had shown that chloroquine inhibited the recovery of a variety of tissues from radiation injury. This early finding is, herein, verified and explained by an important role of autophagy in post-radiation survival, as chloroquine is now known to be an inhibitor of lysosomal function (Kimura et al, 2013).
In conclusion, the ability of lung cancer cells to survive after being exposed to therapeutic levels of ionising radiation depends on their ability to sustain a functional autophagic flux. Abrogation of such ability results in increased radiosensitivity and susceptibility to various chemotherapeutic agents. The identification of compounds or biological agents that are able to inhibit the autophagic function of cancer cells may prove of importance for the eradication of locally advanced lung cancer, and may prolong survival of lung cancer patients with metastatic disease.
Change history
26 July 2016
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Abatzoglou I, Zois CE, Pouliliou S, Koukourakis MI (2013) Establishment and validation of a method for multi-dose irradiation of cells in 96-well microplates. Biochem Biophys Res Commum 431: 456–459.
Bezjak A, Temin S, Franklin G, Giaccone G, Govindan R, Johnson ML, Rimner A, Schneider BJ, Strawn J, Azzoli CG (2015) Definitive and adjuvant radiotherapy in locally advanced non-small-cell lung cancer: American Society of Clinical Oncology Clinical Practice Guideline Endorsement of the American Society for Radiation Oncology Evidence-Based Clinical Practice Guideline. J Clin Oncol 33: 2100–2105.
Bradley JD, Paulus R, Komaki R, Masters G, Blumenschein G, Schild S, Bogart J, Hu C, Forster K, Magliocco A, Kavadi V, Garces YI, Narayan S, Iyengar P, Robinson C, Wynn RB, Koprowski C, Meng J, Beitler J, Gaur R, Curran W Jr, Choy H (2015) Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage IIIA or IIIB non-small-cell lung cancer (RTOG 0617): a randomised, two-by-two factorial phase 3 study. Lancet Oncol 16: 187–199.
Byhardt RW, Scott C, Sause WT, Emami B, Komaki R, Fisher B, Lee JS, Lawton C (1998) Response, toxicity, failure patterns, and survival in five Radiation Therapy Oncology Group (RTOG) trials of sequential and/or concurrent chemotherapy and radiotherapy for locally advanced non-small-cell carcinoma of the lung. Int J Radiat Oncol Biol Phys 42: 469–478.
Chen N, Wu L, Yuan H, Wang J (2015) ROS/Autophagy/Nrf2 pathway mediated low-dose radiation induced radio-resistance in human lung adenocarcinoma A549 cell. Int J Biol Sci 11: 833–844.
Datta K, Suman S, Fornace AJ Jr (2014) Radiation persistently promoted oxidative stress, activated mTOR via PI3K/Akt, and downregulated autophagy pathway in mouse intestine. Int J Biochem Cell Biol 57: 167–176.
Fridman R, Benton G, Aranoutova I, Kleinman HK, Bonfil RD (2012) Increased initiation and growth of tumor cell lines, cancer stem cells and biopsy material in mice using basement membrane matrix protein (Cultrex or Matrigel) co-injection. Nat Protoc 7: 1138–1144.
Giatromanolaki A, Kalamida D, Sivridis E, Karagounis IV, Gatter KC, Harris AL, Koukourakis MI (2015) Increased expression of transcription factor EB (TFEB) is associated with autophagy, migratory phenotype and poor prognosis in non-small cell lung cancer. Lung Cancer 90: 98–105.
Gorbunov NV, Kiang JG (2009) Up-regulation of autophagy in small intestine Paneth cells in response to total-body gamma-irradiation. J Pathol 219: 242–252.
Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y (2005) Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol 26: 1401–1410.
Kalamida D, Karagounis IV, Giatromanolaki A, Koukourakis MI (2014) Important role of autophagy in endothelial cell response to ionizing radiation. PLoS One 9: e102408.
Karagounis IV, Abatzoglou IM, Koukourakis MI (2016) Partial body irradiation of mice using a customized PMMA apparatus and a clinical 3D planning/LINAC radiotherapy system. Med Phys 43: 2200–2206.
Karpathiou G, Sivridis E, Koukourakis MI, Mikroulis D, Bouros D, Froudarakis ME, Giatromanolaki A (2011) Light-chain 3A autophagic activity and prognostic significance in non-small cell lung carcinomas. Chest 140: 127–134.
Kim KW, Moretti L, Mitchell LR, Jung DK, Lu B (2010) Endoplasmic reticulum stress mediates radiation-induced autophagy by perk-eIF2alpha in caspase-3/7-deficient cells. Oncogene 29: 3241–3251.
Kimura T, Takabatake Y, Takahashi A, Isaka Y (2013) Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res 73: 3–7.
Koukourakis MI, Koukouraki S, Giatromanolaki A, Archimandritis SC, Skarlatos J, Beroukas K, Bizakis JG, Retalis G, Karkavitsas N, Helidonis ES (1999) Liposomal doxorubicin and conventionally fractionated radiotherapy in the treatment of locally advanced non-small-cell lung cancer and head and neck cancer. J Clin Oncol 17: 3512–3521.
