Abstract
Background:
Invasion and metastasis are the distinct biologic characteristics of cancer, resulting in an exceptionally low 5-year survival rate in pancreatic ductal adenocarcinoma (PDAC). Understanding in detail the mechanisms underlying PDAC metastasis is critical for prevention and effective interventions. Long non-coding RNAs (lncRNAs) have been documented as having a critical role in cancer development and progression.
Methods:
We examined the expression levels of lncRNA ENST00000480739 and osteosarcoma amplified-9 (OS-9) mRNA in a cohort of 35 PDAC patients. Cell proliferation, invasion and migration were examined with and without ENST00000480739 overexpression in PDAC cells.
Results:
We determined that the ENST00000480739 expression level was remarkably decreased in tumorous tissues compared with their corresponding non-tumorous tissues. The expression of ENST00000480739 was negatively associated with tumour node metastasis stage and lymph node metastasis. In addition, ENST0000048073 was an independent prognostic factor of survival time in PDAC patients following surgery. Besides, enforced expression of ENST00000480739 suppressed PDAC cells’ invasion in vitro. Overexpression of ENST00000480739 significantly increased both mRNA and protein levels of OS-9, and the luciferase assays confirmed that ENST00000480739 positively regulates OS-9 by activating the transcription level of the OS-9 promoter. We further found that ENST00000480739 may target hypoxia-inducible factor-1α (HIF-1α) expression by upregulating OS-9.
Conclusions:
These findings suggest that the frequently downregulated ENST00000480739 in PDAC contributes to tumour metastasis and progression by regulating HIF-1α. Long non-coding RNA ENST00000480739 may provide not only a therapeutic potential to suppress metastasis but it may also be a novel biomarker for risk prognostication and personal therapy screening of PDAC patients.
Similar content being viewed by others
Main
Pancreatic cancer is the fourth leading cause of death by cancer (Siegel et al, 2012). Pancreatic ductal adenocarcinoma (PDAC) is the most common form of pancreatic cancer and its incidence is rising every year (Hezel et al, 2006). Despite recent aggressive treatment approaches (Yokoyama et al, 2009), and great strides in discovering its biology and the underlying molecular mechanisms (Jones et al, 2008), successful therapeutic strategies are limited and long-term survival rates remain unsatisfactory.
In most patients, PDAC is diagnosed at an advanced stage, one accompanied by extensive invasion and lymphatic metastasis, and thus there is an exceptionally low 5-year survival rate of only 5% (Jemal et al, 2009). Invasion and metastasis are the distinct biologic characteristics of cancer. They are the most important factors influencing tumour patients’ therapeutic effect and prognosis. Metastasis-associated genes and proteins have therefore attracted much attention. Yachida and co-workers, 2009 pronounced that in pancreatic cancer the initial primary tumour proliferates for several years before producing metastatic clones (Luebeck, 2010). However, patients with clinically undetectable primary tumours still have a high risk of developing metastases. There is an urgent need to elucidate how pancreatic cancer cells give rise to distant metastasis to provide not only a better understanding of the pathophysiologic mechanisms but also to reveal novel targets that can be used to greatly improve molecular diagnosis and therapeutic intervention.
Pancreatic ductal adenocarcinoma has been expansively studied at the genetic level. There are four hallmark mutations of PDAC (KRAS >90% (Qiu et al, 2011), p16INK4A >90% (Mazur and Siveke, 2012), TP53 70% (Muller et al, 2009) and SMAD4 55% (Iacobuzio-Donahue et al, 2009)) that have all previously been implicated in the metastatic process in human samples. However, the current studies do not fully explain the mechanism of metastasis comprehensively. New theories and methods are urgently needed in the study of molecular mechanisms in PDAC. Recently, large-scale complementary DNA cloning projects have identified that the majority of the mammalian genome is transcribed into long non-coding RNAs (lncRNAs), which do not code for proteins (Wilusz et al, 2009). These lncRNAs represent a subgroup of non-coding RNAs that are longer than 200 nucleotides (Nagano and Fraser, 2011). Multiple studies have indicated that significant numbers of lncRNAs are involved and may have central roles in a variety of biologic processes through complicated mechanisms (Guttman et al, 2009; Huarte et al, 2010; Ørom et al, 2010; Tian et al, 2010; Hung et al, 2011). Notably, the deregulation of lncRNAs has been shown to result in aberrant gene expression that contributes to the progression of a variety of human tumours including PDAC (Gupta et al, 2010; Tahira et al, 2011). Compared with protein-coding genes, however, the clinical significance and functions of most deregulated lncRNAs in the progression and aggressiveness of PDAC remains unknown.
