Abstract
Background:
Tongue squamous cell carcinoma (TSCC) is highly diverse, even in its early stages. This cancer is classified into three subtypes (superficial, exophytic, and endophytic) based on macroscopic appearance. Of these subtypes, the endophytic tumours have the worst prognosis because of their invasiveness and higher frequency of metastasis.
Methods:
To understand the molecular mechanism underlying the endophytic subtype and to identify biomarkers, we performed a comprehensive gene expression microarray analysis of clinical biopsy samples and also confirmed the clinical relevance of differential gene expression.
Results:
Expression of the parvin-beta (PARVB) gene and its encoded protein was significantly upregulated in endophytic-type TSCC. PARVB is known to play a critical role in actin reorganization and focal adhesions. Knockdown of PARVB expression in vitro caused apparent decreases in cell migration and wound healing, implying that PARVB has a crucial role in cell motility. Moreover, metastasis-free survival was significantly lower in patients with higher tumour expression of PARVB.
Conclusions:
These findings suggest that PARVB overexpression is a candidate biomarker for endophytic tumours and metastasis. This protein may be a clinically useful target for adjuvant TSCC therapy.
Similar content being viewed by others
Main
Head and neck squamous cell carcinoma occurs at a variety of sites, including the oral cavity, tongue, pharynx, and larynx (Döbrossy, 2005; Layland et al, 2005). Tongue squamous cell carcinoma (TSCC) is one of the most common and aggressive types of oral squamous cell carcinoma in terms of local invasion and cell spreading (Ye et al, 2008). Risk factors such as alcohol consumption and tobacco smoking increase its incidence by up to 75% (Mackenzie et al, 2000). Oral cavity cancers commonly cause lymph node metastasis, which is a negative prognostic indicator for head and neck squamous cell carcinoma treatment (Layland et al, 2005; Ahmed et al, 2007). In fact, the main causes of death related to TSCC are local relapse and metastasis. At the time of diagnosis, 40% of TSCC patients have neck metastasis; 20–40% of patients with stage I and II TSCC have occult nodal metastasis (Teichgraeber and Clairmont, 1984; Cunningham et al, 1986; Layland et al, 2005).
TSCC tumours are classified as superficial, exophytic, or endophytic based on their macroscopic appearance; among them, endophytic tumours have the worst prognosis (Kirita et al, 1994; Nakagawa et al, 2003). The physical appearance of TSCC can be described in terms of tumour depth, infiltration of peritumoural lymphocytes, and vascular invasion (Li et al, 2003; Bier-Laning et al, 2009; Jung et al, 2009). Although these parameters are useful for predicting occult cervical lymph node metastasis at first diagnosis, the risk of prediction error remains. Thus, there is an urgent need for additional diagnostic tools to support clinical decision-making to determine the best management and most appropriate treatment for each patient. More accurate diagnoses using sensitive biomarkers can improve the management of cancer treatments and increase survival rates (Estilo et al, 2009). Such a biomarker for TSCC could provide better predictions of tumour responses to treatment. The general tumour markers currently available include SCCA, CEA, and CA19-9. They are clinically used to support the diagnosis of oral and metastatic squamous cell carcinoma, particularly in the lymph nodes (Kurokawa et al, 1993). However, there is still no specific and clinically relevant biomarker for metastatic tongue cancer.
Molecular techniques examining gene mutations and expression patterns have the potential to reveal unique biological profiles of cancer cells. Gene expression analysis by high-throughput microarray technology is a promising method to predict the outcome of human transcriptomic abnormalities in malignancies. Because endophytic TSCC cells are more likely to migrate, identifying genes related to the endophytic subtype could potentially define the molecular biomarkers for TSCC migration and metastasis. Therefore, the purpose of this study was to identify predictive or prognostic biomarkers related to TSCC metastasis by investigating correlations between aberrant gene expression and endophytic TSCC comparing with non-endophytic TSCC (superficial and exophytic), which shows better prognosis with lower frequency of metastasis.
Materials and methods
Clinical tissue samples
Clinical biopsy tissues were obtained from patients who were candidates for TSCC brachytherapy between 2006 and 2010 and had not received radiation therapy before treatment at the University Hospital of Medicine, Tokyo Medical and Dental University (TMDU; Tokyo, Japan). This study was approved by the ethical review board at TMDU, and informed consent was obtained from all patients. Samples were analysed using microarray (27 cases) and immunohistochemistry (IHC) (29 cases). Of these 56 cases, 10 were analysed by both methods (Table 1). Tissue sections were prepared on glass slides, stained with haematoxylin and eosin, and pathologically examined to distinguish clusters of tumour cells. Tumour samples were classified into three subtypes based on the macroscopic appearance of cellular growth patterns (Nakagawa et al, 2003) as follows: 10 superficial, 9 exophytic, and 8 endophytic-type samples analysed by microarray; and 9 superficial, 11 exophytic, and 9 endophytic-type samples analysed by IHC.
