Abstract
Background:
Ribosome-binding protein 1 (RRBP1) has been implicated in the regulation of unfolded protein response, which is involved in almost every aspect of cancer development. We aimed to explore the significance of RRBP1 in the progression and prognosis of colorectal cancer (CRC).
Methods:
The study population consisted of 856 patients with stage I–III CRC from two hospitals. RRBP1 expression was examined by immunohistochemisty (IHC) in colorectal tissues. The correlation of RRBP1 expression and CRC occurrence was assessed in paired cancer-adjacent tissues. Factors contributing to prognosis were evaluated in a training-validation design with univariate and multivariate Cox analysis. Colorectal cancer aggressiveness caused by RRBP1 knockdown or overexpression was evaluated in CRC cells.
Results:
RRBP1 was aberrantly overexpressed in CRC. Compared with low-RRBP1 patients, high-RRBP1 patients had shorter disease-specific survival in the training (hazard ratio (HR), 2.423; 95% confidence interval (CI), 1.531–3.835) and validation cohorts (HR, 3.749; 95% CI, 2.166–6.448) in multivariate Cox analysis. High-RRBP1 independently predicted a shorter disease-free survival (HR, 4.821; 95% CI, 3.220–7.218) in the validation cohort. RRBP1 knockdown reduced the aggressiveness of CRC cells in vitro and inhibited the growth of CRC xenografts in vivo.
Conclusions:
High RRBP1 expression facilitates CRC progression and predicts an unfavourable post-operative prognosis.
Similar content being viewed by others
Main
Colorectal cancer (CRC) is the third most common cancer in the world, with more than one million newly diagnosed patients annually worldwide (Cunningham et al, 2010). Of those, approximately 14–25% are diagnosed with distant metastasis (Jemal et al, 2011), and usually have a poor prognosis. Surgical resection is especially effective for patients with localised and regional CRC (Jemal et al, 2011). However, 30–50% of those patients after surgery will develop local recurrence and metachronous metastases, and many of whom will die from the recurrence (Schmoll et al, 2012). Post-operative chemotherapy is beneficial to those patients who are more likely to develop recurrence. However, overtreatment of patients who may not develop recurrence tends to be harmful, owing to many side effects of chemotherapy (Iinuma et al, 2011; Yothers et al, 2013). Therefore, it is of great importance to find patients in the early stage and develop certain selection criteria, such as biomarkers for prognostic classification of CRC.
So far, pathological staging is the most reliable method for routine prognostication and selection of adjuvant therapy. However, clinical outcomes differ even among patients within the same stage (Nagtegaal et al, 2011). About 10–20% of patients with stage II CRC and 30–40% of those with stage III CRC will develop recurrence after therapeutic intervention. Among the biomarkers investigated for CRC, microsatellite instability is the only reproducible marker proven to be useful for prognostic prediction in early CRC (Popat et al, 2005; Van Schaeybroeck et al, 2011). Although several molecular signatures have been established for CRC characterisation and prognostic prediction in recent years (Van Schaeybroeck et al 2011; Yothers et al, 2013; Chang et al, 2014), no specific signature has yet been used in the clinical practice. Therefore, effective biomarkers are needed for the classification of clinical outcomes in CRC.
Cancer usually develops in a stressful microenvironment, such as rapid proliferation and chronic inflammation, which may disrupt the protein-folding machinery of endoplasmic reticulum (ER) to cause accumulation of unfolded or misfolded proteins, referred to as ER stress (Verfaillie et al, 2013; Wang and Kaufman, 2014). To cope with the stress, unfolded protein response (UPR) will be evoked to maintain a productive protein-folding environment in ER. UPR signalling molecules may interact with oncogenes and tumour-suppressor genes to provide a survival strategy for transformed cells even in a stressful microenvironment (Wang and Kaufman, 2014). Both ER stress and UPR activation are involved in multiple aspects of cancer development (Suh et al, 2012; Verfaillie et al, 2013; Wang and Kaufman, 2014). Moreover, ER stress may also trigger autophagy progression (Yorimitsu et al, 2006; Ciechomska et al, 2013). Unfolded protein response signalling molecules, such as immunoglobulin heavy-chain binding protein (BIP; also known as GRP78 and HSP5A), heat shock protein 90 kDa beta (HSP90B1; also known as GRP94) and C/EBP homologous protein (CHOP; also known as DDIT3 and GADD153), have been reported to regulate the chemo- and radio-sensitivities of CRC cells (Mhaidat et al, 2014; Drake et al, 2015), and to be correlated with CRC (Rask et al, 2000; Xing et al, 2006; Takahashi et al, 2011). Notably, BIP, a key molecule in UPR signalling, has been identified as a novel predictive biomarker for CRC patients to receive adjuvant chemoradiotherapy (Thornton et al, 2013; Lee et al, 2015). However, the exact mechanisms and new members still need to be clarified for a better understanding of UPR molecules in CRC development and progression.
Ribosome-binding protein 1 (RRBP1) is mainly localised on the ER membrane, and is critical for the transportation and secretion of nascent proteins in mammalian cells (Savitz and Meyer, 1993). Recently, RRBP1 has been reported to be associated with the regulation of UPR signalling molecules, such as BIP and activating transcription factor 6 (ATF6) (Tsai et al, 2013). A strong correlation was observed between RRBP1 and BIP in lung cancer (Tsai et al, 2013). It is also suggested that RRBP1 may be involved in the accumulation of perinuclear autophagosomes in cancer cells via interaction with kinesin family member 5B (KIF5B) (Diefenbach et al, 2004; Cardoso et al, 2009). The above evidence support RRBP1 may be a key molecule in the signalling network of ER stress, UPR and autophagy. Recently, it is reported that elevated RRBP1 is frequently observed in CRC (Krasnov et al, 2009), breast cancer (Telikicherla et al, 2012) and lung cancer (Tsai et al, 2013). Moreover, RRBP1 expression in Her-2-positive breast cancer correlates with shorter survival (Liang et al, 2015). However, the study of RRBP1 expression in CRC and its correlation to clinicopathological features of CRC only included a small sample size and provided inconclusive results (Krasnov et al, 2009). Furthermore, the role of RRBP1 in CRC progression is largely unknown. The present study aimed to investigate the association between RRBP1 expression in tumour tissues from CRC patients and adjacent normal tissues. The relationship between RRBP1 expression and CRC characteristics, and its role as a potential prognostic biomarker in patients with early CRC, were also evaluated. Moreover, we conducted in vitro and in vivo studies to evaluate the effects of RRBP1 alterations on the aggressiveness of CRC cells.
Materials and Methods
Genomic data mining
Five CRC microarray datasets (GSE37364, GSE23878, GSE22598, GSE9348 and GSE8671) were retrieved from Gene Expression Ominous (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Each of those datasets contained more than 10 cancerous and 10 non-cancerous specimens and was examined by Affymetrix plus 2.0 array platform (Santa Clara, CA, USA). After downloading the raw datasets, mRNA profiling was then extracted using the fRMA package (McCall et al, 2010) under R 3.0.1 statistic environment (http://www.r-project.org/). Values of RRBP1 mRNA expression were manually obtained and then evaluated. Differences between cancerous and non-cancerous specimens were compared using paired or unpaired student-t tests corresponding to the original study designs (Sabates-Bellver et al, 2007; Hong et al, 2010; Uddin et al, 2011; Okazaki et al, 2012; Valcz et al, 2014).
Patients
Pathologically proven, formalin-fixed paraffin-embedded specimens of 856 patients with stage I–III CRC were included in this study. Of those, 174 received curative surgery in Shuguang Hospital, Shanghai University of Traditional Chinese Medicine (Shanghai, China) between January 2005 and December 2007; and 682 received curative surgery in Changhai Hospital, Second Military Medical University (Shanghai, China) between January 2001 and December 2009. Formalin-fixed paraffin-embedded specimens were obtained from primary tumours of all patients in the two hospitals, and adjacent pathologically normal mucosa tissues from 174 patients in Shuguang Hospital were also collected in this study. Donor information including age, sex, disease location, grade, tumour size, depth of invasion, number of examined lymph nodes and TNM stage (determined according to the American Joint Committee on Cancer staging manual, seventh edition) were documented. For the Changhai cohort, additional information of rule-based post-operative chemotherapy (FOLFOX regimen), serum CEA, and CA 199 levels were also obtained. All participants are self-reported Han Chinese. This study was approved by the institutional review boards of Shuguang Hospital and Changhai Hospital. A written informed consent was obtained from each patient.
Immunohistochemistry
Tissue microarrays (TMAs) containing the formalin-fixed paraffin-embedded specimens from Shuguang and Changhai hospitals were commercially developed (Outdo Biotech, Shanghai, China). For each donor block, a 1.2-mm diameter tissue core was punched from the identified cancerous or noncancerous area and transferred into a recipient block. The construction of TMAs has been described previously (Chang et al, 2014). Immunohistochemistry (IHC) examination was carried out on 4-μm-thick sections. Deparaffinisation and rehydration were performed according to the standard procedures. Endogenous peroxidase activity was blocked for 20 min with 0.3% hydrogen peroxide solution. Subsequently, the antigen of RRBP1 was retrieved at 98 °C for 25 min with 10 mM citrate buffer (pH 6.0). Tissue microarrays were incubated overnight with rabbit polyclonal antibodies to human RRBP1 (1 : 6000; ab95983, Abcam, Cambridge, UK). Specificity of the antibody has been verified with whole-cell lysate of HepG2 by the manufacturer. After incubation for 50 min with EnVision+Dual Link System-HRP (Dako, Carpinteria, CA, USA), TMAs were visualised using DAB reagent (Vector Laboratories, Redwood City, CA, USA) for 3–5 min. The arrays were then rinsed with running water and counter-stained with haematoxylin. All arrays were stained simultaneously to eliminate interassay variation. RRBP1 was semi-quantitated using the H-score method as previously reported (Finn et al, 2009; Dziadziuszko et al, 2010). The intensity of the staining (0, 1+, 2+, 3+) and the total percentage of positive epithelial cells were independently assessed by two observers (WC and SG) who were blinded to the clinical status. A minimum of 100 cells were evaluated to calculate the IHC-score. An equation, IHC-score=(%1+ × 1)+(%2+ × 2)+(%3+ × 3), was used to calculate the IHC-score for each specimen. The average IHC-score from the two observers was applied in further analysis. Controversial cases (defined by the difference of IHC-scores more than 10% of the average score) were re-evaluated jointly until a consensus was reached. The interobserver concordance in our study was high with a correlation coefficient of 0.912 (P<0.001).
Follow-up and survival analysis
Follow-up information of the 856 patients was collected in the two hospitals, following a standard procedure as previously described (Chang et al, 2014). The primary outcome of interest was disease-specific survival (DSS), which was defined as months from the date of receiving surgery to the date that patient died of CRC. Disease-free survival (DFS) was measured in months from the date of receiving surgery to the date of first relapse. Only DSS information was obtained in the Shuguang cohort. An optimal cut-off value of RRBP1 IHC-scores was selected to define patient subgroups (high vs low RRBP1 subgroup), which can most effectively discriminate the survival differences of patients in Shuguang cohort as a training set. Subsequently, the Changhai cohort was used as an external validation set to evaluate the prognostic value of RRBP1 protein using the same cut-off value of IHC-score identified in the training set.
Gene dosage analysis
To assess the gene dosage effect (DNA copy number status correlated with mRNA expression levels) of RRBP1 on CRC, the dataset of GSE40967 (Marisa et al, 2013) from GEO and the CRC dataset of The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Network, 2012) (https://tcga-data.nci.nih.gov/docs/publications/coadread_2012/) were downloaded for further use. The GSE40967 dataset contained 463 patients with comparison genome hybridisation data, and 160 of those had corresponding mRNA expression profiling examined by Affymetrix plus 2.0 arrays. The status of ‘GNL’ (G: gain; N: normal; L: loss) was directly retrieved from the ‘gnl’ file in the original study (Marisa et al, 2013) and the mRNA profile was extracted using the fRMA package (McCall et al, 2010) from the raw data. The TCGA CRC dataset contained 564 patients with comparison genome hybridisation data, and 252 of those had corresponding mRNA profile by RNA-Seq examination. The status of ‘GNL’ in the CRC study from TCGA was described in GISTIC marker file. Polynomial regression was applied to explore the correlation of RRBP1 mRNA expression and corresponding chromosomal status (gain, normal or loss).
Cell lines, quantitative RT–PCR, RNA interference and overexpression
SW480, HT29, HCT116 and LoVo CRC cells (obtained from American Type Culture Collection, Manassas, VA, USA) were grown in Dulbecco’s Modified Eagle Medium (GIBCO, Grand Island, NY, USA) with 10% heat-inactivated foetal calf serum (GIBCO), 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin in 5% CO2 incubator. RRBP1 mRNA was examined by quantitative RT–PCR (Q–PCR) using the following primers: 5′ GAG ATG GCG AAA ACT CAC CAC 3′ (forward) and 5′ CTC GAA GGA GGA CAG TCA CAT 3′ (reverse). Human GAPDH was used as internal control using primers 5′ GGA GCG AGA TCC CTC CAA AAT 3′ (forward) and 5′ GGC TGT TGT CAT ACT TCT CAT GG 3 (reverse). The relative expression of RRBP1 mRNA was detected by LightCycler480 II (Roche, Basel, Switzerland) and normalised to the corresponding GAPDH mRNA level. The negative control siRNA and siRNA targeting RRBP1 were designed and synthesised at Shanghai Invitrogen (Shanghai, China). The sequences of RRBP1 siRNA were: 5′-GCU CUG UAG UGA AUU CCA UTT-3′ (sense) and 5′-AUG GAA UUC ACU ACA GAG CTT-3′ (antisense). The sequences of the control siRNA were: 5′-UUC UCC GAA CGU GUC ACG UTT-3′ (sense) and 5′-ACG UGA CAC GUU CGU AGA ATT-3′ (antisense). With Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA, USA), siRNAs were transfected into CRC cells with a final concentration of 20 nM as previously described (Chang et al, 2014). After 48 h of the transfection, Q–PCR and western blot were performed to validate the efficiency of RRBP1 knockdown. The expression vector encoding Myc-tagged RRBP1 under CMV promoter was obtained from OriGene Technologies (Beijing, China), and transfected into CRC cells using Lipofectamine 2000 (Invitrogen). Western blot examination was performed with rabbit polyclonal antibodies to human RRBP1 (1 : 1000, ab95983, Abcam), Myc tag antibody (1 : 500, ab18185, Abcam) and rabbit polyclonal antibodies to human GAPDH (1 : 1000, AP0063, Bioworld Technology, St Louis Park, MN, USA) as previously described (Chang et al, 2014).
Cell proliferation assay
Colorectal cancer cells were seeded in triplicate in 96-well plates at 5000 cells per well, and after 12 h were transfected with RRBP1 siRNA and control siRNA. The number of viable cells at 24, 48 and 72 h after transfection was assayed using Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions. The absorbance at 450 nm was measured to reflect the viable cell population.
Colony formation assay
RRBP1 siRNA and control siRNA transfected cells were cultured in 6-well plates (Corning, Corning, NY, USA) at a density of 2.0 × 103/well. After culturing for 2–3 weeks, the resulting colonies were fixed with ice-cold methanol, stained with crystal violet solution for counting. The assay was performed in triplicate.
Cell invasion assay
Twenty-four-well transwell chambers coated with Matrigel (BD Pharmingen, San Jose, CA, USA) was used to determine the invasiveness of the CRC cells. Transfected CRC cells in serum-free medium were added to the upper chamber at the concentration of 5 × 104 cells, from where the cells were allowed to invade through a polycarbonate membrane with 8-μm pore (Costar, Cambridge, MA, USA) toward the lower chamber. The lower chamber was filled with DMEM supplemented with 10% foetal bovine serum. After the chambers were incubated for 24 h, invaded cells on the lower chamber were fixed with 4% paraformaldehyde (Sigma Aldrich, St Louis, MO, USA), stained with crystal violet (Beyotime, Shanghai, China), and counted under a microscope (Leica, London, UK).
In vivo tumour growth and in vivo siRNA treatments
For the establishment of human tumours in Balb/c nude mice, 5 × 106 CRC cells were subcutaneously injected in the proximal midline of the dorsa. For in vivo treatments of CRC tumours, 50 μ M RRBP1 siRNAs or control siRNAs were incubated with 100 μl Interfering siRNA reagents (Polyplus-transfection Company, Illkirch, France) for 10 min, and then the complexes were rapidly injected into the tumour after the inoculation of tumour cells at multiple sites on days 7, 10, 12, 14 and 17. After siRNA injection, the efficiency of RRBP1 silencing was examined by western blot of tissue lysates after excision of the tumour. Tumour sizes were measured every 3–5 days and the tumour volumes were determined by measuring the maximal (a) and minimal (b) diameters using a calibre and calculated by using the formula a × b2.
Statistical analysis
Paired student t-test was used to evaluate the discrimination power of IHC scores of RRBP1 toward paired cancer and normal tissues. Independent sample t-test was used to compare the differences of IHC scores of RRBP1 between CRC specimens. The optimal cut-off value of the IHC score to define risk subgroups was identified by X-tile software (http://medicine.yale.edu/lab/rimm/research/software.aspx) (Camp et al, 2004). Kaplan–Meier method was used to estimate the DSS and DFS, and the log-rank test was used to compare the survival curves. Cox hazard proportion models were used to estimate the hazard ratios and corresponding 95% confidence intervals. All statistical tests were two-sided and performed with Statistical Program for Social Sciences (SPSS 16.0 for Windows, SPSS, Chicago, IL, USA). P<0.05 was considered as statistically significant.
Results
Elevated RRBP1 expression in CRC
To explore the potential significance of RRBP1 in CRC, we initially used five publicly available microarray datasets totally composing of 181 CRC and 123 adjacent normal tissues to assess the mRNA expression of RRBP1. Excluding the dataset of GSE37364 (Valcz et al 2014), the remaining four datasets (Sabates-Bellver et al, 2007; Hong et al, 2010; Uddin et al, 2011; Okazaki et al, 2012) consistently showed that mRNA expression of RRBP1 was significantly upregulated in CRC (all P values<0.05) as compared with adjacent normal tissues (Supplementary Figure 1).
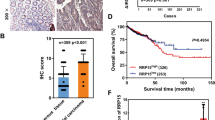
The expression pattern of RRBP1 was further investigated by IHC analysis on 174 paired cancer-adjacent specimens from Shuguang Hospital. RRBP1 staining was mainly distributed in the cytoplasm of colorectal epithelial cells and stromal cells (Figure 1A), and only the staining pattern in epithelial cells was further investigated. The median of IHC-score in CRC tissues was 181 (interquartile range, 131–214), and the median of IHC-score in normal tissues was 32 (interquartile range, 17–53). RRBP1 protein showed slight variation in normal tissues but great heterogeneity in cancer tissues. Results from the paired student t-test showed that the expression of RRBP1 protein was significantly higher in colon cancer and rectum cancer than that in corresponding adjacent specimens (all P<0.001) (Figure 1B). No differences were found between IHC-scores of stage CRC (I+II) and stage III CRC (Figure 1B) and between IHC-scores of colon cancer and rectum cancer specimens.
RRBP1 protein is elevated in colorectal cancer. (A) Representative examples of immunostaining of RRBP1 in CRC and adjacent normal tissues. RRBP1 is expressed in the cytoplasm of colorectal epithelial cells and stromal cells, and corresponding IHC-score was marked. Bar, 50 μm. (B) Expression pattern of RRBP1 protein in formalin-fixed paraffin-embedded specimens of CRC and adjacent normal tissues. The differences of RRBP1 protein expression between cancer vs normal, and stage (I+II) vs stage III are evaluated (stratified by lesion location).
RRBP1 expression predicts an unfavourable survival
Immunohistochemisty-scores of CRC specimens from Shuguang Hospital were further analysed to investigate the prognostic value of RRBP1. When tested as a continuous variable, the IHC-score of RRBP1 was significantly associated with DSS (P<0.001). In exploratory analysis with different cut-off values to define patient subgroups with high RRBP1 or low RRBP1, we found that the cut-off value of 175 of IHC-score identified by X-tile software could most effectively discriminate the differences of DSS. Using the cut-off value, we also found that RRBP1 expression may be uncorrelated to many clinicopathologic characteristics of CRC patients, such as age, sex, tumour location, differentiation grade and TNM stage (Table 1). Kaplan–Meier analysis showed that patients with high RRBP1 (IHC-score >175) were significantly associated with a shorter DSS than those with low RRBP1 (IHC score ⩽175) (Figure 2). Including high expression of RRBP1, other factors such as TNM stage and differentiation grade, were also significantly associated with DSS of CRC patients in univariate Cox analysis (Table 2). Multivariate Cox regression analysis showed high RRBP1 in CRC remained a statistically significant predictor for DSS (hazard ratio, 2.423; 95% confidence interval, 1.531–3.835) after adjusting for variables as stage, grade, age, sex, tumour size, depth of tumour invasion and tumour location (Table 2). Notably, patients with high RRBP1 were significantly associated with a poor DSS in both stage (I+II) CRC and stage III CRC (Figure 2).
High IHC-score of RRBP1 predicts poor survivals of patients with CRC. Patients with stage I–III, stage I–II or stage III tumour were dichotomised into the subgroups with high- or low-RRBP1 protein expression according to RRBP1 IHC-score (cut-off value=175) in the Shuguang and Changhai cohorts, respectively. Disease-specific survival and DFS are presented. Log-rank P values and hazard ratios (HRs) from univariate Cox regression analysis are shown.
Validation of the prognostic value of RRBP1 in an independent cohort
The cut-off value of IHC-scores of RRBP1 identified in the Shuguang cohort was then used to dichotomise patients into high- or low-RRBP1 subgroups in the Changhai cohort. Patients with high RRBP1 (IHC-score >175) showed significantly shorter DSS and shorter DFS than those patients with low RRBP1 at stage I–III (Figure 2). Multivariate Cox regression analysis demonstrated that high RRBP1 remained an independent factor for DSS (hazard ratio, 3.749; 95% confidence interval, 2.166–6.448) and for DFS (hazard ratio, 4.821; 95% confidence interval, 3.220–7.218) (Table 3). When included more covariates (e.g., CEA and CA 199), multivariate Cox regression analysis still showed that high RRBP1 was an independent factor for CRC survival in Changhai cohort (Supplementary Table 1). Importantly, high RRBP1 expression predicted poor DSS and poor DFS in patients with stage (I+II) or stage III CRC (Figure 2), which was consistent with the results obtained in the Shuguang cohort. We further assessed prognostic value of RRBP1 protein for patients with or without post-operative chemotherapy in the Changhai cohort. Patients with high RRBP1 expression were significantly associated with shorter DSS and shorter DFS in patients with or without post-operative chemotherapy (Supplementary Figure 2).
RRBP1 expression is related to chromosomal aberrations
As chromosomal imbalance may cause gene expression alteration, we hypothesised that there might be a correlation between chromosome aberrations and mRNA alterations of RRBP1. To determine the relationship, two publicly available datasets (GSE40967 and a dataset from TCGA) were used. We found that chromosomal gains of RRBP1 and chromosomal losses of RRBP1 in CRC were 40.8% (189/463) and 15.8% (73/463), respectively, in the dataset of GSE40967. The frequency of gains was significantly higher than that of losses (P<0.001). In this dataset, only 160 patients had data for RRBP1 mRNA expression and corresponding chromosomal aberration. In patients with chromosomal gains, RRBP1 mRNA level was the highest, followed by chromosomal normal, and was the lowest in patients with chromosomal loss (Ptrend<0.001), which showed a dose–response between RRBP1 mRNA levels and chromosomal aberration, as shown in Supplementary Figure 3. Similar analysis was also performed for the TCGA dataset. Chromosomal gains and chromosomal losses of RRBP1 in 564 TCGA CRC specimens was 41.7% and 15.3%, respectively, which was consistent with the results obtained from the dataset of GSE40967. Among TCGA CRC patients, 252 had RRBP1 mRNA expression data. A dose–response between RRBP1 mRNA levels and chromosomal aberration (Ptrend<0.001) was observed again in the TCGA dataset (Supplementary Figure 3).
RRBP1 promotes the growth and aggressiveness of CRC cells
To explore the roles of RRBP1 in CRC progression, we examined the effects of RRBP1 expression on growth and aggressiveness of CRC cells. RRBP1 is highly expressed in SW480, HT29 and HCT116 cells, but is relatively low in LoVo cells (Figure 3A). Therefore, we decreased RRBP1 expression in SW480, HT29 and HCT116 cells, and overexpressed RRBP1 in LoVo cells. We synthesised three pairs of siRNA specific for RRBP1, and found that the second siRNA pair (referred to as RRBP1 siRNAs) could most efficiently downregulate RRBP1 expression, as examined by both Q–PCR and western blot (Figure 3B). Then, we examined the proliferation, clone formation and invasion of CRC cells transfected with RRBP1 siRNA in vitro (Figure 3C). The results showed that RRBP1 knockdown significantly inhibited the proliferation, colony formation and invasiveness of SW480, HT29 and HCT116 cells (Figure 3D–F). Meanwhile, RRBP1 overexpression significantly promoted the proliferation, colony formation and invasiveness of LoVo cells (Figure 3D–F). To exclude the possible off-target effects, we transiently overexpressed RRBP1 in SW480 cells after RRBP1 knockdown, and found that RRBP1 could rescue the effects of RRBP1 knockdown on cell proliferation (Supplementary Figure 4). These data suggest that RRBP1 may promote the growth and aggressiveness of CRC cells in vitro. To further investigate the effects of RRBP1 on CRC progression, we inoculated SW480, HT29 and HCT116 cells in Balb/c nude mice and treated the neoplasm with RRBP1 siRNAs (Figure 3G). The results showed that RRBP1 siRNA could significantly inhibit the growth of CRC xenografts in vivo (Figure 3H). Consistently, RRBP1-overexpressed LoVo xenografts demonstrated increased growth in nude mice (Figure 3H). These data indicate that RRBP1 is involved CRC progression in vivo and knockdown of RRBP1 may be a potential therapeutic for CRC.
RRBP1 promotes the growth and aggressiveness of CRC cells. (A) Relative levels of RRBP1 mRNA in CRC cells examined by Q–PCR (upper panel) and western blot (lower panel). (B) Efficiencies of RRBP1 siRNA pairs 1–3 in downregulating RRBP1 expression in SW480 cells were examined by Q–PCR (upper panel) and western blot (lower panel). (C) Similar to (B), CRC cells were transfected with RRBP1-siRNAs and examined for RRBP1 expression by western blot. Otherwise, LoVo cells were transfected with Mock or RRBP1-Myc vector and examined by western blot. IB, immunoblot. (D–F) Colorectal cancer cells in (C) were examined for cell proliferation (D), colony formation (E) and invasiveness (F). (G, H) Colorectal cancer xenografts in Balb/c nude mice. On indicated days after inoculation, RRBP1 expression in representative tumours was examined by western blot (G) and tumour volumes were monitored and presented as mean±s.e.m. (H). (**P<0.01; ***P<0.001; as compared with control cells, ANOVA or unpaired t tests was used.)
Discussion
In this study, we applied a genomic data-mining strategy to explore the expression pattern of RRBP1 mRNA in CRC. Among five published microarray datasets, four studies consistently supported that RRBP1 mRNA is higher in CRC than adjacent normal tissues. This result is consistent with a previous proteomic study (Krasnov et al, 2009), in which only 11 CRC and corresponding normal specimens were used. Thus, the bioinformatics approach has been proven to be a fast and effective measure to identify candidate molecules. Coupled with our IHC analysis using 174 paired CRC and adjacent normal tissues, we showed that the expression of RRBP1 protein was elevated in CRC tissues as compared with adjacent normal tissues, which was quite consistent with that from RRBP1 mRNA. Moreover, we did not observe any gradation of RRBP1 protein expression in the tumour specimens of CRC patients in different TNM stages, which indicate that the roles of RRBP1 in CRC progression may be different to many oncogenes that usually correlated to TNM stages. Similar results are also reported in lung cancer (Tsai et al, 2013) and breast cancer (Telikicherla et al, 2012). Therefore, RRBP1 expression is distinctly different between CRC and non-cancerous tissue, and might serve as a potential biomarker for CRC diagnosis, even in early stage tumours.
In exception of a recent study (Liang et al, 2015) in breast cancer, previous studies (Krasnov et al, 2009; Telikicherla et al, 2012; Tsai et al, 2013) have not clearly established the association between RRBP1 expression and the prognosis of cancer patients. With an optimal cut-off value of RRBP1 IHC-score (175), we found that CRC patients with high RRBP1 expression had shorter DSS than those patients with low RRBP1 expression in the training cohort. Further, RRBP1 expression also efficiently discriminated CRC patients with distinct prognosis (both DSS and DFS) in the validation cohort. Colorectal cancer prognosis is usually dependent on tumour stage and grade. After balancing TNM stage and grade in both cohorts, high RRBP1 expression still independently predicted an unfavourable prognosis. These results strongly support that RRBP1 expression is independently correlated with clinical outcomes of CRC patients. Notably, RRBP1 expression was significantly associated with post-operative prognosis of stage (I+II) and stage III CRC patients (Figure 2). Dichotomisation of CRC patients within the same stage by biomarkers like RRBP1 is of great promise in the field of prognostic classification. GRP78 (BIP), a key player in UPR, has been reported to be correlated with TNM stage and 5-year overall survival of CRC patients (Thornton et al, 2013). It may be better to examine the expression of RRBP1 together with GRP78 for the understanding of UPR in CRC.
Recently, the studies in cancer systems biology hold the notion that genes with somatic mutations or chromosome aberrations are more likely to drive cancer development and progression, especially for those genes with dosage effects (Akavia et al, 2010; Mine et al, 2013; Tang and Amon, 2013). Through integrating with published data, we found chromosomal regions of RRBP1 were more frequently gained than lost in CRC, and RRBP1 chromosomal gains were positively associated with the mRNA levels of RRBP1. These results suggested that RRBP1 might serve as a driver in the initiation and/or progression of CRC. Importantly, our in vitro and in vivo data of RRBP1 knockdown or overexpression in CRC cells suggested that RRBP1 may participate in the control of growth and progression of CRC. It also has been reported that knockdown of RRBP1 significantly reduced the in vivo tumourigenicity of lung cancer cells (Tsai et al, 2013). These results, together with the results from the CRC cohorts, suggest that RRBP1 may function as an oncogene in CRC. However, underlying mechanisms for RRBP1 in regulating CRC tumourigenicity need further investigation.
On the basis of a large study population and a training-validation study design, we effectively evaluated the prognostic value of RRBP1 protein. The current study also has a few limitations. First, we cannot exclude the selection bias in our cohorts because of the loss of patients to follow-up or tissue samples owing to the unappreciated factors and the potential differences when the specimens for original objects were collected. Second, cancer-adjacent normal tissues were used, which could not completely mimic the differences between cancer and real normal tissues. Third, some important prognostic factors such as microsatellite instability and extramural venous invasion was not included (data unavailable for the present study owing to the damage of tissues during the preparation of TMAs), which led to an incomplete inclusion of variants in multivariate Cox analysis.
In summary, our study suggests that RRBP1 expression is a poor prognostic marker in early colorectal cancer (stage I–III) and potentially a useful IHC marker for the diagnosis of CRC. Further prospective studies are required for better understanding of RRBP1 as a prognostic marker in CRC.
Change history
01 September 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA, Pe’er D (2010) An integrated approach to uncover drivers of cancer. Cell 143: 1005–1017.
Camp RL, Dolled-Filhart M, Rimm DL (2004) X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 10: 7252–7259.
Cancer Genome Atlas Network (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337.
Cardoso CM, Groth-Pedersen L, Hoyer-Hansen M, Kirkegaard T, Corcelle E, Andersen JS, Jaattela M, Nylandsted J (2009) Depletion of kinesin 5B affects lysosomal distribution and stability and induces peri-nuclear accumulation of autophagosomes in cancer cells. PLoS One 4: e4424.
Chang W, Gao X, Han Y, Du Y, Liu Q, Wang L, Tan X, Zhang Q, Liu Y, Zhu Y, Yu Y, Fan X, Zhang H, Zhou W, Wang J, Fu C, Cao G (2014) Gene expression profiling-derived immunohistochemistry signature with high prognostic value in colorectal carcinoma. Gut 63: 1457–1467.
Ciechomska IA, Gabrusiewicz K, Szczepankiewicz AA, Kaminska B (2013) Endoplasmic reticulum stress triggers autophagy in malignant glioma cells undergoing cyclosporine a-induced cell death. Oncogene 32: 15118–15129.
Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N (2010) Colorectal cancer. Lancet 375: 1030–1047.
Diefenbach RJ, Diefenbach E, Douglas MW, Cunningham AL (2004) The ribosome receptor, p180, interacts with kinesin heavy chain, KIF5B. Biochem Biophys Res Commun 319: 987–992.
Drake TM, Ritchie JE, Kanthou C, Staves JJ, Narramore R, Wyld L (2015) Targeting the endoplasmic reticulum mediates radiation sensitivity in colorectal cancer. Exp Mol Pathol 98: 532–539.
Dziadziuszko R, Merrick DT, Witta SE, Mendoza AD, Szostakiewicz B, Szymanowska A, Rzyman W, Dziadziuszko K, Jassem J, Bunn PA Jr, Varella-Garcia M, Hirsch FR (2010) Insulin-like growth factor receptor 1 (IGF1R) gene copy number is associated with survival in operable non-small-cell lung cancer: a comparison between IGF1R fluorescent in situ hybridization, protein expression, and mRNA expression. J Clin Oncol 28: 2174–2180.
Finn RS, Press MF, Dering J, Arbushites M, Koehler M, Oliva C, Williams LS, Di Leo A (2009) Estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2), and epidermal growth factor receptor expression and benefit from lapatinib in a randomized trial of paclitaxel with lapatinib or placebo as first-line treatment in HER2-negative or unknown metastatic breast cancer. J Clin Oncol 27: 3908–3915.
Hong Y, Downey T, Eu KW, Koh PK, Cheah PY (2010) A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis 27: 83–90.
Iinuma H, Watanabe T, Mimori K, Adachi M, Hayashi N, Tamura J, Matsuda K, Fukushima R, Okinaga K, Sasako M, Mori M (2011) Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J Clin Oncol 29: 1547–1555.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90.
Krasnov GS, Oparina N, Khankin SL, Mashkova TD, Ershov AN, Zatsepina OG, Karpov VL, Beresten SF (2009) Colorectal cancer 2D-proteomics: identification of altered protein expression. Mol Biol (Mosk) 43: 348–356.
Lee HY, Jung JH, Cho HM, Kim SH, Lee KM, Kim HJ, Lee JH, Shim BY (2015) GRP78 protein expression as prognostic values in neoadjuvant chemoradiotherapy and laparoscopic surgery for locally advanced rectal cancer. Cancer Res Treat e-pub ahead of print 30 January 2015; doi:10.4143/crt.2014.121.
Liang X, Sun S, Zhang X, Wu H, Tao W, Liu T, Wei W, Geng J, Pang D (2015) Expression of ribosome-binding protein 1 correlates with shorter survival in Her-2 positive breast cancer. Cancer Sci 106: 740–746.
Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, Kirzin S, Chazal M, Fléjou JF, Benchimol D, Berger A, Lagarde A, Pencreach E, Piard F, Elias D, Parc Y, Olschwang S, Milano G, Laurent-Puig P, Boige V (2013) Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 10: e1001453.
McCall MN, Bolstad BM, Irizarry RA (2010) Frozen robust multiarray analysis (fRMA). Biostatistics 11: 242–253.
Mhaidat NM, Alzoubi KH, Khabour OF, Banihani MN, Al-Balas QA, Swaidan S (2014) GRP78 regulates sensitivity of human colorectal cancer cells to DNA targeting agents. Cytotechnology e-pub ahead print 16 November 2014; doi:10.1007/s10616-014-9799-8.
Mine KL, Shulzhenko N, Yambartsev A, Rochman M, Sanson GF, Lando M, Varma S, Skinner J, Volfovsky N, Deng T, Brenna SM, Carvalho CR, Ribalta JC, Bustin M, Matzinger P, Silva ID, Lyng H, Gerbase-DeLima M, Morgun A (2013) Gene network reconstruction reveals cell cycle and antiviral genes as major drivers of cervical cancer. Nat Commun 4: 1806.
Nagtegaal ID, Quirke P, Schmoll HJ (2011) Has the new TNM classification for colorectal cancer improved care? Nat Rev Clin Oncol 9: 119–123.
Okazaki S, Ishikawa T, Iida S, Ishiguro M, Kobayashi H, Higuchi T, Enomoto M, Mogushi K, Mizushima H, Tanaka H, Uetake H, Sugihara K (2012) Clinical significance of UNC5B expression in colorectal cancer. Int J Oncol 40: 209–216.
Popat S, Hubner R, Houlston RS (2005) Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 23: 609–618.
Rask K, Thörn M, Pontén F, Kraaz W, Sundfeldt K, Hedin L, Enerbäck S (2000) Increased expression of the transcription factors CCAAT-enhancer binding protein-beta (C/EBBeta) and C/EBzeta (CHOP) correlate with invasiveness of human colorectal cancer. Int J Cancer 86: 337–343.
Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA, Bujnicki JM, Menigatti M, Luz J, Ranalli TV, Gomes V, Pastorelli A, Faggiani R, Anti M, Jiricny J, Clevers H, Marra G (2007) Transcriptome profile of human colorectal adenomas. Mol Cancer Res 5: 1263–1275.
Savitz AJ, Meyer DI (1993) 180-kD ribosome receptor is essential for both ribosome binding and protein translocation. J Cell Biol 120: 853–863.
Schmoll HJ, Van Cutsem E, Stein A, Valentini V, Glimelius B, Haustermans K, Nordlinger B, van de Velde CJ, Balmana J, Regula J, Nagtegaal ID, Beets-Tan RG, Arnold D, Ciardiello F, Hoff P, Kerr D, Köhne CH, Labianca R, Price T, Scheithauer W, Sobrero A, Tabernero J, Aderka D, Barroso S, Bodoky G, Douillard JY, El Ghazaly H, Gallardo J, Garin A, Glynne-Jones R, Jordan K, Meshcheryakov A, Papamichail D, Pfeiffer P, Souglakos I, Turhal S, Cervantes A (2012) ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol 23: 2479–2516.
Suh DH, Kim MK, Kim HS, Chung HH, Song YS (2012) Unfolded protein response to autophagy as a promising druggable target for anticancer therapy. Ann N Y Acad Sci 1271: 20–32.
Takahashi H, Wang JP, Zheng HC, Masuda S, Takano Y (2011) Overexpression of GRP78 and GRP94 is involved in colorectal carcinogenesis. Histol Histopathol 26: 663–671.
Tang YC, Amon A (2013) Gene copy-number alterations: a cost-benefit analysis. Cell 152: 394–405.
Telikicherla D, Marimuthu A, Kashyap MK, Ramachandra YL, Mohan S, Roa JC, Maharudraiah J, Pandey A (2012) Overexpression of ribosome binding protein 1 (RRBP1) in breast cancer. Clin Proteomics 9: 7.
Thornton M, Aslam MA, Tweedle EM, Ang C, Campbell F, Jackson R, Costello E, Rooney PS, Vlatković N, Boyd MT (2013) The unfolded protein response regulator GRP78 is a novel predictive biomarker in colorectal cancer. Int J Cancer 133: 1408–1418.
Tsai HY, Yang YF, Wu AT, Yang CJ, Liu YP, Jan YH, Lee CH, Hsiao YW, Yeh CT, Shen CN, Lu PJ, Huang MS, Hsiao M (2013) Endoplasmic reticulum ribosome-binding protein 1 (RRBP1) overexpression is frequently found in lung cancer patients and alleviates intracellular stress-induced apoptosis through the enhancement of GRP78. Oncogene 32: 4921–4931.
Uddin S, Ahmed M, Hussain A, Abubaker J, Al-Sanea N, AbdulJabbar A, Ashari LH, Alhomoud S, Al-Dayel F, Jehan Z, Bavi P, Siraj AK, Al-Kuraya KS (2011) Genome-wide expression analysis of Middle Eastern colorectal cancer reveals FOXM1 as a novel target for cancer therapy. Am J Pathol 178: 537–547.
Valcz G, Patai AV, Kalmár A, Péterfia B, Fűri I, Wichmann B, Műzes G, Sipos F, Krenács T, Mihály E, Spisák S, Molnár B, Tulassay Z (2014) Myofibroblast-derived SFRP1 as potential inhibitor of colorectal carcinoma field effect. PLoS One 9: e106143.
Van Schaeybroeck S, Allen WL, Turkington RC, Johnston PG (2011) Implementing prognostic and predictive biomarkers in CRC clinical trials. Nat Rev Clin Oncol 8: 222–232.
Verfaillie T, Garg AD, Agostinis P (2013) Targeting ER stress induced apoptosis and inflammation in cancer. Cancer Lett 332: 249–264.
Wang M, Kaufman RJ (2014) The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer 14: 581–597.
Xing X, Lai M, Wang Y, Xu E, Huang Q (2006) Overexpression of glucose-regulated protein 78 in colon cancer. Clin Chim Acta 364: 308–315.
Yorimitsu T, Nair U, Yang Z, Klionsky DJ (2006) Endoplasmic reticulum stress triggers autophagy. J Biol Chem 281: 30299–30304.
Yothers G, O’Connell MJ, Lee M, Lopatin M, Clark-Langone KM, Millward C, Paik S, Sharif S, Shak S, Wolmark N (2013) Validation of the 12-gene colon cancer recurrence score in NSABP C-07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol 31: 4512–4519.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81372671 to WC; 81402005 to FC; 81201936 to XG; and 81272561 to CF).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Pan, Y., Cao, F., Guo, A. et al. Endoplasmic reticulum ribosome-binding protein 1, RRBP1, promotes progression of colorectal cancer and predicts an unfavourable prognosis. Br J Cancer 113, 763–772 (2015). https://doi.org/10.1038/bjc.2015.260
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2015.260
Keywords
This article is cited by
-
CircVPS13C promotes pituitary adenoma growth by decreasing the stability of IFITM1 mRNA via interacting with RRBP1
Oncogene (2022)
-
RRBP1 rewires cisplatin resistance in oral squamous cell carcinoma by regulating Hippo pathway
British Journal of Cancer (2021)
-
Genome-wide scan for common variants associated with intramuscular fat and moisture content in rainbow trout
BMC Genomics (2020)
-
Predicting distant metastasis and chemotherapy benefit in locally advanced rectal cancer
Nature Communications (2020)
-
RRBP1 overexpression is associated with progression and prognosis in endometrial endometrioid adenocarcinoma
Diagnostic Pathology (2019)