According to the result of Arraystar lncRNA and mRNA microarray, we found that the lncRNA ENST00000480739 was markedly decreased in PDAC tumorous tissues. Analogously, we found in a cohort of PDAC patients (n=35) that the expression of ENST0000048073 was significantly decreased in cancerous tissues than in adjacent normal tissues. ENST0000048073 expression was negatively associated with tumour node metastasis (TNM) stage and lymph node metastasis in PDAC. In addition, ENST0000048073 was an independent risk factor for overall patient survival following surgery. Our research found that ENST00000480739 could suppress PDAC cell invasion in vitro by directly upregulating osteosarcoma amplified-9 (OS-9) expression. Moreover, hypoxia-inducible factor-1α (HIF-1α), which regulates a large number of genes that are involved in tumorigenesis, metabolism, angiogenesis, oncogene expression metastasis (Semenza, 2003), was identified as a indirect and functional target of ENST0000048073.
Materials and methods
Patients and clinical samples
A total of 35 primary cancer tissues and the paired adjacent non-cancerous tissue were collected from patients who underwent surgery for pancreatic cancer at Renji Hospital. These tissues were flash frozen in liquid nitrogen immediately after surgery and subsequently stored at −80 °C. All the resected nodules were identified as PDAC by pathologic examination. No patients received anticancer treatments before surgery in this study. All samples were staged in accordance with the tumour node metastasis (TNM) classification and criteria of the World Health Organisation (WHO), and tumour grade was assessed according to the WHO criteria. Written informed consent was obtained from all patients. The Ethics Committee of Shanghai Jiao-Tong University School of Medicine Renji Hospital approved the use of tissues for this study
Cell culture
Human PDAC cell lines (ASPC-1, BXPC-3, CFPAC-1, PANC-1 and SW1990) and normal human pancreatic HPNE cell lines were purchased from the Cell Resource Center, Shanghai Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM)+10% fetal bovine serum (FBS) containing 4.5 g l glucose (Life Technologies, Carlsbad, CA, USA). Cells were cultured under either normoxic (5% CO2 in air) or hypoxic (1% O2, 5% CO2 and 94% N2) conditions at 37 °C.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from tumour tissues and purchased cell lines using TRIzol (Life Technologies) according to supplier’s instructions. RNA concentration was measured with a NanoDrop ND-2000 spectrophotometer (Life Technologies). cDNA was synthesised by using a Prime Script RT Reagent Kit (Takara, Otsu, Shiga, Japan) according to the manufacturer’s instructions. RNA expression in tumour tissues and cell lines was measured by quantitative real-time PCR (qRT–PCR) using a SYBR Green method according to the manufacturer’s instructions. The qRT–PCR was performed using SYBR Premix Ex Taq (Takara) and StepOne Real-Time PCR System (Applied Biosystems, Grand Island, NY, USA). β-Actin was used as an internal control. The primers used for the amplification were as follows: ENST00000480739 forward primer, 5′-GCGGATGGTGCTTCGTGTT-3′ and reverse primer, 5′-GGACCTCCCTGTATTTATCTATT-3′; OS-9 forward primer, 5′-GGGAGAAUUUGAGAAGGAATT-3′ and reverse primer, 5′-UUCCUUCUCAAAUUCUCCCTT-3′; β-actin forward primer, 5′-ATCGTGGGGCGCCCCAGGCACC-3′ and reverse primer, 5′-CTCCTTAATGTCACGCACGATTTC-3′; MXI-1 