RNA isolation and microarray analysis
Cancer cells were isolated from sectioned tissues by laser microdissection using the LMD6000 (Leica microsystems, Wetzlar, Germany). Total RNA was extracted using the RNeasy Micro Kit (QIAGEN, Hilden, Germany) and was amplified, biotinylated, and fragmented using the MessageAmp II-Biotin Enhanced Kit (Ambion, Austin, TX, USA) according to the manufacturers’ instructions. The fragmented aRNA was applied to GeneChip Human Genome U133 Plus 2.0 (Affymetrix, Santa Clara, CA, USA) and incubated at 45 °C for 16 h, followed by washing and staining using Fluidic Station 450 (Affymetrix). The hybridized probe arrays were scanned with the GeneChip Scanner 3000 7G (Affymetrix). Microarray data used in this study are available at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE52915.
Quantitative expression analysis by real-time RT–PCR
Twelve total RNAs from a microarray sample set were converted to first-strand cDNAs using the High-Capacity cDNA Reveres Transcription kit (Applied Biosystems, Foster City, CA, USA). Quantitative real-time RT–PCR (qRT–PCR) was performed using the Applied Biosystems 7500 Fast real-time RT–PCR system with IQ SYBR-Green Supermix (Bio-Rad, Hercules, CA, USA). The specific primer sets for PARVB were as follows: Forward, 5′-CATCCGCCTTCCTGAGCAT-3′; Reverse, 5′-AGCAGGCCTTCCCGTTTC-3′. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. Polymerase chain reactions were run in duplicate, and relative gene expression levels were calculated as fold-change values by DataAssist v3.0 (Applied Biosystems). Significant differences in PARVB expression between endophytic and non-endophytic types were statistically analysed.
Immunohistochemical assay of PARVB in endophytic and non-endophytic tumours
To investigate PARVB protein expression levels, we immunohistochemically stained 29 samples (9 endophytic and 20 non-endophytic). Endogenous peroxidase activity was blocked by soaking the sectioned tissues on glass slides in methanol-hydrogen peroxide solution (100 : 1) for 30 min. Tissue sections were then washed with PBS and incubated for 1 h with blocking solution from the Histofine SAB-PO (M) kit (Nichirei, Tokyo, Japan). Sections were then covered with mouse monoclonal anti-PARVB antibody solution (1 : 30 dilution, Abnova, Taipei, Taiwan) and incubated overnight at 4 °C. Slides were then incubated for 20 min at room temperature with biotinylated goat anti-mouse secondary antibody, followed by a 15-min incubation with biotin-streptavidin. The peroxidase reaction was performed by incubation in DAB solution for 5 min at room temperature. Finally, all sections were stained with haematoxylin for 30 s. Stained cancer cells were evaluated by the pathologist. To quantitate the IHC results, positively and negatively stained tumour cells were counted in five different areas (5 × 103 μm2 each) that were randomly selected in each sample; the mean positive ratio was calculated as follows: mean positive ratio=positive cell count/(positive cell count+negative cell count). The expression of PARVB was compared between endophytic and non-endophytic TSCCs using the Wilcoxon rank sum test.
Difference in metastasis-free survival with respect to PARVB expression
Immunohistochemistry data were analysed for a potential correlation between metastasis-free survival (MFS) and PARVB overexpression using the Kaplan–Meier method. Patients with an IHC-positive ratio <50% were classified as ‘low expression,’ whereas those with an IHC-positive ratio ⩾50% were classified as ‘high expression’. The follow-up period for MFS was calculated as the interval between the first day of brachytherapy and the date when lymph node metastasis was observed or the last follow-up date.
SAS cell culture and PARVB expression
SAS cells (TKG 0470, an invasive type of TSCC) were purchased from the Cell Resource Center for Biomedical Research of Tohoku University and cultured in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich, St Louis, MO, USA) with 10% foetal bovine serum and 1% penicillin at 37 °C in a humidified, 5% CO2 environment. To measure the gene expression of PARVB, total RNA was extracted using ISOGEN (NIPPON GENE, Tokyo, Japan) and reverse-transcribed into first strand cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). Polymerase chain reaction amplification was performed using the Taq PCR master mix kit (QIAGEN) with PARVB primers. GAPDH primers were also used to detect endogenous GAPDH expression as a control.
PARVB knockdown and cell growth assay
The knockdown assay was conducted with small interference RNA (siRNA) targeting human PARVB (siPARVB; sense sequence: 5′-AAGCUGAAUUUGGAGGUGACG-3′), as previously described (Yamaji et al, 2004). Approximately 1 × 105 SAS cells were seeded into each well of a 24-well plate and cultured at 37 °C for 24 h. Cells not transfected with siRNA (untransfected cells) or those transfected with non-silencing and scramble siRNA (AllStars Negative Control siRNA, QIAGEN) or siPARVB were used as controls. All transfections were conducted with the RNAi Human/Mouse starter kit (QIAGEN). The optimal incubation time for siPARVB was determined by incubating the cultures at 37 °C for 24 h, 48 h, 72 h, or 96 h, followed by total RNA extraction with ISOGEN and reverse-transcription to cDNAs. The efficacy of RNA interference by siPARVB was quantitatively confirmed by time-course measurements of PARVB expression using real-time qRT–PCR with Fast SYBR Green Master Mix (Applied Biosystems). The possible side-effects of siPARVB on cell proliferation were evaluated using the Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) using the same incubation conditions as the time optimisation assay. The absorbance of culture supernatants was measured at 0 h, 24 h, 48 h, 72 h, and 96 h with CCK-8 reagent at 450 nm using the iMark Microplate Absorbance Reader (Bio-Rad). All assays were performed in triplicate. The growth rates of PARVB knockdown cells were compared with those of control untransfected SAS cells and cells treated with negative control siRNA.
Migration and wound-healing assays
To analyse the migration capacity of SAS cells under PARVB suppression, migration and wound healing were assayed. SAS cells were cultured to 100% confluence in 24-well plates. The migration assay was performed using transparent polyethylene membrane cell culture inserts with an 8.0 μm pore size (BD Biosciences, Franklin Lakes, NJ, USA) following the manufacturer’s instructions. Cells (5 × 104) were seeded in 500 μl serum-free medium per well in the upper chamber. Negative control siRNA and siPARVB were separately transfected into cells and control cells were also prepared without transfection. Medium (750 μl) containing 10% foetal bovine serum was added to the lower chamber. After 72 h incubation at 37 °C, cells were fixed and stained with Diff-Quik (BD Biosciences). Stained cells that migrated through the membrane were counted in five random areas (5 × 104 μm2 each) under the microscope.
To observe the wound-healing response, two parallel scratches were made on the confluent culture cells using a sterile 1 ml micropipette tip, followed by washing and the addition of 500 μl fresh foetal bovine serum-free medium. The wound areas were monitored under the microscope at 0 and 72 h after siPARVB transfection, and the ratios of the mean distances were calculated from measurements at five randomly chosen areas.
Statistical analysis
For microarray analysis, raw data were processed using the robust multi-array average method with R statistical software version 2.12.1 (http://www.r-project.org/) and the ‘affy’ Bioconductor package (http://www.bioconductor.org/). Probe sets corresponding to known genes were selected based on the NetAffx annotation file version 32 (available at http://www.affymetrix.com/analysis/index.affx). Probe sets marked as ‘present’ by Expression Console Software version 1.1 (Affymetrix) in more than 50% of the patients were then selected. Differentially expressed genes were identified using the Wilcoxon rank sum test with the Wilcox exact function provided by the ‘coin’ package available through the Comprehensive R Archive Network (http://cran.r-project.org/). Probe sets that satisfied P<0.01 and a change >two-fold between endophytic and non-endophytic TSCCs were considered significant. The genes differently expressed among the superficial, exophytic, and endophytic types were identified by Kruskal–Wallis test with a significance level of P<0.01. The Wilcoxon rank sum test was applied to qRT–PCR results to identify differences in relative gene expression between endophytic and non-endophytic cancer tissues. The Wilcoxon rank sum test was also used to determine differences between positive and negative ratios of IHC-stained cancer cells in each sample. The Kaplan–Meier method was applied to estimate the MFS curve; the difference between groups was calculated using the log-rank test. P<0.05 was considered statistically significant.
Results
Comparison of gene expression between TSCC subtypes
Microarray data were used to identify genes differentially expressed between endophytic and non-endophytic TSCC. Out of 57 613 probe sets on the Human Genome U133 Plus 2.0 array, 41 479 were known human genes. Of these, 23 264 probe sets were present in more than 50% of the patients. Using the Wilcoxon rank sum test and two-fold change threshold, we identified 26 differentially expressed genes corresponding to 32 probe sets (Supplementary Table 1). The robustness of the sample classification was also confirmed by hierarchical clustering of the 26 genes exhibiting significantly differential expression (Supplementary Figure 1). Most of the 26 genes were highly expressed among endophytic samples. Between the superficial, exophytic, and endophytic types, 149 probe sets were differentially expressed (P<0.01 by Kruskal–Wallis test). Hierarchical cluster analysis indicated that the endophytic type was distinguished from superficial and exophytic types except in three patients with the exophytic type (Supplementary Figure 2). Of the differentially expressed genes, nine were identified according to Gene Ontology terms as mediating functions involved in invasion or metastasis, such as cell adhesion, cell migration, cell–cell adhesion, cytoskeletal organization, positive regulation of cell-matrix adhesion, regulation of cell migration, and regulation of cell motility (Supplementary Table 2). We selected PARVB for further study based on a literature survey indicating a major role for PARVB in cell adhesion and cancer invasion. An association between PARVB and TSCC has not been previously reported.
Quantitative expression analysis by real-time RT–PCR
Expression of the PARVB gene was confirmed quantitatively by real-time RT–PCR. Quantitative RT–PCR results showed that PARVB expression was significantly higher in endophytic TSCC (P<0.005) (Figure 1), indicating a similar profile as the microarray experiments.
Immunohistochemical assay quantitatively confirms differential expression of PARVB
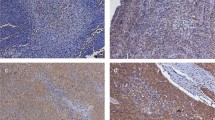
Immunohistochemistry was performed to determine whether the protein encoded by the PARVB gene was highly expressed in endophytic-type TSCC (n=29). Positive staining of PARVB was mostly observed in the cytoplasm of endophytic-type tumour cells (Figure 2A). Statistical analysis revealed PARVB protein expression was significantly higher in endophytic- than non-endophytic-type TSCC (P<0.0001) (Figure 2B). This result confirmed that the overexpression of PARVB occurred at both the mRNA and protein levels.
Immunohistochemistry analysis of PARVB protein expression in 29 samples. (A) Left panel, non-endophytic tissue with weak staining in a few cells; right panel, endophytic tissue with strong staining in most of the cells. Scale bar: 100 μm. (B) Result of IHC analysis showing expression of PARVB was significantly higher in endophytic tumours than in non-endophytic tumours (*P<0.0001).
Influence of PARVB expression on lymph node metastasis
Metastasis-free survival analysis was performed to determine the correlation between PARVB expression and lymph node metastasis. We observed that patients with higher expression of PARVB had significantly lower MFS rates than did those with lower expression (log-rank test, P<0.05) (Figure 3). PARVB thus correlated positively with metastasis.
Effect of PARVB knockdown on cell proliferation, cell migration, and wound-healing rates in SAS cells
The inhibitory effect of siPARVB and optimal incubation time were demonstrated by the quantitative analysis showing that PARVB expression was most effectively inhibited by siPARVB at 72 h (Supplementary Figure 3) although negative siRNAs had a slight suppressive effect. More than 50% of the gene expression was suppressed for at least 48 h. Although PARVB expression was significantly suppressed in PARVB knockdown cells, we observed no remarkable difference in cell proliferation rates between SAS control cells, negative siRNA, and PARVB knockdown cells (Figure 4A), indicating that suppression of PARVB mRNA expression did not affect cell proliferation. The migration assay showed a marked decrease in the migration rate of PARVB knockdown cells compared with that of untransfected control and negative siRNA cells (Figure 4B and C). This result implies that the expression of PARVB is a key factor conferring migratory ability to TSCC. To determine whether PARVB expression has an effect on migration rates during wound repair, wound healing was compared between SAS cells with and without siPARVB transfection. Wounds repaired faster in cells expressing PARVB than in PARVB knockdown cells (Figure 5A). Moreover, PARVB knockdown cells located at the wound edges changed from needle-shaped, similar to fibroblasts, to round-shaped. The appearance of SAS cells expressing PARVB did not change (Figure 5B). Therefore, PARVB expression was shown to play a role in conferring migration capability during the wound-healing process.
Migration of SAS cells was evaluated by migration assays of cells with or without PARVB expression. (A) Growth assays of untransfected control, negative siRNA, and siPARVB cells using CCK-8 siPARVB did not affect SAS cell proliferation compared with either control or negative siRNA cells. (B) Stained cells that migrated to the bottom side of the PET membranes; left panel, control cells exhibited a high migration rate; centre panel, control cells with negative siRNA; right panel, PARVB knockdown cells exhibit a lower migration rate. Arrows show migrated cells. Scale bar: 100 μm. (C) The Wilcoxon rank sum test showed that the migration rate was significantly lower in cells with siPARVB than in those without siRNA (control) or with negative siRNA (Wilcoxon rank sum test, *P=0.67 and **P<0.0001, respectively).
Wound-healing assay to investigate the effect of PARVB knockdown on cell migration. (A) SAS cells without siRNA (control) and siPARVB SAS cells incubated for 0 and 72 h scratching the surface of confluent cells. PARVB knockdown decreased the migration rate and altered the cell morphology from spindle to round. (B) Magnified images of control and siPARVB cells at 72 h display the difference in cell morphologies. Left panel, SAS cells with expression of PARVB had sharp extensions of the cells at the edge of the wound; right panel, PARVB knockdown cells display a rounded morphology, possibly indicating cell transformation due to loss of PARVB expression.
Discussion
Expression of the parvin-beta (PARVB) gene and its encoded protein was significantly upregulated in endophytic-type TSCC. PARVB is known to play a critical role in actin reorganization and focal adhesions. Knockdown of PARVB expression in vitro caused apparent decreases in cell migration and wound healing, implying that PARVB has a crucial role in cell motility. Metastasis-free survival was significantly lower in patients with higher tumour expression of PARVB. These findings suggest that PARVB overexpression may be a biomarker for endophytic tumours and metastasis.
Numerous studies have investigated molecular biomarkers and their relationships to the pathogenesis of oral cancer. Abnormal expression of MAGED4B and KLK13 was reported to correlate with metastasis in oral cancer (Chong et al, 2012; Ishige et al, 2013). In addition, copy number aberrations, including 11q13 amplification and NKX3-1 loss, have been reported as potential biomarkers associated with metastasis (Sugahara et al, 2011; Miyaguchi et al, 2012). However, macroscopic subtypes have rarely been considered for studying tumour molecular signatures. Understanding endophytic TSCC is particularly important, as this is the most invasive TSCC subtype, with the highest risk of lymph node metastasis and consequently lower survival (Kirita et al, 1994; Sunakawa et al, 2002; Nakagawa et al, 2003).
In the present study, we performed comprehensive gene expression analysis to find predictive or prognostic biomarkers related to the endophytic subtype. Microarray analysis identified 26 genes with significant differences in expression between endophytic and non-endophytic subtypes. Quantitative RT–PCR and IHC assays confirmed significant overexpression of the PARVB gene and its encoded protein in endophytic TSCC. Although the remaining genes may have important associations with the endophytic phenotype and should be further investigated, we focused this study on the significance of PARVB overexpression.
PARVB encodes an actin-binding protein, also known as affixin, that is highly expressed in the skeletal muscles, spleen, heart, and kidneys (Korenbaum et al, 2001; Yamaji et al, 2004; Matsuda et al, 2008). Although the molecular mechanism of PARVB in healthy cells has been described, the role of PARVB in cancer progression remains unclear. PARVB is known to play important roles in cell spreading and motility and is also essential for extracellular matrix adhesion (Sepulveda and Wu, 2006; Wu et al, 2010). Disruptions in cell–cell and cell–extracellular matrix adhesions are necessary for the migration and invasion processes. Therefore, cell-surface adhesion molecules play key roles in tumour cell migration, the capacity of epithelial cells to invade adjacent tissue, to drive metastasis (Hanahan and Weinberg, 2000).
PARVB contains two calponin homology domains: an N-terminal domain (CH1) and a C-terminal domain (CH2) (Gagné et al, 2010). The CH1 domain is phosphorylated by α-PIX, leading to the activation of Cdc42/Rac1, followed by actin reorganization, lamellipodium formation, and cell spreading (Yamaji et al, 2001, 2004; Mishima et al, 2004; Matsuda et al, 2008). The CH2 domain is phosphorylated by integrin-linked kinase; located in focal adhesions, this scaffold protein activates α-actinin and causes the actin cytoskeletal reorganization essential for lamellipodium formation and cell motility (Yamaji et al, 2004; Matsuda et al, 2008; Gagné et al, 2010). PARVB is known to localize nearby at the leading edges of cells where the lamellipodium forms (Liu et al, 2000; Condeelis et al, 2001). Additional studies are required to identify the CH domain(s) and downstream elements interacting with PARVB signalling in endophytic TSCC to promote tumour metastasis.
Lamellipodia are mature actin-based structures involved in cell invasion and metastasis. They function by generating force on the extracellular matrix to promote cell migration through the following series of steps: extension (assembly of actin filaments), adhesion (attachment of actin filaments and cytoskeletal integrins), contraction (bilateral cell contraction toward the nucleus), and release (dissociation of receptor agonist at the back of cell) (Sheetz et al, 1998). If cell extensions fail to attach to the substratum, they fold back and movement stops. Lamellipodia also induce distinct morphological changes. In the present study, the sharp extensions observed in control cells may have been formed by lamellipodia because they were absent in PARVB knockdown cells. The disruption of lamellipodia formation by PARVB suppression supports a key role for PARVB in actin polymerization (Yamaji et al, 2004; Matsuda et al, 2008). In addition, the main function of lamellipodia is the quick repair of tissue damage (Tse et al, 2012). We confirmed that the rates of migration and wound healing increased with PARVB expression in human TSCC cells. These may be partially explained by the promotion of lamellipodia formation regulated by PARVB-induced actin polymerization (Yamaji et al, 2004). Moreover, PARVB knockdown suppressed cell migration and wound healing with a loss of lamellipodia in a tongue cancer cell line. Taken together, these observations suggest that PARVB overexpression plays an important role in the initiation of lamellipodia formation, possibly through signalling, inducing the reorganization of the actin cytoskeleton and ultimately leading to enhanced cell migration and wound healing. Studies with larger cohorts and further investigations of the specific molecular functions of PARVB are required to gain a full understanding of its role in lamellipodia formation.
Previous studies have reported that PARVB is downregulated in urothelial cell carcinoma and breast cancer (Mongroo et al, 2004; Wu et al, 2010). Cell growth and migration in upper urinary tract urothelial cell carcinoma were significantly increased by the downregulation of PARVB (Wu et al, 2010). Mongroo et al (2004) showed that PARVB expression was downregulated in advanced breast tumours and breast cancer cell lines and also suggested that the loss of PARVB expression promotes integrin-linked kinase activity in tumours. Although these authors discussed the interaction between PARVB and integrin-linked kinase, a correlation between PARVB expression and cell motility was not examined. To the best of our knowledge, no existing empirical research has shown a relationship between the overexpression of PARVB and TSCC or any other type of oral cancer. This is the first study reporting that PARVB is overexpressed in endophytic TSCC. Elucidation of the specific mechanism whereby PARVB affects cell invasion requires further study.
In conclusion, we comprehensively examined the gene expression profiles of tongue cancers based on macroscopic growth subtypes. Overexpression of PARVB was significantly associated with the endophytic tumour morphology. A significant correlation was also observed between PARVB expression and metastasis. In PARVB knockdown cells, migration and wound healing declined markedly, and the fibroblast-like structure was lost at the edge of wounds. Overall, these results suggest that the overexpression of PARVB increases cell migration and is a promising biomarker for endophytic growth and lymph node metastasis in TSCC.
Accession codes
Change history
20 January 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Ahmed MU, Khawar A, Ahmed J, Ajmal M, Bangash WK, Akhter MR (2007) Occult metastasis in carcinoma of oral cavity. J Coll Physicians Surg Pak 17: 313–315.
Bier-Laning CM, Durazo-Arvizu R, Muzaffar K, Petruzzelli GJ (2009) Primary tumor thickness as a risk factor for contralateral cervical metastases in T1/T2 oral tongue squamous cell carcinoma. Laryngoscope 119: 883–888.
Chong CE, Lim KP, Gan CP, Marsh Ca, Zain RB, Abraham MT, Prime SS, Teo S-H, Silvio Gutkind J, Patel V, Cheong SC (2012) Over-expression of MAGED4B increases cell migration and growth in oral squamous cell carcinoma and is associated with poor disease outcome. Cancer Lett 321: 18–26.
Condeelis JS, Wyckoff JB, Bailly M, Pestell R, Lawrence D, Backer J, Segall JE (2001) Lamellipodia in invasion. Semin Cancer Biol 11: 119–128.
Cunningham MJ, Johnson JT, Myers EN, Schramm VL, Thearle PB (1986) Cervical lymph node metastasis after local excision of early squamous cell carcinoma of the oral cavity. Am J Surg 152: 361–366.
Döbrossy L (2005) Epidemiology of head and neck cancer: magnitude of the problem. Cancer Metastasis Rev 24: 9–17.
Estilo CL, O-charoenrat P, Talbot S, Socci ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y, Boyle JO, Kraus DH, Patel S, Shaha AR, Wong RJ, Huryn JM, Shah JP, Singh B (2009) Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer 9: 11.
Gagné D, Groulx J-F, Benoit YD, Basora N, Herring E, Vachon PH, Beaulieu J-F (2010) Integrin-linked kinase regulates migration and proliferation of human intestinal cells under a fibronectin-dependent mechanism. J Cell Physiol 222: 387–400.
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70.
Ishige S, Kasamatsu A, Ogoshi K, Saito Y, Usukura K, Yokoe H, Kouzu Y, Koike H, Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K (2013) Decreased expression of kallikrein-related peptidase 13: Possible contribution to metastasis of human oral cancer. Mol Carcinog 53: 557–565.
Jung J, Cho NH, Kim J, Choi EC, Lee SY, Byeon HK, Park YM, Yang WS, Kim SH (2009) Significant invasion depth of early oral tongue cancer originated from the lateral border to predict regional metastases and prognosis. Int J Oral Maxillofac Surg 38: 653–660.
Kirita T, Okabe S, Izumo T, Sugimura M (1994) Risk factors for the postoperative local recurrence of tongue carcinoma. J Oral Maxillofac Surg 52: 149–154.
Korenbaum E, Olski TM, Noegel AA (2001) Genomic organization and expression profile of the parvin family of focal adhesion proteins in mice and humans. Gene 279: 69–79.
Kurokawa H, Tsuru S, Okada M, Nakamura T, Kajiyama M (1993) Evaluation of tumor markers in patients with squamous cell carcinoma in the oral cavity. Int J Oral Maxillofac Surg 22: 35–38.
Layland MK, Sessions DG, Lenox J (2005) The influence of lymph node metastasis in the treatment of squamous cell carcinoma of the oral cavity, oropharynx, larynx, and hypopharynx: N0 versus N+. Laryngoscope 115: 629–639.
Li QL, Chen FJ, Zeng ZY, Yang AK, Wu QL, Zhang HZ, Wu GH, Xu GP, Guo ZM, Zhang Q (2003) [Clinical and pathological related factors of occult cervical lymph node metastasis in squamous cell carcinoma of tongue]. Ai Zheng 22: 66–70.
Liu S, Calderwood DA, Ginsberg MH (2000) Integrin cytoplasmic domain-binding proteins. J Cell Sci 113: 3563–3571.
Mackenzie J, Ah-See K, Thakker N, Sloan P, Maran AG, Birch J, Macfarlane GJ (2000) Increasing incidence of oral cancer amongst young persons: what is the aetiology? Oral Oncol 36: 387–389.
Matsuda C, Kameyama K, Suzuki A, Mishima W, Yamaji S, Okamoto H, Nishino I, Hayashi YK (2008) Affixin activates Rac1 via betaPIX in C2C12 myoblast. FEBS Lett 582: 1189–1196.
Mishima W, Suzuki A, Yamaji S, Yoshimi R, Ueda A, Kaneko T, Tanaka J, Miwa Y, Ohno S, Ishigatsubo Y (2004) The first CH domain of affixin activates Cdc42 and Rac1 through alphaPIX, a Cdc42/Rac1-specific guanine nucleotide exchanging factor. Genes Cells 9: 193–204.
Miyaguchi K, Uzawa N, Mogushi K, Takahashi K-I, Michikawa C, Nakata Y, Sumino J, Okada N, Mizushima H, Fukuoka Y, Tanaka H (2012) Loss of NKX3-1 as a potential marker for an increased risk of occult lymph node metastasis and poor prognosis in oral squamous cell carcinoma. Int J Oncol 40: 1907–1914.
Mongroo PS, Johnstone CN, Naruszewicz I, Leung-Hagesteijn C, Sung RK, Carnio L, Rustgi AK, Hannigan GE (2004) Beta-parvin inhibits integrin-linked kinase signaling and is downregulated in breast cancer. Oncogene 23: 8959–8970.
Nakagawa T, Shibuya H, Yoshimura R, Miura M, Okada N, Kishimoto S, Amagasa M, Omura K (2003) Neck node metastasis after successful brachytherapy for early stage tongue carcinoma. Radiother Oncol 68: 129–135.
Sepulveda JL, Wu C (2006) The parvins. Cell Mol Life Sci 63: 25–35.
Sheetz MP, Felsenfeld DP, Galbraith CG (1998) Cell migration: regulation of force on extracellular-matrix-integrin complexes. Trends Cell Biol 8: 51–54.
Sugahara K, Michikawa Y, Ishikawa K, Shoji Y, Iwakawa M, Shibahara T, Imai T (2011) Combination effects of distinct cores in 11q13 amplification region on cervical lymph node metastasis of oral squamous cell carcinoma. Int J Oncol 39: 761–769.
Sunakawa K, Nonoyama M, Fujii R, Iwai N, Sakata H, Shirai M, Sato T, Kajino M, Toyonaga Y, Sano T, Naito A, Minagawa K, Niida Y, Oda T, Yokozawa M, Asanuma H, Shimura K, Fujimura M, Kitajima H, Fujinami K, Numazaki K, Fujikawa T, Kobayashi Y, Sato Y, Nishimura T, Iwata S, Tsuchihashi N, Oishi T, Matsumoto S, Motohiro T, Osawa M, Sunahara M, Shirakawa S, Nishida H, Takahashi N, Nakano R, Sai N, Iyoda K, Yoshimitsu K, Ogawa K, Okazaki T, Tsukimoto I, Motoyama O, Takada Y, Kawasaki M, Sunaoshi W, Nakamura S, Ueda Y, Kamata M, Kato T, Chiba M, Ouchi K, Sato S, Horiuchi T, Suzuki K, Shimoyama T, Masaki H, Aikyo M, Kawada M, Banba M, Furukawa S, Okada T, Yamaguchi S, Hirota O, Koizumi S, Wada H, Ohta K, Uehara T, Yukitake K, Mori T, Takakuwa S, Matsuyama K (2002) [Pharmacokinetic and clinical studies on teicoplanin for sepsis by methicillin-cephem resistant Staphylococcus aureus in the pediatric and neonate field]. Jpn J Antibiot 55: 656–677.
Teichgraeber JF, Clairmont AA (1984) The incidence of occult metastases for cancer of the oral tongue and floor of the mouth: treatment rationale. Head Neck Surg 7: 15–21.
Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK, Munn LL (2012) Mechanical compression drives cancer cells toward invasive phenotype. Proc Natl Acad Sci U S A 109: 911–916.
Wu CF, Ng KF, Chen CS, Chang PL, Chuang CK, Weng WH, Liao SK, Pang ST (2010) Expression of parvin-beta is a prognostic factor for patients with urothelial cell carcinoma of the upper urinary tract. Br J Cancer 103: 852–860.
Yamaji S, Suzuki A, Kanamori H, Mishima W, Yoshimi R, Takasaki H, Takabayashi M, Fujimaki K, Fujisawa S, Ohno S, Ishigatsubo Y (2004) Affixin interacts with alpha-actinin and mediates integrin signaling for reorganization of F-actin induced by initial cell-substrate interaction. J Cell Biol 165: 539–551.
Yamaji S, Suzuki A, Sugiyama Y, Koide Y, Yoshida M, Kanamori H, Mohri H, Ohno S, Ishigatsubo Y (2001) A novel integrin-linked kinase-binding protein, affixin, is involved in the early stage of cell-substrate interaction. J Cell Biol 153: 1251–1264.
Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X (2008) Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics 9: 69.
Acknowledgements
We thank Dr Mayuko Ishikawa for kind help in microarray experiments. This work was supported in part by grant-in-aids from Science and Technology Promotion Adjustment Expenses of Japan (No. 08005234) and Integrated Database Project, Ministry of Education, Culture, Sports, Science and Technology in Japan.
Author information
Authors and Affiliations
Corresponding authors
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Eslami, A., Miyaguchi, K., Mogushi, K. et al. PARVB overexpression increases cell migration capability and defines high risk for endophytic growth and metastasis in tongue squamous cell carcinoma. Br J Cancer 112, 338–344 (2015). https://doi.org/10.1038/bjc.2014.590
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.590
Keywords
This article is cited by
-
NET-related gene signature for predicting AML prognosis
Scientific Reports (2024)
-
A novel NET-related gene signature for predicting DLBCL prognosis
Journal of Translational Medicine (2023)
-
Roles of IGFBP-3 in cell migration and growth in an endophytic tongue squamous cell carcinoma cell line
Scientific Reports (2022)
-
Integrin-linked kinase (ILK) regulates KRAS, IPP complex and Ras suppressor-1 (RSU1) promoting lung adenocarcinoma progression and poor survival
Journal of Molecular Histology (2020)
-
ROCK inhibitor Y-27632 inhibits the growth, migration, and invasion of Tca8113 and CAL-27 cells in tongue squamous cell carcinoma
Tumor Biology (2016)