Koukourakis MI, Kalamida D, Mitrakas A, Pouliliou S, Kalamida S, Sivridis E, Giatromanolaki A (2015) Intensified autophagy compromises the efficacy of radiotherapy against prostate cancer. Biochem Biophys Res Commun 461: 268–274.
Liu M, Ma S, Liu M, Hou Y, Liang B, Su X, Liu X (2014) Synergistic killing of lung cancer cells by cisplatin and radiation via autophagy and apoptosis. Oncol Lett 7: 1903–1910.
Moretti L, Attia A, Kim KW, Lu B (2007) Crosstalk between Bak/Bax and mTOR signalling regulates radiation-induced autophagy. Autophagy 3: 142–144.
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145.
Parzych KR, Klionsky DJ (2014) An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 20: 460–473.
Pazmiño NH, Yuhas JM (1974) Chloroquine: nonselective inhibition of recovery from radiation injury in tumors and normal tissues. Radiat Res 60: 54–61.
Santana-Davila R, Devisetty K, Szabo A, Sparapani R, Arce-Lara C, Gore EM, Moran A, Williams CD, Kelley MJ, Whittle J (2015) Cisplatin and etoposide versus carboplatin and paclitaxel with concurrent radiotherapy for stage III non-small-cell lung cancer: an analysis of Veterans Health Administration data. J Clin Oncol 33: 567–574.
Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A (2011) TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433.
Socinski MA, Stinchcombe TE, Moore DT, Gettinger SN, Decker RH, Petty WJ, Blackstock AW, Schwartz G, Lankford S, Khandani A, Morris DE (2012a) Incorporating bevacizumab and erlotinib in the combined-modality treatment of stage III non-small-cell lung cancer: results of a phase I/II trial. J Clin Oncol 30: 3953–3959.
Socinski MA, Bondarenko I, Karaseva NA, Makhson AM, Vynnychenko I, Okamoto I, Hon JK, Hirsh V, Bhar P, Zhang H, Iglesias JL, Renschler MF (2012b) Weekly nab-paclitaxel in combination with carboplatin versus solvent-based paclitaxel plus carboplatin as first-line therapy in patients with advanced non-small-cell lung cancer: final results of a phase III trial. J Clin Oncol 30: 2055–2062.
Stathopoulos GP, Antoniou D, Dimitroulis J, Stathopoulos J, Marosis K, Michalopoulou P (2011) Comparison of liposomal cisplatin versus cisplatin in non-squamous cell non-small-cell lung cancer. Cancer Chemother Pharmacol 68: 945–950.
Toulany M, Mihatsch J, Holler M, Chaachouay H, Rodemann HP (2014) Cisplatin-mediated radio-sensitization of non-small cell lung cancer cells is stimulated by ATM inhibition. Radiother Oncol 111: 228–236.
Wang EH, Corso CD, Rutter CE, Park HS, Chen AB, Kim AW, Wilson LD, Decker RH, Yu JB (2015) Postoperative radiation therapy is associated with improved overall survival in incompletely resected stage II and III non-small-cell lung cancer. J Clin Oncol 33: 2727–2734.
World Cancer Research Fund International (2012) Available from http://www.wcrf.org/int/cancer-facts-figures/worldwide-data.
Zachari MA, Chondrou PS, Pouliliou SE, Mitrakas AG, Abatzoglou I, Zois CE, Koukourakis MI (2014) Evaluation of the alamarblue assay for adherent cell irradiation experiments. Dose Response 12: 246–258.
Zois CE, Giatromanolaki A, Sivridis E, Papaiakovou M, Kainulainen H, Koukourakis MI (2011) "Autophagic flux" in normal mouse tissues: focus on endogenous LC3A processing. Autophagy 7: 1371–1378.
Zois CE, Koukourakis MI (2009) Radiation-induced autophagy in normal and cancer cells: towards novel cytoprotection and radio-sensitization policies? Autophagy 5: 442–450.
Acknowledgements
The study has been financially supported by the Special Account Democritus University of Thrace, code number 81006 and by the «TRAINING AND LIFELONG LEARNING - ARISTEIA project, code no 520, ESPA 2007–2013, GGET decision number 12605/26.09.2012. We thank Ralph A. Pietrofesa, for his valuable help in editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Karagounis, I., Kalamida, D., Mitrakas, A. et al. Repression of the autophagic response sensitises lung cancer cells to radiation and chemotherapy. Br J Cancer 115, 312–321 (2016). https://doi.org/10.1038/bjc.2016.202
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2016.202
Keywords
This article is cited by
-
A new glance at autophagolysosomal-dependent or -independent function of transcriptional factor EB in human cancer
Acta Pharmacologica Sinica (2023)
-
Role of lysosomes in physiological activities, diseases, and therapy
Journal of Hematology & Oncology (2021)
-
Opposite effects of the triple target (DNA-PK/PI3K/mTOR) inhibitor PI-103 on the radiation sensitivity of glioblastoma cell lines proficient and deficient in DNA-PKcs
BMC Cancer (2021)
-
Cisplatin remodels the tumor immune microenvironment via the transcription factor EB in ovarian cancer
Cell Death Discovery (2021)
-
Suppressed PLIN3 frequently occurs in prostate cancer, promoting docetaxel resistance via intensified autophagy, an event reversed by chloroquine
Medical Oncology (2021)