forward primer, 5′-CAAAGTGAATGAAGCCAGTGAC-3′ and reverse primer, 5′-TTTCCAGCCCATAATCCTGT-3′; PDGFC forward primer, 5′-ATTCCTCCCTCGCCTTCAC-3′ and reverse primer, 5′-GCCCTCTTCTGTGTCTCCAG-3′; FN1 forward primer, 5′-ATGCTCCCACTAACCTCCAG-3′ and reverse primer, 5′-GGCTGTCCTCTTCGGGTAA-3′; MMP28 forward primer, 5′-AAACTGGTGGTGCTCTCAGG-3′ and reverse primer, 5′-GGCTGTCCTCTTCGGGTAA-3′; ANGPTL4 forward primer, 5′-TGGCTCAGTGGACTTCAACC-3′ and reverse primer, 5′-GATGCTATGCACCTTCTCCAG-3′; L1CAM forward primer, 5′-CCCTGGAGAGTGACAACGA-3′ and reverse primer, 5′-CCAATGAACGAACCATCCTC-3′; and MT1-MMP forward primer, 5′-AGGGTCTTCGTTGCTCAGTC-3′ and reverse primer, 5′-AGGGTTTCTTCTGCCCACTT-3′. The relative mRNA levels were calculated based on the Ct values and normalised using the β-actin expression.
Plasmid, siRNA and cell transfection
Plasmid pcDNA3.1-ENST00000480739 and pcDNA3.1-NC were purchased from BioYare Technologies (Shanghai, China). Small interfering RNAs were bought from Genepharm Technologies (Shanghai, China). The OS-9 promoter reporter plasmid, pGL3-enhancer-lncPRNA and control plasmid were bought from Generay Biotechnology. Plasmid and siRNA transfections were performed using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. Cells were incubated for 24 h before use in experiments.
Transwell invasion assay
The invasiveness of pancreatic cancer cells was assessed based on the invasion of cells through Matrigel-coated transwell inserts, as described previously (Zhang et al, 2000). In brief, 3 × 105 cells were seeded into 24-well plate-sized inserts (a chamber containing an 8-μm pore size; Millipore, Darmstadt, Germany) with Matrigel (BD Biosciences, San Jose, CA, USA). Cells were plated in customised DMEM medium without serum, with the lower chamber containing the medium plus 30% FBS, and thus serving as a chemoattractant. After incubation for 24 h, the cells that did not invade through the pores were carefully wiped out with cotton wool. All cells that had migrated from the upper to the lower side of the filter were stained with 1% crystal violet in 20% methanol solution, and then counted and imaged. The assay was conducted three separate times.
The ELISA assay
The MMP2/9 ELISA assay was conducted using an MMP2/9 ELISA Quantitative Kit (WestTang Bio-Tech Co., Ltd, Shanghai, China), following the manufacturer’s protocol. The supernatant of PDAC cells was prepared 48 h after transfection, and analyzed at 1 : 1/1 : 20 dilution defined by the pretests and the manufacturer’s instructions. All the steps were repeated three times.
Western blot analysis
Western blotting were conducted under standard procedures (Cai et al, 2010). Briefly, cells were broken open to obtain proteins using RIPA. Proteins were separated by 8% SDS–PAGE and then transferred to PVDF membrane (Bio-Rad, Hercules, CA, USA). After blocking in 5% nonfat milk, the membranes were incubated with the following primary antibodies: OS-9 (1 : 1000; Novus Biologicals, Littleton, CO, USA), HIF-1α (1 : 1000; BD Biosciences), GAPDH (1 : 10000; KangChen, Shanghai, China) and tubulin (1 : 1000; Beyotime, Beijing, China), and then the secondary antibodies: rabbit anti-mouse IgG and goat anti-rabbit IgG (1 : 10000; SAB, Bethesda, MD, USA). All antibodies were diluted in nonfat dry milk. The immunoreactive protein bands were visualised by ECL Kit (Pierce, Thermo Fisher Scientific, IL, USA). The experiment was performed three separate times.
Immunohistochemistry analysis
To expand the numbers of patients in the sample set, tissue microarrays were constructed from the archival formalin-fixed paraffin-embedded tissue blocks of 120 surgically resected primary PDACs. Immunohistochemistry was performed as previously described (Muders et al, 2006). Primary antibodies were OS-9 (1 : 100; Novus Biologicals) and HIF-1α (1 : 250; Epitomics, Burlingame, CA, USA). Scoring of the immunohistochemical staining was based on the percentage of positive cells and staining intensity under a light microscope at × 200 magnification. Four categories were denoted (0, 1, 2 and 3) as negative, weak, moderate and strong. Groups 2 and 3 were considered high-expression groups and groups 0 and 1 as low-expression groups.
Luciferase reporter assay
For analysis of luciferase reporters, cells were plated in 24-well plates, and a mixture of 500 ng indicated PGL3 reporter plasmids for OS-9 promoter region, 600 ng pcDNA-ENST00000480739 or pcDNA3.1, 25 ng Renilla plasmid and pGL3 enhancer lncPRNA plasmid were transfected into cells with Lipofectamine 2000 (Life Technologies). After 48 h, luciferase activities were determined using the Luciferase assay system (Promega) according to the manufacturer’s instructions. Renilla luciferase activity was used as an internal control for transfection efficiency.
Animal experiments
The lenti-control virus and lenti-lncENST00000480739 virus were constructed by Shanghai SBO Medical Biotechnology Company (Shanghai, China). SW1990 cells were infected with lenti-lncENST00000480739 virus or lenti-control virus to construct the SW1990-lncENST00000480739, SW1990-control stable cell lines, respectively. We injected SW1990-lncENST00000480739 and SW1990-control cells into the tail veins of nude mice, respectively, once a week for 10 weeks. Then, the livers and lungs of the nude mice were excised post mortem for hematoxylin–eosin staining. All experimental procedures were approved by the Institutional Animal Care and Use Committee.
ChIP and RT ChIP
Chromatin immunoprecipitation (ChIP) assays were performed using the ChIP Assay Kit (Upstate, Charlottesville, VA, USA) according to the manufacturer’s instructions. The protein–DNA complexes were precipitated using 5 μg antibodies against acetyl-histone H3 (Lys27) antibody (no. 4353; CST, Danvers, MA, USA). Approximately 2–5 μl of ChIP-enriched chromatin was subjected to a standard ChIP-PCR reaction, and the enrichment of specific genomic regions was assessed relative to either control IgG or control cells.
Real-time PCR was performed using an Applied Biosystems 7900 quantitative PCR system (Applied Biosystems). Each PCR reaction was carried out in a 10 μl reaction volume using 3 μl of the eluted immunoprecipitated DNA. The amount of genomic DNA co-precipitated with the specific antibody was calculated in comparison with the total input DNA used for each immunoprecipitation, as follows: CBTB=CBTB (genomic input)−CBTB (specific antibody), where CBTB (genomic input) and CBTB (specific antibody) were the mean threshold cycles of PCR performed in triplicate on DNA samples from the genomic input samples and the specific antibody samples, respectively. The ChIP primers for the OS9 gene promoter were: OS9P-F (5′-ATCATGAAGAGATGTTGAAT-3′) and OS9P-R (5′-TTATATACCACGACCAAGT-3′). A ChIP primers pair for GAPDH was used as negative control for STAT3 binding: (5′-CACCGTGTGCCCAAGACCTC-3′) and (5′-CAGCCCTGTAGCCTGGACCT-3′).
Statistical analysis
All statistical analyses were performed with SPSS version 19.0 software (SPSS software, Armonk, NY, USA). For statistical comparisons, a one-way analysis of variance, Fisher’s exact test or two-tailed Student’s t-test was performed. Data are presented as mean±s.d. The survival curves were calculated using the Kaplan–Meier method, and the differences were assessed by a log-rank test. Cox regression analysis was used to determine the independent factors, which were based on the variables selected by a univariate analysis. Results with a P-value <0.05 were considered statistically significant.
Results
The novel lncRNA ENST00000480739 is downregulated in PDAC
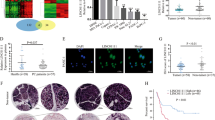
We previously found systemic variations in the expression of lncRNAs between PDAC and the paired non-tumour samples using a microarray analysis. From that study, we noted the lncRNA ENST00000480739 expression was remarkably decreased in PDAC tissues than in adjacent normal tissues. (The microarray data mentioned in this article can be viewed at GEO as GSE57144.) The lncRNA ENST00000480739 can be found in the UCSC genome database, and the sequence of this lncRNA is available in the Supplementary Figure 1. Furthermore, the lncRNA ENST00000480739 does not contain a valid Kozak sequence and it is identified to be an lncRNA rather than a protein-coding transcript by CNCI software (The Broad Institute, Cambridge, MA, USA) (Sun et al, 2013). We also performed a codon substitution frequency analysis using PyhloCSF (Lin et al, 2011). Long non-coding RNA ENST00000480739 showed very low codon substitution frequency scores (−0.252658), indicating that it is a non-coding RNA. To further corroborate our prior findings, we detected ENST00000480739 expression by using qRT–PCR in 35 pairs of PDAC tissues and in their adjacent normal tissues. The data showed that ENST00000480739 expression was remarkably decreased, reaching at least a 50% reduction in 66% (23 of 35) of PDAC tissues (Figure 1A), and its average intensity was reduced more than three-fold in tumours than in adjacent normal tissues (Figure 1B). The RNA level of ENST00000480739 in pancreatic cancer cell lines detected by qRT–PCR was significantly decreased in all five pancreatic cancer cell lines examined than in the normal pancreatic epithelial-like cell line HPNE (Figure 1C). To detect whether the silencing of lncRNA expression in PDAC occurs at the transcriptional or posttranscriptional level, we cloned the 5′ flanking DNA fragment (∼1 kbp) of ENST00000480739 promoter region. This DNA fragment was then inserted into pGL3 enhancer plasmid to construct the pGL3 enhancer lncPRNA plasmid. Furthermore, this recombination plasmid was transfected into SW1990 and normal pancreatic epithelial HPNE cells. As Figure 1D showed that the luciferase activity of pGL3 enhancer lncPRNA was significantly decreased in SW1990 cells when compared with that in HPNE normal pancreatic cells at the same condition, suggesting that the decreasing expression of lncRNA ENST00000480739 may be regulated at the transcriptional level, but not at the posttranscriptional level. Based on these data, we confirmed that this novel lncRNA was significantly downregulated in PDAC tissues as well as in PDAC cell lines.
The novel lncRNA ENST00000480739 is downregulated in PDAC. (A) Downregulation of ENST00000480739 was found in 92% (32 of 35) of PDAC tissues. A relative fold change (2−ΔΔCT) <1/2 was defined as significantly low expression, and >1 was defined as high expression. (B) Expression of mature ENST00000480739 determined by qRT–PCR in 35 paired human PDAC and their corresponding non-tumorous samples (NT) normalised by endogenous β-actin. Paired t-test, P=0.029. (C) Expression level of ENST00000480739 in five pancreatic cancer cell lines and normal pancreatic cell line HPNE. Long non-coding abundance was normalised by endogenous β-actin. Statistical analysis was conducted by one-way analysis of variance, *P<0.05. (D) Relative luciferase assays were performed in SW1990 and HPNE pancreatic cells. Data are shown as mean±s.d., n=3, *P<0.05 (Student’s t-test), compared with the control.
The association between lncRNA ENST00000480739, clinicopathologic characteristics and prognosis after surgery
To determine whether ENST00000480739 expression level was correlated with any general clinical characteristics of PDAC, its expression was evaluated in 35 PDAC patients with different clinicopathologic features (Table 1). Results revealed that ENST00000480739 expression was negatively correlated with the TNM stage (P=0.0354; Figure 2A) and with lymph node metastasis (P=0.0004; Figure 2B). There was no significant correlation between ENST00000480739 expression with age, gender, diameter or degree of differentiation. The data suggest that loss of ENST00000480739 expression may have an important role in PDAC progression and metastasis.
The association between lncRNA ENST00000480739, clinicopathologic characteristics and prognosis after surgery. (A) ENST00000480739 expression in different clinical stages of PDAC. Patients were staged in accordance with the 7th Edition of the AJCC Cancer‘s TNM Classification. (B) Downregulation of ENST00000480739 in PDAC was associated with lymph node metastasis; patients were classified into lymph node metastasis-negative group (LN-negative) and -positive group (LN-positive). (C) Kaplan–Meier curves of overall survival. Patients were scored as low expression level (below the median value, n=17) or high expression level (above the median value, n=18) according to ENST00000480739 expression value. ENST00000480739 downregulation was significantly correlated with an overall shorter survival time. The P-values are shown with the use of log-rank test in Table 2.
We also compared the survival time following surgery in 35 cases of PDAC patients with higher-ENST00000480739-expressing tumours (above the median value, n=18) than those with lower-ENST00000480739-expressing tumours (below the median value, n=17). The cumulative survival rate was significantly better in patients with higher-ENST00000480739-expressing tumours than in those with lower-ENST00000480739-expressing tumours (P=0.005, HR (95% CI)=0.182 (0.056–0.596); Figure 2C). In addition, the univariate analysis showed that patients with lower expression of ENST00000480739, lymph node metastasis, TNM stage and the lower expression of OS-9 were significantly associated with an increased risk of cancer-related death (Table 2). The multivariate analysis revealed that TNM stage (P=0.017) and lymph node metastasis (P=0.001) are key prognostic factors. Furthermore, the lower expression of ENST00000480739 (P=0.005) was found to be significantly associated with poor survival in PDAC patients independently of the TNM stage and lymph node metastasis. These data indicate that ENST00000480739 expression is a new prognostic factor in PDAC patients.
The lncRNA ENST00000480739 inhibits tumour cell invasion in vitro and in vivo
Because the statistical analysis indicated that downegulation of ENST00000480739 in PDAC frequently occurred and was closely associated with tumour metastasis, we hypothesised that overexpression of ENST00000480739 in PDAC cells may exert inhibitory effects on cell invasion. To verify this hypothesis, cells were transfected with either pcDNA-ENST00000480739 or pcDNA3.1. Real-time PCR detected that ENST00000480739 was successfully overexpressed in PDAC cells SW1990 and PANC-1. Successful overexpression of mature ENST00000480739 was confirmed by qRT–PCR. Transwell cell migration assay confirmed that overexpression of ENST00000480739 significantly inhibited cell invasion (Figure 3A). Similarly, the ELISA assay result supported this conclusion (Figures 3B and C). The protein levels of MMP2/9, which are important factors in invasion (Brooks et al, 1996; Ramos-DeSimone et al, 1999), were significantly lower in supernatants of ENST00000480739-overexpressing cells than in control cells. We next developed a pancreatic cancer metastasis model in nude mice, and explored the functional impact of lncRNA ENST00000480739 overexpression in vivo using a nude mouse metastatic tumour model. Different stable cells were transplanted into BALB/c-nu/nu mice via tail vein injection. Hematoxylin–eosin staining showed that less metastatic pancreatic cancer SW1990 cells were observed in the lungs of nude mice at 10 weeks after injection of SW1990-lncENST00000480739 control cells, whereas most of the mice injected with SW1990 control cells displayed significant lung metastases (Figures 3E and F). These data suggest that lncRNA ENST00000480739 may have a critical role in pancreatic cancer cell motility and metastasis. We also assessed whether ENST00000480739 could affect tumour cell proliferation, apoptosis or cell cycle. However, the consequence of CCK-8 assay and flow cytometry assay suggested a negative verdict (Supplementary Figure 3).
The lncRNA ENST00000480739 inhibits tumour invasion in vitro. (A–C) Overexpression of ENST00000480739 significantly impeded abilities of cell invasion in SW1990 and PANC-1 cells. (a) SW1990 cells were transfected with vector; (b) SW1990 cells were transfected with ENST00000480739; (c) PANC-1 cells were transfected with vector; (d) PANC-1 cells were transfected with ENST00000480739. (D and E) Concentrations of MMP2/9 were lower in supernatants of the ENST00000480739 overexpression group. Statistical analysis conducted by paired t-test. *P<0.05 and **P<0.01. (E) Representative haematoxylin–eosin staining and (F) summarised data showed that less metastatic CRC cells were observed in the lungs of nude mice at 10 weeks after injection of SW1990-lncENST00000480739 stable cells, whereas most of the mice injected with SW1990 control cells displayed significant lung metastases.
The lncRNA ENST00000480739 associates with OS-9
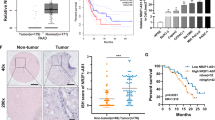
Bioinformatics analysis revealed that ENST00000480739 is located on chromosome 12 upstream of the OS-9 promoter region and the lncRNA ENST00000480739 and OS9 are transcribed from the same DNA strand in UCSC genome database (Figure 4A). Thus, we conjectured that ENST00000480739 might associate with OS-9. To test this prediction biochemically, we examined the mRNA level of OS-9 in our clinical samples. Real-time PCR data showed that the expression of OS-9 was significantly decreased in tumour tissues than in non-cancerous tissue in 35 pairs of primary cancer tissues and the paired adjacent samples (Figure 4B). Furthermore, we found that high expression of OS-9 was more likely to be detected in PDAC tissues with high levels of ENST00000480739 (Figure 4C). Moreover, we also found the expression of OS-9 was positively correlated with patient survival outcome in Kaplan–Meier survival analysis (P=0.035, HR (95% CI)=0.320 (0.112–0.915); Figure 4D). ENST00000480739 was also an independent risk factor for overall survival (Table 2). The protein level of OS-9 detected in the tissue microarray of 120 patients was markedly decreased in cancerous tissues than in adjacent normal tissues (Figure 4E). Low expression of OS-9 was closely associated with TNM stage and metastasis (Table 3). The data indicated that loss of OS-9 expression might be key to PDAC progression.
The lncRNA ENST00000480739 associates with OS-9. (A) ENST00000480739 is located on chromosome 12 upstream of the promoter region of OS-9. (B) Expression of OS-9 determined by qRT–PCR in 35 paired human PDAC and their corresponding non-tumorous samples (NT) and normalised by endogenous β-actin. P=0.0001, paired t-test. (C) Relationship between OS-9 and ENST00000480739 expression explored by Spearman’s correlation in 35 paired PC tissues and adjacent NP tissues. (D) Kaplan–Meier curves of overall survival. Patients were scored as low expression level (below the median value, n=17) or high expression level (above the median value, n=18) according to the OS-9 expression. The OS-9 downregulation was significantly correlated with an overall shorter survival time. The P-values are shown with the use of log-rank test in Table 2. (E) Immunohistochemistry analysis performed in tissue microarray assay that contained 120 pairs of PDAC and their corresponding non-tumorous tissues ( × 400). The protein level of OS-9 detected in the tissue microarray of 120 patients was markedly decreased in cancerous tissues than in adjacent normal tissues (P<0.05). (F) mRNA levels of OS-9 as determined by qRT–PCR in ENST00000480739 overexpression cells and negative control cells; β-actin served as an internal control. (G) Western blot analysis used to detect the expression level of endogenous OS-9 in SW1990 cells after transfected with ENST00000480739 or control vector; β-actin served as an internal control. (H) Promoter region fragment of OS-9 directly fused downstream of the firefly luciferase gene in the luciferase reporter plasmids, which were co-transfected with Renilla plasmid and pcDNA3.1-ENST00000480739 or pcDNA3.1 into SW1990 cells. Luciferase activity was normalised to Renilla luciferase activity. The normalised luciferase activity of SW1990 vector group was set as 1. *P<0.05 and **P<0.01.
Next, we examined whether ENST00000480739 may regulate OS-9 expression in PDAC cells. Real-time PCR data showed that the OS-9 mRNA level was significantly increased after overexpression of ENST00000480739 in SW1990 cells (Figure 4F). Furthermore, western blot analysis revealed that the protein level of OS-9 was markedly increased in response to ENST00000480739 overexpression in SW1990 cells (Figure 4G). These data suggest that the OS-9 expression is regulated by ENST00000480739 in PDAC cells.
Furthermore, to detect whether OS-9 is a direct target of ENST00000480739, SW1990 cells were co-transfected with the OS-9 promoter reporter plasmid combined with pcDNA-ENST00000480739 or pcDNA3.1. Luciferase data showed that overexpression of ENST00000480739 significantly increased the transcriptional activity of the OS9 promoter (Figure 4H). In addition, we have looked through the UCSC genome database, and found that the promoter region of OS9 may be modified by H3K27 acetylation. We further explored whether the lncRNA upregulated OS9 expression by modifying the H3K27 acetylation level of OS9 gene promoter. Supplementary Figure 2A showed that H3K27 acetylation site may exist in the DNA promoter region of OS9 gene. Real-time ChIP PCR showed that overexpression of lncRNA ENST00000480739 significantly increased the H3K27 acetylation level of OS9 gene promoter (Supplementary Figure 1B). These data indicate that ENST00000480739 may regulate the expression of OS9 by directly regulating the transcriptional activity of the OS9 promoter.
The lncRNA ENST00000480739 inhibits tumour invasion by suppressing HIF-1α expression through modulating OS-9 function
Osteosarcoma amplified-9 has previously been shown to be an important factor in the negative regulation of HIF-1α (Baek et al, 2005), which has a key role in the response of cells to hypoxia. Hypoxia has been suggested as a major cause of the poor prognosis of patients with solid tumours (Hashimoto et al, 2011) and the overexpression of HIF-1α has been reported in tumour metastasis (Fujiwara et al, 2007). Importantly, for our study, PDAC is thought to occur in high levels of hypoxia, and HIF-1α is highly expressed (Koong et al, 2000).
Several studies have demonstrated that lncRNAs usually perform their molecular functions by binding to specific proteins (Maruyama, 2012). Our findings in this study revealed that ENST00000480739 may have an important role in positively regulating OS-9 expression. Based on these observations, we sought to determine whether or not ENST00000480739 had a key role in inhibiting tumour metastasis by suppressing HIF-1α indirectly through the modulation of OS-9.
To examine our hypothesis, we analysed the effects of ENST00000480739 together with OS-9 on invasion using SW1990 cells. Invasion assay data showed that knockdown of OS-9 remarkably increased the ability of invasion in PDAC cells. Furthermore, downregulation of OS9 significantly rescued ENST00000480739-induced decrease in the cell invasion in PDAC cells (Figure 5A). Additionally, results from the ELISA assay were consistent with the transwell invasion assay. Downregulation of OS9 significantly rescued ENST00000480739-induced decrease in MMP2/MMP9 expression in PDAC cells (Figures 5B and C). Taken together, these data suggest that ENST00000480739 may inhibit PDAC cells invasion and motility by directly targeting OS-9.
The lncRNA ENST00000480739 inhibits tumour invasion by suppressing HIF-1 α expression through the modulation of OS-9 function. (A) Knockdown of OS-9 partially improved the ability of invasion in ENST00000480739 overexpression cells. (a) Cells were co-transfected with pcDNA3.1 and control siRNA; (b) cells were co-transfected with pcDNA-ENST00000480739 and control siRNA; (c) cells were co-transfected with pcDNA3.1 and OS-9-specific siRNA; (d) cells were co-transfected with pcDNA-ENST00000480739 and OS-9-specific siRNA. (B and C) The concentrations of MMP2/9 were lowest in cells co-transfected with ENST00000480739 and control siRNA, and were highest in the group co-transfected with NC and OS-9-specific siRNA. