Abstract
Background:
Human papillomavirus (HPV) infection is a powerful prognostic biomarker in a subset of head and neck squamous cell carcinomas, specifically oropharyngeal cancers. However, the role of HPV in non-oropharyngeal sites, such as the larynx, remains unconfirmed.
Methods:
We evaluated a cohort of 324 laryngeal squamous cell carcinoma (LSCC) patients for the expression of p16INK4A (p16) protein by immunohistochemistry (IHC) and for high-risk HPV E6 and E7 mRNA transcripts by RNA in situ hybridisation (ISH). p16 expression and HPV status were correlated with clinicopathological features and outcomes.
Results:
Of 307 patients assessable for p16 IHC, 20 (6.5%) were p16 positive. Females and node-positive patients were more likely to be p16 positive (P<0.05). There were no other significant clinical or demographic differences between p16-positive and -negative cases. There was no difference in overall survival (OS) between p16-positive and -negative patients with 2-year survival of 79% in each group (HR=0.83, 95% CI 0.36–1.89, P=0.65). There was no statistically significant difference in failure-free survival (FFS) with 2-year FFS of 79% and 66% for p16-positive and -negative patients, respectively (HR=0.60, 95% CI 0.26–1.36, P=0.22). Only seven cases were found to be HPV RNA ISH positive, all of which were p16 IHC positive. There was no statistically significant difference in OS between patients with HPV RNA ISH-positive tumours compared with -negative tumours with 2-year survival of 86% and 71%, respectively (HR=0.76, 95% CI 0.23–2.5, P=0.65). The 2-year FFS was 86% and 59%, respectively (HR=0.62, 95% CI 0.19–2.03, P=0.43).
Conclusions:
p16 overexpression is infrequent in LSCC and the proportion of cases with high-risk HPV transcripts is even lower. There are no statistically significant correlations between p16 IHC or HPV RNA ISH status and OS or disease outcomes.
Similar content being viewed by others
Main
Cancer of the larynx, in particular laryngeal squamous cell carcinoma (LSCC), is diagnosed in over 12 500 people in the United States per year, with over 3600 deaths (Siegel et al, 2014). The principal aims of managing laryngeal cancer include maintaining organ function while ensuring high cure rates. Despite successful laryngeal preservation in the majority of locally advanced cancers (Department of Veterans Affairs Laryngeal Cancer Study Group, 1991), the 5-year survival is <50% (Corry et al, 2011). Decision making with regard to larynx preservation vs total laryngectomy is largely based on clinical and radiologic features. Unfortunately, there are no clinically useful prognostic/predictive molecular markers in LSCC.
Human papillomavirus (HPV) is known to have a causative role in a significant proportion of head and neck squamous cell carcinomas (HNSCC), specifically cancers of the oropharynx (Chung and Gillison, 2009). HPV-positive oropharyngeal cancer is distinct from HPV-negative disease epidemiologically, clinically and molecularly (Urban et al, 2014), and importantly, HPV-positive patients have improved outcomes compared with HPV-negative patients (Ang et al, 2010; Rischin et al, 2010; Young et al, 2011). It is not entirely clear why HPV-positive oropharyngeal cancer patients have improved prognosis but it may be due in part to fewer comorbidities, including smoking related diseases, better response to treatment (Fakhry et al, 2008) and fewer second primaries (Morris et al, 2011). The role of HPV in squamous cell carcinomas of head and neck sites outside the oropharynx, and its relationship to clinical outcomes, has not been conclusively established (Isayeva et al, 2012; Li et al, 2013). Although it is generally accepted that the rates of HPV infection in non-oropharyngeal sites is much lower than in the oropharynx, studies continue to report conflicting results of the frequency of HPV infection, ranging from 3 to 97% (Duray et al, 2011; Bishop et al, 2012). In addition, although p16INK4A (p16) immunohistochemistry (IHC) is commonly used as a surrogate marker of HPV infection in oropharyngeal squamous cell carcinomas (Weinberger et al, 2006; Ang et al, 2010), the validity of p16 overexpression as a marker of HPV in LSCC remains to be proven. Nevertheless, we have noted that many pathology laboratories are now reporting on p16 in non-oropharyngeal head and neck cancer as well as oropharyngeal cancer biopsies.
Few studies have investigated the relationship between HPV status and outcomes in non-oropharyngeal sites (Isayeva et al, 2012), and have not yet convincingly proven a link between HPV status and outcome (Vlachtsis et al, 2005; Morshed, 2010; Duray et al, 2011; Stephen et al, 2012; Chernock et al, 2013). To date, the largest study reporting the correlation between HPV status and outcomes in laryngeal cancer included only 130 patients and reported no significant associations (Morshed, 2010).
In the present study we aimed to investigate the frequency of HPV status, both by p16 protein overexpression and transcriptionally active HPV infection, and its correlation to outcome in, to our knowledge, the largest cohort of LSCC reported.
Materials and methods
Patients
Three hundred and twenty-four consecutive patients with LSCC treated at our institution between 2002 and 2012 with available tumour samples were included in this study. Patients were identified from a prospective database of all new patients treated at our institution. Baseline demographics and clinicopathological features were extracted from this prospective database complemented by review of the patient charts while treatment outcomes were extracted retrospectively from patient files. Tumour samples were from primary resections or from biopsies before the commencement of treatment. This study was approved by our Institutional Ethics Committees.
Tissue microarrays
Formalin-fixed paraffin-embedded (FFPE) tumour tissue blocks were sourced for each of the 324 patients. A pathologist identified areas containing tumour on a haematoxylin and eosin-stained section from each tumour block. For those considered to have a large tumour area, up to six 1-mm tumour cores were punched from the identified area for creation of tissue microarray (TMA) blocks. Cases where it was not possible to punch cores for TMA but still had tumour identified had IHC and RNA in situ hybridisation (ISH) performed on whole slides.
p16 IHC
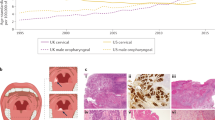
Four micron FFPE sections from the TMAs or whole slides were stained for p16 using Ventana Ultraview Detection reagents on a Benchmark Ultra instrument (Ventana Medical Systems, Tucson, AZ, USA) as previously described (Lim et al, 2014). In brief, antigen retrieval was performed with CC1 high-pH retrieval solution for 52 min followed by incubation at 36 °C for 32 min in primary antibody (mouse monoclonal p16, Clone E6H4; Ventana Medical Systems) and then detection with UltraView Universal DAB detection kit (Ventana Medical Systems). Sections were counterstained with haematoxylin. The intensity and proportion of staining in the cell nucleus and cytoplasm was semiquantitatively scored by a pathologist. Intensity was scored as 0 (none), 1 (weak), 2 (moderate) or 3 (strong). The proportion of cells stained was scored between 0 and 100%. An intensity score of 2 (moderate) or 3 (strong) in ⩾30% of tumour cells was used to define positivity (Figure 1). p16 scoring was performed blinded to the HPV RNA ISH results and to clinical outcomes.
HPV RNA ISH
HPV RNA ISH for high-risk HPV E6/E7 mRNA was performed using the RNAscope HPV assay (Advanced Cell Diagnostics, Hayward, CA, USA) according to the manufacturer’s instructions. In brief, 4-μm FFPE sections from the TMAs or whole slides were dewaxed, heat pretreated and protease digested before hybridisation with target probes for 2 h at 40 °C. Each case was individually probed with a negative control probe (the bacterial gene DapB), a positive control probe (the housekeeping gene PPIB), an HPV 16 specific probe and a high-risk HPV cocktail probe for the detection of 18 high-risk HPV subtypes (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73 and 82). Probe incubation was followed by several amplification steps and colour development with DAB. Each case was only scored for HPV after demonstrating no staining on the negative control slide and at least 2–5 signals/cell were seen on the positive control slide (as evidence of the presence of intact RNA) (Figure 2). HPV was classified in a binary manner as either negative (no cytoplasmic staining) or positive (brown punctate cytoplasmic signals) (Figure 2). HPV RNA ISH scoring was performed blinded to the p16 IHC results and to clinical outcomes.
Examples of HPV RNA ISH. (A) A p16/HPV-positive case showing strong p16 overexpression (1) and brown punctate cytoplasmic signals for HPV RNA ISH (2). The RNA ISH-positive control (3) confirms the presence of intact RNA. (B) a p16-positive/HPV-negative case showing strong p16 overexpression (4) but no signals for HPV RNA ISH (5). The RNA ISH-positive control (6) confirms the presence of intact RNA. (C) a p16-negative/HPV-negative case showing no p16 expression (7) and no HPV RNA ISH signals (8). The RNA ISH-positive control (9) confirms the presence of intact RNA. Images 1, 4 and 7= × 20 objective lens, images 2, 3, 5, 6, 8 and 9= × 60 objective lens.
Statistics
Statistical analyses were performed using the Stata Statistical package (version IC 11.1, Statacorp LP, College Station, TX, USA). Correlations between categorical variables were analysed with χ2-tests. The correlation between p16 IHC score and HPV RNA ISH positivity was performed using Spearman’s rank correlation. All survival curves were calculated using the Kaplan–Meier method. Patients treated with palliative intent were excluded from survival analyses. Overall survival (OS) was defined from the time of initial diagnosis to the date of death, and failure-free survival (FFS) was defined from the date of initial diagnosis to treatment failure, disease progression or death, whichever came first. Time to progression (TTP) was defined as the time from diagnosis to progression of cancer, with deaths from other causes being censored. The effects of demographic and pathologic variables and multivariate analyses were tested using the Cox proportional hazards model. A two-sided P-value <0.05 was considered statistically significant.
Results
Patient characteristics
FFPE tumour blocks were obtained for 324 patients, with 307 having adequate tumour samples for p16 IHC and HPV RNA ISH. The clinicopathological characteristics of the patients are summarised in Table 1. The median age of patients at diagnosis was 66 (range 36–88). Males comprised 94% of the patients, with 95% being current or former smokers. Primary treatment consisted of radiotherapy alone in 109 (36%), chemoradiotherapy in 98 patients (32%), surgery with adjuvant radiotherapy in 39 (13%), surgery with adjuvant chemoradiotherapy in 32 (11%), surgery alone in 23 patients (8%) and surgery with induction chemotherapy in 1 patient (<1%). One additional patient (<1%) was treated with induction chemotherapy but died before starting definitive chemoradiotherapy.
p16 IHC
Twenty patients (6.5%) were scored as p16 positive, as defined by moderate or strong intensity staining in ⩾30% of tumour cells (Table 2). Of the 287 cases scored as p16 negative, 235 had no detectable staining, and the majority of the remaining 52 cases showed only weak or moderate staining in <10% of tumour cells. Patients with p16-positive tumours were more likely to be female (20% vs 5%, P<0.01) and more likely to have involved lymph nodes (60% vs 29%, P<0.01). No other significant clinical or demographic differences were observed between these groups (Table 1).
HPV RNA ISH
HPV RNA ISH was performed on a subset of 307 patients with p16 IHC results, including all 20 of the cases considered p16-positive and 118 of the 287 p16-negative cases. Of these 138 cases, 58 were not assessable because of poor quality RNA, as determined by no or low signals on the positive control slide. Eighty cases showed strong signals on the positive control and were evaluated for HPV RNA ISH. Seven samples were positive for HPV RNA ISH (Table 2), all of which were p16 positive. All seven cases were positive for HPV subtype 16. HPV RNA ISH was negative in 73 cases, all of which were p16 negative. Of note, three cases scored for p16 as moderate staining in 10–20% of tumour cells tested RNA ISH negative. Similarly, three cases with weak p16 staining in 90–100% of the tumour cells also tested RNA ISH negative. HPV RNA ISH significantly correlated with the proportion of p16 staining (Spearman’s rho=0.695, P<0.001). There were no significant clinical or demographic differences between HPV RNA-positive and HPV RNA-negative patients.
Patient outcomes
OS and FFS
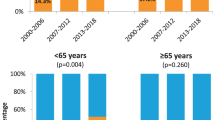
The OS and FFS were available for 288 patients. Median follow-up for patients who did not die was 41 months (range 2–116). There was no significant difference in OS between p16-positive compared with p16-negative patients (Figure 3a), on both univariate and multivariate analyses (Table 3). The 2-year survival of patients with p16-positive and -negative disease was 79% in each group (HR=0.83, 95% CI 0.36–1.89, P=0.65). There was no statistically significant difference in OS between patients with HPV RNA ISH-positive tumours compared with -negative tumours with 2-year survival of 86% and 71%, respectively (HR=0.76, 95% CI 0.23–2.5, P=0.65, Figure 3b). Similarly, there was no significant difference in OS by p16 status in 194 patients treated with primary radiotherapy/chemoradiotherapy (HR 1.28, 95% CI 0.55–2.96, P=0.57).
There was no statistically significant difference in FFS between patients with p16-positive compared with p16-negative tumours (Figure 3c). The 2-year FFS was 79% and 66% in the p16-positive and -negative groups, respectively (HR=0.60, 95% CI 0.26–1.36, P=0.22). There was no statistically significant difference in FFS between patients with RNA ISH-positive tumours compared with -negative tumours with 2-year FFS of 86% and 59%, respectively (HR=0.62, 95% CI 0.19–2.03, P=0.43) (Figure 3d).
There was no statistically significant difference in OS or FFS between p16-positive compared with p16-negative patients, irrespective of the proportion of positively staining cells used to define p16 positivity (i.e., ⩾70% or ⩾50%, data not shown).
TTP and patterns of first failure
The TTP was available for 276 patients. There were no statistically significant differences in TTP between RNA ISH-positive tumours compared with -negative tumours (HR 0.35, 95% CI 0.05–2.63, P=0.31) but the results favoured the p16-positive patients (HR=0.30, 95% CI 0.08–1.23, P=0.10). Two of 18 patients (11%) with p16 positive tumours progressed compared with 82 out of 258 (32%) in patients with p16-negative tumours and 1 out of 7 (14%) patients with HPV RNA ISH-positive tumour progressed compared with22 out of 67 (33%) with RNA ISH-negative tumours.
There was no statistically significant difference in the patterns of first failure by p16 or HPV status (both P>0.4). However, neither of the two p16-positive patients failed distantly, whereas 21 out of 82 (26%) patients with p16-negative tumours that failed progressed with distant metastasis as part of the first site of failure. Of these 11 out of 21 presented with distant metastasis as the only site of failure.
Discussion
This is to our knowledge the largest single study investigating the frequency and prognostic significance of HPV infection in LSCC. We have demonstrated that the incidence of p16 positivity in LSCC is low at 6.5%. We found an even lower incidence of transcriptionally active HPV infection with only 7 out of 14 evaluable p16-positive cases found to be HPV positive in our cohort. Importantly, our data show no statistically significant correlation between p16 or HPV status and disease outcomes in LSCC.
Our finding of a low incidence of HPV infection in LSCC is similar to several recent smaller studies that reported HPV positivity rates in LSCC between 1.6 and 6.5% (Bishop et al, 2012; Lewis et al, 2012b; Bussu et al, 2013; Chernock et al, 2013; Halec et al, 2013). Of note, as with the current study, these recent reports all determined the presence of transcriptionally active HPV, using either RNA ISH (Bishop et al, 2012; Lewis et al, 2012b; Chernock et al, 2013) or RT–PCR (Bussu et al, 2013; Halec et al, 2013). In comparison, older studies have reported higher incidences of HPV infection (Baumann et al, 2009; Morshed, 2010; Duray et al, 2011; Stephen et al, 2012); however, they used HPV DNA PCR, which by detecting both active and inactive HPV infection, is known to overestimate HPV infection (Wang et al, 2013). Therefore, we believe the incidence of active HPV infection in LSCC is consistently and markedly lower than the reported 70% of HPV-associated oropharyngeal cancers (Chaturvedi et al, 2011). There is a distinct but rare subgroup of patients who have recurrent respiratory papillomatosis, which is caused by ‘low-risk’ HPV genotypes 6 and 11 (Draganov et al, 2006) and is associated with malignant transformation (Omland et al, 2014). Although our HPV RNA ISH assay does not detect these subtypes, we are not aware of any of our cases occurring in patients with respiratory papillomatosis.
Even though the correlation between p16 and HPV infection in oropharyngeal cancer is well established, the most reliable definition of p16 positivity is not unanimous (Lewis et al, 2012a). However, the correlation between p16 overexpression and HPV infection in non-oropharyngeal head and neck sites remains unclear. Some have suggested that p16 may act as a surrogate marker for HPV infection in non-oropharyngeal sites (Doxtader & Katzenstein, 2012) and LSCC specifically (Gheit et al, 2014), whereas others report a poor correlation and discourage the use of p16 expression in non-oropharyngeal SCC (Bussu et al, 2013; Chernock et al, 2013). Given this lack of a clear relationship between p16 overexpression and HPV infection in LSCC, and based on a report where p16 staining in <30% of head and neck SCC cells was an accurate negative predictor for HPV infection (Chen et al, 2012), we chose to use this less stringent cutoff for p16 positivity. Indeed, in our cohort this 30% cutoff clearly separated the p16-positive and p16-negative cases. Furthermore, there were no significant differences in our results by using the often recommended positive staining of p16 in >70% of cells. Overall, our results suggest that p16 negativity is highly predictive for the lack of HPV infection in LSCC and are concordant with others (Chernock et al, 2013). However, the specificity of p16 is poor, with only 7 out of 14 (50%) evaluable cases with p16 overexpression having transcriptionally active HPV infection, suggesting that mechanisms other than HPV infection are responsible for p16 overexpression in LSCC.
As p16 positivity and HPV infection are both associated with better survival in oropharyngeal SCC (Ang et al, 2010), we evaluated survival and disease outcomes in our cohort by both p16 status and transcriptionally active HPV status. In contrast to oropharyngeal SCC, our results do not show a significant difference in OS according to p16 status or HPV status in LSCC. However, there were numerically fewer patients who progressed in both the p16-and HPV-positive patients. Isayeva et al (2012), in a recent review, found only four published studies examining the relationship between HPV and patient outcome in a combined total of 319 patients with LSCC and concluded that there was no evidence of an association. However, all of these studies used PCR to evaluate HPV status. Similarly, Torrente et al (2011), in a review of HPV and laryngeal cancer, concluded that the clinical significance and implications of HPV infection were unclear and required further investigation. Chung et al (2014) recently reported better survival in p16-positive non-oropharyngeal cancers when combining the oral cavity, hypopharynx and larynx subsites. Although prior to the HPV era this grouping of mucosal head and neck sites was considered standard, it is now acknowledged that each site may have different risk factors and natural history. In fact, when Chung et al (2014) analysed the different subsites independently they showed no survival advantage in 140 LSCC patients by p16 status. Therefore, until significantly larger studies are performed specifically in LSCC, the prognostic significance of p16 or HPV status in LSCC remains unclear. Furthermore, given the publicity surrounding HPV-associated ‘throat cancer’ and the potential for confusion among patients and oncologists alike, we believe p16 results should not be routinely reported in LSCC unless compelling evidence of clinical utility emerges.
Our study has some strengths and limitations. The most significant strength of this study is the large cohort consisting of a single site of head and neck cancer only, namely the larynx. Historically, head and neck cancers of different subsites have been grouped together, both in the clinic and the laboratory. This classification needs to be reviewed in light of the different clinical features and risk factors associated with specific sites, for example, HPV in the oropharynx and Betel quid chewing in the oral cavity (Petti, 2009). We observed that the rate of smoking in our laryngeal cohort is significantly higher than what we have found in an oral tongue cohort from our institution (Young et al, 2013). There are a number of limitations associated with the retrospective nature of the study, including the accuracy of FFS and TTP and the poor quality of RNA in many of our FFPE samples. The detection of HPV infection in HNSCC by the RNAscope RNA ISH assay is well validated (Masand et al, 2011; Ukpo et al, 2011; Schache et al, 2013). The advantage of this assay is that it detects transcriptionally active E6 and E7 mRNA and can be used on FFPE samples. However, we found a high failure rate with 42% of samples failing. This failure rate is most likely due to poor RNA quality, resulting from variations in storage, fixation and the age of the FFPE blocks used. Of note, attempts to extract RNA from these same blocks for separate studies also failed, indicating that the failures suggest poor RNA quality (data not shown) as opposed to failure of the RNAscope assay per se. However, the large cohort size of over 320 patients offsets some of these limitations. Given the poor quality of the RNA in some of our samples, use of the gold standard RT–PCR assay to determine HPV status would also have been compromised.
In conclusion, we have shown that in LSCC p16 overexpression is infrequent, the proportion of cases with high-risk HPV transcripts is even lower and there are no statistically significant correlations with survival outcomes. Given the very low frequency of HPV infection in LSCC, much larger studies would be required to definitively establish if p16 overexpression and/or HPV status have a prognostic role in LSCC. At present, there is no role for routinely evaluating p16 or HPV status in LSCC.
Change history
17 March 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, Silverman CC, Redmond KP, Gillison ML (2010) Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 363 (1): 24–35.
Baumann JL, Cohen S, Evjen AN, Law JH, Vadivelu S, Attia A, Schindler JS, Chung CH, Wirth PS, Meijer CJ, Snijders PJ, Yarbrough WG, Slebos RJ (2009) Human papillomavirus in early laryngeal carcinoma. Laryngoscope 119 (8): 1531–1537.
Bishop JA, Ma XJ, Wang H, Luo Y, Illei PB, Begum S, Taube JM, Koch WM, Westra WH (2012) Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol 36 (12): 1874–1882.
Bussu F, Sali M, Gallus R, Vellone VG, Zannoni GF, Autorino R, Dinapoli N, Santangelo R, Martucci R, Graziani C, Micciche F, Almadori G, Galli J, Delogu G, Sanguinetti M, Rindi G, Valentini V, Paludetti G (2013) HPV infection in squamous cell carcinomas arising from different mucosal sites of the head and neck region. Is p16 immunohistochemistry a reliable surrogate marker? Br J Cancer 108 (5): 1157–1162.
Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML (2011) Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 29 (32): 4294–4301.
Chen ZW, Weinreb I, Kamel-Reid S, Perez-Ordonez B (2012) Equivocal p16 immunostaining in squamous cell carcinoma of the head and neck: staining patterns are suggestive of HPV status. Head Neck Pathol 6 (4): 422–429.
Chernock RD, Wang X, Gao G, Lewis JS Jr., Zhang Q, Thorstad WL, El-Mofty SK (2013) Detection and significance of human papillomavirus, CDKN2A(p16) and CDKN1A(p21) expression in squamous cell carcinoma of the larynx. Mod Pathol 26 (2): 223–231.
Chung CH, Gillison ML (2009) Human papillomavirus in head and neck cancer: its role in pathogenesis and clinical implications. Clin Cancer Res 15 (22): 6758–6762.
Chung CH, Zhang Q, Kong CS, Harris J, Fertig EJ, Harari PM, Wang D, Redmond KP, Shenouda G, Trotti A, Raben D, Gillison ML, Jordan RC, Le QT (2014) p16 protein expression and human papillomavirus status as prognostic biomarkers of nonoropharyngeal head and neck squamous cell carcinoma. J Clin Oncol 32 (35): 3930–3938.
Corry J, Rischin D, Cotton S, D'Costa I, Chua M, Vallance N, Lyons B, Kleid S, Sizeland A, Peters LJ (2011) Larynx preservation with primary non-surgical treatment for loco-regionally advanced larynx cancer. J Med Imaging Radiat Oncol 55 (2): 229–235.
Department of Veterans Affairs Laryngeal Cancer Study Group (1991) Induction chemotherapy plus radiation compared with surgery plus radiation in patients with advanced laryngeal cancer. N Engl J Med 324 (24): 1685–1690.
Doxtader EE, Katzenstein AL (2012) The relationship between p16 expression and high-risk human papillomavirus infection in squamous cell carcinomas from sites other than uterine cervix: a study of 137 cases. Hum Pathol 43 (3): 327–332.
Draganov P, Todorov S, Todorov I, Karchev T, Kalvatchev Z (2006) Identification of HPV DNA in patients with juvenile-onset recurrent respiratory papillomatosis using SYBR Green real-time PCR. Int J Pediatr Otorhinolaryngol 70 (3): 469–473.
Duray A, Descamps G, Arafa M, Decaestecker C, Remmelink M, Sirtaine N, Ernoux-Neufcoeur P, Mutijima E, Somja J, Depuydt CE, Delvenne P, Saussez S (2011) High incidence of high-risk HPV in benign and malignant lesions of the larynx. Int J Oncol 39 (1): 51–59.
Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML (2008) Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 100 (4): 261–269.
Gheit T, Abedi-Ardekani B, Carreira C, Missad CG, Tommasino M, Torrente MC (2014) Comprehensive analysis of HPV expression in laryngeal squamous cell carcinoma. J Med Virol 86 (4): 642–646.
Halec G, Holzinger D, Schmitt M, Flechtenmacher C, Dyckhoff G, Lloveras B, Hofler D, Bosch FX, Pawlita M (2013) Biological evidence for a causal role of HPV16 in a small fraction of laryngeal squamous cell carcinoma. Br J Cancer 109 (1): 172–183.
Isayeva T, Li Y, Maswahu D, Brandwein-Gensler M (2012) Human papillomavirus in non-oropharyngeal head and neck cancers: a systematic literature review. Head Neck Pathol 6 Suppl 1: S104–S120.
Lewis JS Jr., Chernock RD, Ma XJ, Flanagan JJ, Luo Y, Gao G, Wang X, El-Mofty SK (2012a) Partial p16 staining in oropharyngeal squamous cell carcinoma: extent and pattern correlate with human papillomavirus RNA status. Mod Pathol 25 (9): 1212–1220.
Lewis JS Jr., Ukpo OC, Ma XJ, Flanagan JJ, Luo Y, Thorstad WL, Chernock RD (2012b) Transcriptionally-active high-risk human papillomavirus is rare in oral cavity and laryngeal/hypopharyngeal squamous cell carcinomas—a tissue microarray study utilizing E6/E7 mRNA in situ hybridization. Histopathology 60 (6): 982–991.
Li X, Gao L, Li H, Gao J, Yang Y, Zhou F, Gao C, Li M, Jin Q (2013) Human papillomavirus infection and laryngeal cancer risk: a systematic review and meta-analysis. J Infect Dis 207 (3): 479–488.
Lim AM, Do H, Young RJ, Wong SQ, Angel C, Collins M, Takano EA, Corry J, Wiesenfeld D, Kleid S, Sigston E, Lyons B, Fox SB, Rischin D, Dobrovic A, Solomon B (2014) Differential mechanisms of CDKN2A (p16) alteration in oral tongue squamous cell carcinomas and correlation with patient outcome. Int J Cancer 135 (4): 887–895.
Masand RP, El-Mofty SK, Ma XJ, Luo Y, Flanagan JJ, Lewis JS Jr. (2011) Adenosquamous carcinoma of the head and neck: relationship to human papillomavirus and review of the literature. Head Neck Pathol 5 (2): 108–116.
Morris LG, Sikora AG, Patel SG, Hayes RB, Ganly I (2011) Second primary cancers after an index head and neck cancer: subsite-specific trends in the era of human papillomavirus-associated oropharyngeal cancer. J Clin Oncol 29 (6): 739–746.
Morshed K (2010) Association between human papillomavirus infection and laryngeal squamous cell carcinoma. J Med Virol 82 (6): 1017–1023.
Omland T, Lie KA, Akre H, Sandlie LE, Jebsen P, Sandvik L, Nymoen DA, Bzhalava D, Dillner J, Brondbo K (2014) Recurrent respiratory papillomatosis: HPV genotypes and risk of high-grade laryngeal neoplasia. PLoS One 9 (6): e99114.
Petti S (2009) Lifestyle risk factors for oral cancer. Oral Oncol 45 (4-5): 340–350.
Rischin D, Young RJ, Fisher R, Fox SB, Le QT, Peters LJ, Solomon B, Choi J, O'Sullivan B, Kenny LM, McArthur GA (2010) Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J Clin Oncol 28 (27): 4142–4148.
Schache AG, Liloglou T, Risk JM, Jones TM, Ma XJ, Wang H, Bui S, Luo Y, Sloan P, Shaw RJ, Robinson M (2013) Validation of a novel diagnostic standard in HPV-positive oropharyngeal squamous cell carcinoma. Br J Cancer 108 (6): 1332–1339.
Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64 (1): 9–29.
Stephen JK, Chen KM, Shah V, Havard S, Lu M, Schweitzer VP, Gardner G, Worsham MJ (2012) Human papillomavirus outcomes in an access-to-care laryngeal cancer cohort. Otolaryngol Head Neck Surg 146 (5): 730–738.
Torrente MC, Rodrigo JP, Haigentz M Jr., Dikkers FG, Rinaldo A, Takes RP, Olofsson J, Ferlito A (2011) Human papillomavirus infections in laryngeal cancer. Head Neck 33 (4): 581–586.
Ukpo OC, Flanagan JJ, Ma XJ, Luo Y, Thorstad WL, Lewis JS Jr. (2011) High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol 35 (9): 1343–1350.
Urban D, Corry J, Rischin D (2014) What is the best treatment for patients with human papillomavirus-positive and -negative oropharyngeal cancer? Cancer 120 (10): 1462–1470.
Vlachtsis K, Nikolaou A, Markou K, Fountzilas G, Daniilidis I (2005) Clinical and molecular prognostic factors in operable laryngeal cancer. Eur Arch Otorhinolaryngol 262 (11): 890–898.
Wang H, Sun R, Lin H, Hu WH (2013) P16INK4A as a surrogate biomarker for human papillomavirus-associated oropharyngeal carcinoma: consideration of some aspects. Cancer science 104 (12): 1553–1559.
Weinberger PM, Yu Z, Haffty BG, Kowalski D, Harigopal M, Brandsma J, Sasaki C, Joe J, Camp RL, Rimm DL, Psyrri A (2006) Molecular classification identifies a subset of human papillomavirus—associated oropharyngeal cancers with favorable prognosis. J Clin Oncol 24 (5): 736–747.
Young RJ, Lim AM, Angel C, Collins M, Deb S, Corry J, Wiesenfeld D, Kleid S, Sigston E, Lyons B, Russell PA, Wright G, McArthur GA, Fox SB, Rischin D, Solomon B (2013) Frequency of Fibroblast Growth Factor Receptor 1 gene amplification in oral tongue squamous cell carcinomas and associations with clinical features and patient outcome. Oral Oncol 49 (6): 576–581.
Young RJ, Rischin D, Fisher R, McArthur GA, Fox SB, Peters LJ, Corry J, Lim A, Waldeck K, Solomon B (2011) Relationship between Epidermal Growth Factor Receptor Status, p16INK4A, and Outcome in Head and Neck Squamous Cell Carcinoma. Cancer Epidemiol Biomarkers Prev 20 (6): 1230–1237.
Acknowledgements
This project was funded by a Peter MacCallum Foundation Grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Young, R., Urban, D., Angel, C. et al. Frequency and prognostic significance of p16INK4A protein overexpression and transcriptionally active human papillomavirus infection in laryngeal squamous cell carcinoma. Br J Cancer 112, 1098–1104 (2015). https://doi.org/10.1038/bjc.2015.59
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2015.59
Keywords
This article is cited by
-
Human papillomavirus infection and non-oropharyngeal head and neck cancers: an umbrella review of meta-analysis
European Archives of Oto-Rhino-Laryngology (2023)
-
p16INK4a and pRb expression in laryngeal squamous cell carcinoma with and without infection by EBV or different genotypes of HPV: a retrospective study
Infectious Agents and Cancer (2023)
-
High-risk type human papillomavirus infection and p16 expression in laryngeal cancer
Infectious Agents and Cancer (2019)
-
The role of high-risk human papillomavirus infections in laryngeal squamous cell carcinoma
European Archives of Oto-Rhino-Laryngology (2017)