Abstract
Background:
Epidemiological studies have indicated significant associations of leukocyte mitochondrial DNA (mtDNA) copy number with risk of several malignancies, including glioma. However, whether mtDNA content can predict the clinical outcome of glioma patients has not been investigated.
Methods:
The mtDNA content of peripheral blood leukocytes from 336 glioma patients was examined using a real-time PCR-based method. Kaplan–Meier curves and Cox proportional hazards regression model were used to examine the association of mtDNA content with overall survival (OS) and progression-free survival (PFS) of patients. To explore the potential mechanism, the immune phenotypes of peripheral blood mononuclear cells (PBMCs) and plasma concentrations of several cytokines from another 20 glioma patients were detected by flow cytometry and enzyme-linked immunosorbent assay (ELISA), respectively.
Results:
Patients with high mtDNA content showed both poorer OS and PFS than those with low mtDNA content. Multivariate Cox regression analysis demonstrated that mtDNA content was an independent prognostic factor for both OS and PFS. Stratified analyses showed that high mtDNA content was significantly associated with poor prognosis of patients with younger age, high-grade glioma or adjuvant radiochemotherapy. Immunological analysis indicated that patients with high mtDNA content had significantly lower frequency of natural killer cells in PBMCs and higher plasma concentrations of interleukin-2 and tumour necrosis factor-α, suggesting an immunosuppression-related mechanism involved in mtDNA-mediated prognosis.
Conclusions:
Our study for the first time demonstrated that leukocyte mtDNA content could serve as an independent prognostic marker and an indicator of immune functions in glioma patients.
Similar content being viewed by others
Main
Glioma is the most common form of primary brain tumour, accounting for 7% of cancer-related death before the age of 70 years (Furnari et al, 2007; Gladson et al, 2010). Despite significant improvement in treatment for glioma patients, the median survival remains not optimistic, particularly for those with glioblastoma (Wen and Kesari, 2008). Patients with newly diagnosed glioblastoma exhibit a median survival of ∼1 year, with generally poor responses to all therapeutic modalities. Thus, the elucidation of glioma survival factors is imperative and could potentially give support to the treatment of glioma patients.
Mitochondria are organelles generally found in all nucleated cells. Its major role is to generate cellular adenosine triphosphate (ATP) and reactive oxygen species (ROS) through oxidative respiration, to synthesise organic molecules and regulate the concentration of intracellular calcium and cellular apoptosis (Chan, 2006). Mitochondria have their own genome (mitochondrial DNA (mtDNA)), an ∼16.6 kb circular double-stranded DNA that encodes 13 polypeptide subunits of the respiratory chain complexes, 2 rRNAs and a set of 22 tRNAs (Chen and Butow, 2005). Generally, each human cell contains several hundred to 1000 mitochondria, and each mitochondrion has 2 to 10 copies of mtDNA. Under physiological conditions, intracellular mtDNA content has a steady-state level in each specific tissue that is related to the energy demand of the host cells (Capps et al, 2003). Interindividual variations of mtDNA copy number in cells exist in the general population (Moraes, 2001).
Recent experimental and epidemiological studies have demonstrated that alterations of mtDNA content are involved in the development of cancer (Yu, 2011). It has been found that alterations of mtDNA content occur early in premalignant lesions (Lin et al, 2010; Cormio et al, 2012). Continuous changes of mtDNA content may last all through the procedure of carcinogenesis. When compared with their corresponding normal tissues, tumour tissues have the altered (increased or decreased) mtDNA content in many types of malignancies, including glioma (Wang et al, 2005; Yu et al, 2007; Mizumachi et al, 2008). Moreover, mtDNA content in cancer tissues are associated with the progression and chemosensitivity (Yu et al, 2007; Hsu et al, 2010; Lin et al, 2010; Cui et al, 2013; Guo et al, 2013) as well as the prognosis of patients (Yu et al, 2007; Cui et al, 2013; Cheau-Feng Lin et al, 2014). A series of epidemiological evidence suggest that mtDNA content variations in peripheral leukocytes can be used as a biomarker to predict the risks of cancers, such as renal, breast, lung, colorectal and liver cancers as well as non-Hodgkin’s lymphoma (NHL) (Lan et al, 2008; Hosgood et al, 2010; Shen et al, 2010; Qu et al, 2011; Zhao et al, 2011). Our previous study has also demonstrated that higher peripheral leukocyte mtDNA content is associated with increased risk of glioma (Zhang et al, 2014). However, whether leukocyte mtDNA content can predict the prognosis of glioma patients has never been investigated.
In this study, we measured the leukocyte mtDNA content using a real-time PCR-based method and assessed its association with the overall survival (OS) and progression-free survival (PFS) of glioma patients after tumour resection. Furthermore, we explored the potential immunological mechanisms of glioma patients with different mtDNA content. To the best of our knowledge, this is the first study to assess the value of mtDNA content in predicting prognosis of glioma patients.
Materials and Methods
Patients
A total of 336 glioma patients were enrolled at the Department of Neurosurgery, Tangdu Hospital in Xi’an, China, from January 2000 to April 2012. Patient eligibility criteria were as follows: (1) with histologically confirmed primary glioma, (2) received surgical resection, (3) availability of complete clinical and follow-up data, (4) no preoperative anticancer treatment, (5) no history of other synchronous malignancy and (6) alive at least 3 months after surgical operation. Moreover, 20 additional glioma patients were enrolled for the detection of peripheral blood mononuclear cell (PBMC) immunophenotypes and plasma cytokine concentrations at Tangdu Hospital from October to December in 2013. Before surgery, 5 ml venous blood sample was collected from each patient and subjected to genomic DNA extraction using the RelaxGene Blood DNA System (TIANGEN, Beijing, China) according to the manufacturer’s instructions. The study was approved by the Ethic Committee of Fourth Military Medical University and informed consents were obtained from all participants. All experimental procedures were performed according to the Helsinki Declaration.
Patient data collection
Epidemiological information was collected using a standardised questionnaire through in-person interviewing by well-trained staffs. Clinical data were obtained by medical record review. Follow-up data were obtained through on-site interview, telephone calling or medical chart review. The latest follow-up date was March 2014. Overall survival time was defined as the interval from surgery to death or last follow-up. Progression-free survival time was defined as the interval from surgery to the date of the first magnetic resonance imaging (MRI)-confirmed tumour progression according to the RANO criteria (Brandsma and van den Bent, 2009).
Determination of mtDNA content by quantitative real-time PCR
Relative mtDNA content was measured by a quantitative real-time PCR-based method in the Mx3005P QPCR System (Agilent, Santa Clara, CA) as previously described, with the same primers that were used for the mitochondrial ND1 gene (ND1-F and ND1-R) and the single-copy nuclear gene human globulin (HGB-1and HGB-2) (Xing et al, 2008). In short, two pairs of primers were used in the two steps of relative quantification for mtDNA copy number. The primer sequences were as follows: ND1 forward (ND1-F), 5′-CCCTAAAACCCGCCACATCT-3′; ND1 reverse (ND1-R), 5′-GAGCGATGGTGAGAGCTAAGGT-3′; HGB forward (HGB-1), 5′-GTGCACCTGACTCCTGAGGAGA-3′; HGB reverse (HGB-2), 5′-CCTTGATACCAACCTGCCCAG-3′. In the first step, the ratio of mtDNA copy number to HGB copy number was calculated for each sample from standard curves. In the second step, the ratio for each sample was normalised to a calibrator DNA in order to standardise between different runs, and then defined as the measurement of relative mtDNA content. The calibrator DNA is a genomic DNA sample from a healthy control subject. It was measured in each run and used to standardise different real-time quantitative PCR runs. The value of this only scaling factor varied from 0.92 to 1.14 across different runs. The intra-assay or interassay variations were evaluated by assaying one sample in eight replicates or in three different runs, respectively.
The PCR reaction system (20 μl) consisted of 1 × SYBR green mastermix (TaKaRa, Dalian, China), 10 nM ND1-R (or HGB-1) primer, 10 nM ND1-F (or HGB-2) primer and 8 ng of genomic DNA. The thermal cycling conditions for both primer pairs were 95 °C for 30 s, followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 50 s with signal acquisition. The PCRs were always performed on separate 96-well plates, with the same samples in the same well positions. All samples were assayed in duplicate using the Mx3005P QPCR System (Agilent, Santa Clara, CA, USA). In each run, negative and positive controls, a calibrator DNA and a standard curve were included. For each standard curve, one reference DNA sample (the same DNA sample for all runs) was diluted with a three-fold increment per dilution to produce a 5-point standard curve between 0.593 and 48 ng DNA in each reaction. The R2 for each standard curve was ≥0.99, with acceptable s.d. values set at 0.25 (for the Ct values). Otherwise, the test was repeated.
Determination of lymphocyte immunophenotypes by flow cytometry
The PBMCs were isolated by density gradient centrifugation over Ficoll-Hypaque (Amersham Pharmacia Biotech, NJ, USA). The cells were then fixed and stained with isotype control immunoglobulins or fluorescence-conjugated antibodies against the following immune markers: CD3, CD4, CD8, CD25, FOXP3, CD19 and CD56 (eBioscience, San Diego, CA). Finally, subtype analysis of PBMCs was carried out on a FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
Cytokine detection by enzyme-linked immunoassay (ELISA)
Blood plasma from CRC patients was by centrifugation at 4000 r.p.m. under 4 °C. The concentrations of interleukin (IL)-2, IL-4, transforming growth factor (TGF)-β1, tumour necrosis factor (TNF)-α and interferon (IFN)-γ were examined by ELISA using commercial kits according to the manufacturer’s instructions (eBioscience).
Statistical analysis
Because of the nonlinearity in the effects of mtDNA content on the progression of cancer, either higher or lower cutoff point would affect the statistical power. Therefore, mtDNA content should be converted into category variable by the optimal cutoff point. Finally, we found that patients in the second and third tertile had similar median OS (21 and 25 months, respectively) and PFS (14 and 15 months, respectively) time, whereas those in the first tertile showed much longer OS (39 months) and PFS (29 months) time. We therefore converted mtDNA content into category variable by the first tertile value. Mann–Whitney U-test was used to examine the frequencies of immune cell subsets and plasma cytokine concentrations between patients with different mtDNA content. Mann–Whitney U-test was also used to examine the difference in mtDNA between two subgroups and Kruskal–Wallis test was used to test difference in mtDNA among more than three groups. Kaplan–Meier survival curve was plotted and compared by log-rank test. Multivariate Cox proportional hazards regression model was used to calculate the hazard ratio (HR) and 95% confidence interval (CI) for prognosis evaluation. All statistical analyses were performed using the SPSS Statistics 22.0 software (IBM, Armonk, NY, USA), and P<0.05 was considered to be statistically significant.
Results
Host characteristics and mtDNA content distribution of glioma patients
The demographic and clinical characteristics of the study population are summarised in Table 1. Among the total 336 patients, 212 were diagnosed with low-grade glioma (LGG, WHO grade 1 or 2) including 142 astrocytomas, 3 oligodendrogliomas, 53 oligoastrocytomas and 14 ependymomas, whereas 125 were diagnosed with high-grade glioma (HGG, WHO grade 3 or 4), including 81 anaplastic astrocytomas, 13 anaplastic oligodendrogliomas, 2 anaplastic oligoastrocytomas, 4 anaplastic ependymomas and 23 glioblastomas. During the follow-up period (median, 25 months; range, 3–99 months), 201 patients died of glioma and 217 developed recurrence or progression. The median OS and PFS time of the total population was 26.8 and 22.3 months, respectively. We further analysed the prognosis of patients with different grade gliomas. Generally, the prognosis pattern in our study is very similar as previously summarised (Ricard et al, 2012). In grade II gliomas, ependymoma and oligoastrocytoma showed similar prognosis that was better than that of astrocytoma. In grade III gliomas, anaplastic oligoglioma showed better prognosis than anaplastic astrocytoma (detailed data not shown).
We measured mtDNA content using a real-time PCR-based method in all samples. The mean inter-assay coefficient variation (CV) of real-time PCR reaction was 7.8% (range, 4.4–10.1%), whereas intra-assay CV was 4.7% (range, 2.9–7.4%), indicating good assay reproducibility. We then explored the distributions of mtDNA by characteristics of glioma patients. As shown in Table 1, there was no significant difference in mtDNA between patients with different age, gender, lesion location, WHO grade or treatment regimen.
Prognostic analysis of mtDNA content in glioma patients
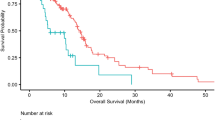
We first compared the OS and PFS between patients with different mtDNA content. As shown in Figure 1, patients with high mtDNA content had poorer OS and PFS than those with low mtDNA content (log-rank P=0.023 for OS and P=0.027 for PFS). Univariate and multivariate Cox regression analyses revealed that age, WHO grade, resection degree and adjuvant therapy regimen were all independent prognostic factors for both OS and PFS of patients (Table 2). Moreover, univariate Cox regression analysis showed that both risks of death and progression decreased in patients with high mtDNA content when compared with those with low mtDNA content. Multivariate Cox regression analysis further confirmed mtDNA content as an independent prognostic factor for both OS and PFS (HR 1.477, 95% CI 1.075–2.029, P=0.016 for OS; HR 1.528, 95% CI 1.130–2.066, P=0.006 for PFS; Table 2). To exclude the confounding factors that might affect the prognostic value of mtDNA content, we further analysed the effects of mtDNA content on the OS and PFS of glioma patients stratified by clinical characteristics. As shown in Table 3, high mtDNA content was significantly associated with poor prognosis in patients with older age, HGG or concomitant radiochemotherapy, but not in those with younger age, LGG or radiotherapy only. Kaplan–Meier curve analysis further confirmed the results of Cox regression model (Figure 2 and Supplementary Figure S1).
Kaplan–Meier curves stratified by age, WHO grade or adjuvant therapy. (A, C and E) Overall survival (OS) of younger patients, patients with HGG and receiving radiochemotherapy, respectively. (B, D and F) Progression-free survival (PFS) of younger patients, patients with HGG and receiving radiochemotherapy, respectively.
Immunophenotypes of lymphocytes and concentration of plasma cytokines in glioma patients with different mtDNA content
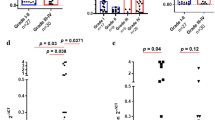
To explore the underlying mechanisms that account for the different prognosis of glioma patients with different mtDNA content, we first examined the subtypes of lymphocytes in PBMCs from glioma patients by flow cytometry analysis. Our results showed that patients with high mtDNA content had significantly lower frequency of NK cells (9.8% vs 18.9%, P=0.037, Figure 3G) than those with low mtDNA content. No significant difference on percentage of total, CD4+ or CD8+ T cells, B cells or Tregs was observed between the two patient subgroups (Figure 3).
Previous studies have demonstrated that mtDNA may affect the secretion of cytokines from immune cells, such as IL-2 and IL-4 (Kaminski et al, 2010). Interleukin-2 plays crucial roles in the survival, proliferation and activation of T and NK cells (Abbas et al, 2012). Interferon-γ and IL-4 are the key cytokines for cell differentiation in cell-mediated and humoral immune responses, respectively. Transforming growth factor-β1 is the main mediator of immunosuppressive effects conferred by Tregs. Tumour necrosis factor-α and IFN-γ may exert anticancer effects by directly killing target cells. We therefore examined the association between mtDNA content and plasma concentrations of these cytokines. As shown in Figure 3, high mtDNA content was significantly associated with higher concentration of IL-2 (50.3 vs 38.9 pg ml−1, P=0.009) and TNF-α (63.3 vs 41.6 pg ml−1, P=0.023). No significant concentration difference of IL-4, IFN-γ or TGF-β1 was noted between two patient subgroups.
Discussion
In this study, we explored the prognostic value of leukocyte mtDNA content in 336 glioma patients. We found that patients with high mtDNA content showed significantly poorer OS and RFS than those with low mtDNA. We also evaluated the association of mtDNA content with immune functions of glioma patients and found that high mtDNA content was associated with lower NK cell frequency and higher IL-2 and TNF-α concentrations. These data suggest that high mtDNA content may contribute to the progression of glioma possibly through the abnormal alteration of immune functions.
Alteration of mtDNA content plays an important role in the development of several malignancies (Yu, 2011). High mtDNA content may increase the intracellular bioenergenesis and anabolism, leading to a proliferative advantage and enhanced chemoresistance in cancer cells (Cavalli et al, 1997; Hsu et al, 2010). Progressive increase of mtDNA content has been observed during the malignant transformation from normal epithelia to endometrial, oesophageal or head and neck cancers (Kim et al, 2004; Lin et al, 2010; Cormio et al, 2012). Furthermore, high mtDNA content in cancer tissues has been shown to be associated with progression of oesophageal and laryngeal squamous cell carcinoma (Lin et al, 2010; Guo et al, 2013). Dickinson et al (2013) have demonstrated that higher mtDNA content facilitates the proliferation of glioblastoma cells. Moreover, Cheau-Feng Lin et al (2014) have shown that high mtDNA content predicts poor prognosis in patients with head and neck cancer. In line with these findings, our study demonstrated that higher leukocyte mtDNA content was associated with poor prognosis of glioma patients. All these results collectively suggest the critical role of mtDNA content increase in the initiation and progression of glioma.
Several studies have reported the opposite results, indicating that decreased mtDNA content is associated with progression and worse prognosis of patients with HCC (Yamada et al, 2006), CRC (Cui et al, 2013), breast cancer (Yu et al, 2007), prostate cancer (Koochekpour et al, 2013) and early-stage laryngeal cancer (Dang et al, 2014). These contradictory findings suggest that the homeostasis of mitochondrial content plays important roles in tumourigenesis. When this homeostasis is broken down by various oncogenic factors, either increased or decreased mtDNA content might damage mitochondrial functions and thus promote tumour development and progression. Previous studies have reported both increased and decreased mtDNA content in different cancer types (Yu, 2011). Therefore, it is easy to understand that mtDNA content may affect cancer progression and prognosis of patients in a disease-specific manner. However, the underlying mechanisms that account for mtDNA content variations and their functional roles in different malignancies remain largely unknown. Moreover, differences in histology of samples, ethnic population and experimental procedures might also contribute to these discrepancies. Further investigations are needed to elucidate the mechanisms underlying different effects of mtDNA content on cancer progression.
The underlying mechanisms that account for prognostic effects of leukocyte mtDNA content in glioma remain unknown. One explanation is that mtDNA content may alter the immune functions of glioma patients, especially the functions of NK cells that play important roles in the first-line defense against tumours (Vivier et al, 2008). Recent studies have demonstrated that NK cells may be a promising strategy for the treatment of glioma (Ogbomo et al, 2011). Our immunoassay indicated that glioma patients with high mtDNA content had low frequency of NK cells that may impair the immune defense against tumour cells and lead to poor prognosis.
Cytokines play pivotal roles in controlling the survival, expansion, activation and target cell killing of NK cells (Vivier et al, 2008). Immunological studies have shown that the combination of IL-2 and TNF-α promotes the apoptosis of NK cells (Jewett and Bonavida, 2000; Jewett et al, 2006). In this study, we found that glioma patients with high mtDNA content also had higher IL-2 and TNF-α levels that may contribute to NK cell reduction in these patients. Previous studies have demonstrated that mitochondria-derived ROS play an important role in the cytokine secretion of immune cells, such as IL-2 and TNF-α (Yang et al, 2013; Yu et al, 2014). Clearance of ROS by antioxidants has been shown to inhibit the secretion of IL-2 and TNF-α (Tatla et al, 1999; Li et al, 2014). In mammalian cells, mitochondria are the major source of ROS (Murphy, 2009), and intracellular ROS level is generally proportional to intracellular mtDNA content (Lee et al, 2000). Therefore, it is plausible that glioma patients with high mtDNA content have higher plasma concentration of IL-2 and TNF-α in our study. Collectively, these data suggest that high leukocyte mtDNA content might promote the tumour progression partially via ROS-induced NK cell inhibition.
In the stratified analyses, we found that high mtDNA content was significantly associated with poor prognosis in patients with younger age, HGG or adjuvant radiochemotherapy, but not in those with older age, LGG or adjuvant radiotherapy, indicating the modulating effects of host characteristics through unknown mechanisms. One possible explanation may be the immunodeficiency in patients with these characteristics: HGG patients have severer immunosuppression than LGG patients (Gomez and Kruse, 2006); the combination of radiotherapy and chemotherapy can cause long-lasting immunosuppression in glioma patients (Fadul et al, 2011); younger patients in our population received more adjuvant radiochemotherapy than older ones (81 vs 60 cases). All these factors may synergise the immunological effects of mtDNA content, leading to worse prognosis in patients. However, the detailed mechanisms need to be further investigated.
There are several limitations in our study. First, there were not enough patients receiving surgical treatment alone (n=30), and this may limit the statistical power of our stratified analysis. Thus, our findings from stratified analysis need to be further validated. Second, we only performed association analyses between mtDNA content and immunological parameters of glioma patients. The underlying mechanisms through which mtDNA content affects the immune functions need further investigation. Third, all glioma patients were given standard-of-care treatment as suggested by the guidelines of European Society for Medical Oncology and China Health Ministry. However, because of the evolution of these guidelines in the study course and the differences in the status of patients, the details of adjuvant radiotherapy and chemotherapy such as dose, course and period greatly different among patients. Therefore, we could only classify the adjuvant therapy into radiotherapy and radiochemotherapy without further subdivision. The heterogeneity in adjuvant therapy thus could not be effectively adjusted in our multivariate analysis.
In summary, our study for the first time demonstrates that high mtDNA content is associated with poor OS and PFS in glioma patients partially through the inhibition of NK cells. Once confirmed, leukocyte mtDNA content may serve as a useful biomarker to improve the prognosis prediction for glioma patients. Further studies are needed to explore the mechanisms underlying the effects of mtDNA content on the clinical outcomes and immune functions in glioma patients.
Change history
30 June 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Abbas AK, Lichtman AH, Pillai S (2012) Cellular and Molecular Immunology 7th edn, Saunders/Elsevier: Philadelphia.
Brandsma D, van den Bent MJ (2009) Pseudoprogression and pseudoresponse in the treatment of gliomas. Curr Opin Neurol 22 (6): 633–638.
Capps GJ, Samuels DC, Chinnery PF (2003) A model of the nuclear control of mitochondrial DNA replication. J Theor Biol 221 (4): 565–583.
Cavalli LR, Varella-Garcia M, Liang BC (1997) Diminished tumorigenic phenotype after depletion of mitochondrial DNA. Cell Growth Differ 8 (11): 1189–1198.
Chan DC (2006) Mitochondria: dynamic organelles in disease, aging, and development. Cell 125 (7): 1241–1252.
Cheau-Feng Lin F, Jeng YC, Huang TY, Chi CS, Chou MC, Chin-Shaw Tsai S (2014) Mitochondrial DNA copy number is associated with diagnosis and prognosis of head and neck cancer. Biomarkers 19 (4): 269–274.
Chen XJ, Butow RA (2005) The organization and inheritance of the mitochondrial genome. Nat Rev Genet 6 (11): 815–825.
Cormio A, Guerra F, Cormio G, Pesce V, Fracasso F, Loizzi V, Resta L, Putignano G, Cantatore P, Selvaggi LE, Gadaleta MN (2012) Mitochondrial DNA content and mass increase in progression from normal to hyperplastic to cancer endometrium. BMC Res Notes 5: 279.
Cui H, Huang P, Wang Z, Zhang Y, Zhang Z, Xu W, Wang X, Han Y, Guo X (2013) Association of decreased mitochondrial DNA content with the progression of colorectal cancer. BMC Cancer 13: 110.
Dang S, Qu Y, Wei J, Shao Y, Yang Q, Ji M, Shi B, Hou P (2014) Low copy number of mitochondrial DNA (mtDNA) predicts worse prognosis in early-stage laryngeal cancer patients. Diagn Pathol 9: 28.
Dickinson A, Yeung KY, Donoghue J, Baker MJ, Kelly RD, McKenzie M, Johns TG St, John JC (2013) The regulation of mitochondrial DNA copy number in glioblastoma cells. Cell Death Differ 20 (12): 1644–1653.
Fadul CE, Fisher JL, Gui J, Hampton TH, Cote AL, Ernstoff MS (2011) Immune modulation effects of concomitant temozolomide and radiation therapy on peripheral blood mononuclear cells in patients with glioblastoma multiforme. Neuro Oncol 13 (4): 393–400.
Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21 (21): 2683–2710.
Gladson CL, Prayson RA, Liu WM (2010) The pathobiology of glioma tumors. Annu Rev Pathol 5: 33–50.
Gomez GG, Kruse CA (2006) Mechanisms of malignant glioma immune resistance and sources of immunosuppression. Gene Ther Mol Biol 10(A): 133–146.
Guo W, Yang D, Xu H, Zhang Y, Huang J, Yang Z, Chen X, Huang Z (2013) Mutations in the D-loop region and increased copy number of mitochondrial DNA in human laryngeal squamous cell carcinoma. Molecular Biology Reports 40 (1): 13–20.
Hosgood HD 3rd, Liu CS, Rothman N, Weinstein SJ, Bonner MR, Shen M, Lim U, Virtamo J, Cheng WL, Albanes D, Lan Q (2010) Mitochondrial DNA copy number and lung cancer risk in a prospective cohort study. Carcinogenesis 31 (5): 847–849.
Hsu CW, Yin PH, Lee HC, Chi CW, Tseng LM (2010) Mitochondrial DNA content as a potential marker to predict response to anthracycline in breast cancer patients. Breast J 16 (3): 264–270.
Jewett A, Bonavida B (2000) MHC-Class I antigens regulate both the function and the survival of human peripheral blood NK cells: role of endogenously secreted TNF-alpha. Clin Immunol 96 (1): 19–28.
Jewett A, Cacalano NA, Head C, Teruel A (2006) Coengagement of CD16 and CD94 receptors mediates secretion of chemokines and induces apoptotic death of naive natural killer cells. Clin Cancer Res 12 (7 Pt 1): 1994–2003.
Kaminski MM, Sauer SW, Klemke CD, Suss D, Okun JG, Krammer PH, Gulow K (2010) Mitochondrial reactive oxygen species control T cell activation by regulating IL-2 and IL-4 expression: mechanism of ciprofloxacin-mediated immunosuppression. J Immunol 184 (9): 4827–4841.
Kim MM, Clinger JD, Masayesva BG, Ha PK, Zahurak ML, Westra WH, Califano JA (2004) Mitochondrial DNA quantity increases with histopathologic grade in premalignant and malignant head and neck lesions. Clin Cancer Res 10 (24): 8512–8515.
Koochekpour S, Marlowe T, Singh KK, Attwood K, Chandra D (2013) Reduced mitochondrial DNA content associates with poor prognosis of prostate cancer in African American men. PLoS One 8 (9): e74688.
Lan Q, Lim U, Liu CS, Weinstein SJ, Chanock S, Bonner MR, Virtamo J, Albanes D, Rothman N (2008) A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood 112 (10): 4247–4249.
Lee HC, Yin PH, Lu CY, Chi CW, Wei YH (2000) Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem J 348 (Pt 2): 425–432.
Li J, Li J, Yue Y, Hu Y, Cheng W, Liu R, Pan X, Zhang P (2014) Genistein suppresses tumor necrosis factor alpha-induced inflammation via modulating reactive oxygen species/Akt/nuclear factor kappaB and adenosine monophosphate-activated protein kinase signal pathways in human synoviocyte MH7A cells. Drug Des Devel Ther 8: 315–323.
Lin CS, Chang SC, Wang LS, Chou TY, Hsu WH, Wu YC, Wei YH (2010) The role of mitochondrial DNA alterations in esophageal squamous cell carcinomas. J Thorac Cardiovasc Surg 139 (1): 189–197 e4.
Mizumachi T, Muskhelishvili L, Naito A, Furusawa J, Fan CY, Siegel ER, Kadlubar FF, Kumar U, Higuchi M (2008) Increased distributional variance of mitochondrial DNA content associated with prostate cancer cells as compared with normal prostate cells. Prostate 68 (4): 408–417.
Moraes CT (2001) What regulates mitochondrial DNA copy number in animal cells? Trends Genet 17 (4): 199–205.
Murphy MP (2009) How mitochondria produce reactive oxygen species. Biochem J 417 (1): 1–13.
Ogbomo H, Cinatl J Jr, Mody CH, Forsyth PA (2011) Immunotherapy in gliomas: limitations and potential of natural killer (NK) cell therapy. Trends Mol Med 17 (8): 433–441.
Qu F, Liu X, Zhou F, Yang H, Bao G, He X, Xing J (2011) Association between mitochondrial DNA content in leukocytes and colorectal cancer risk: a case-control analysis. Cancer 117 (14): 3148–3155.
Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY (2012) Primary brain tumours in adults. Lancet 379 (9830): 1984–1996.
Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H (2010) Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion 10 (1): 62–68.
Tatla S, Woodhead V, Foreman JC, Chain BM (1999) The role of reactive oxygen species in triggering proliferation and IL-2 secretion in T cells. Free Radic Biol Med 26 (1-2): 14–24.
Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S (2008) Functions of natural killer cells. Nat Immunol 9 (5): 503–510.
Wang Y, Liu VW, Xue WC, Tsang PC, Cheung AN, Ngan HY (2005) The increase of mitochondrial DNA content in endometrial adenocarcinoma cells: a quantitative study using laser-captured microdissected tissues. Gynecol Oncol 98 (1): 104–110.
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359 (5): 492–507.
Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, de Andrade M, Swan GE, Wu X (2008) Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst 100 (15): 1104–1112.
Yamada S, Nomoto S, Fujii T, Kaneko T, Takeda S, Inoue S, Kanazumi N, Nakao A (2006) Correlation between copy number of mitochondrial DNA and clinico-pathologic parameters of hepatocellular carcinoma. Eur J Surg Oncol 32 (3): 303–307.
Yang Y, Bazhin AV, Werner J, Karakhanova S (2013) Reactive oxygen species in the immune system. Int Rev Immunol 32 (3): 249–270.
Yu M (2011) Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sci 89 (3-4): 65–71.
Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R (2007) Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life 59 (7): 450–457.
Yu Q, Nie SP, Wang JQ, Yin PF, Huang DF, Li WJ, Xie MY (2014) Toll-like receptor 4-mediated ROS signaling pathway involved in Ganoderma atrum polysaccharide-induced tumor necrosis factor-alpha secretion during macrophage activation. Food Chem Toxicol 66: 14–22.
Zhang J, Li D, Qu F, Chen Y, Li G, Jiang H, Huang X, Yang H, Xing J (2014) Association of leukocyte mitochondrial DNA content with glioma risk: evidence from a Chinese case-control study. BMC Cancer 14 (1): 680.
Zhao S, Yang Y, Liu J, Liu H, Ge N, Yang H, Zhang H, Xing J (2011) Association of mitochondrial DNA content in peripheral blood leukocyte with hepatitis B virus-related hepatocellular carcinoma in a Chinese Han population. Cancer Sci 102 (8): 1553–1558.
Acknowledgements
This work was supported by Program for New Century Excellent Talents in University, National Natural Science Foundation (81171966) and National Key Technologies R&D Program (2011ZX09307-001-04) of China.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Chen, Y., Zhang, J., Huang, X. et al. High leukocyte mitochondrial DNA content contributes to poor prognosis in glioma patients through its immunosuppressive effect. Br J Cancer 113, 99–106 (2015). https://doi.org/10.1038/bjc.2015.184
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2015.184
Keywords
This article is cited by
-
High leukocyte mitochondrial DNA copy number contributes to poor prognosis in breast cancer patients
BMC Cancer (2023)
-
Higher leukocyte count predicts 3-month poor outcome of ruptured cerebral aneurysms
Scientific Reports (2018)
-
Altered mitochondrial DNA copy number contributes to human cancer risk: evidence from an updated meta-analysis
Scientific Reports (2016)
-
Elevated Mitochondrial DNA Copy Number in Peripheral Blood and Tissue Predict the Opposite Outcome of Cancer: A Meta-Analysis
Scientific Reports (2016)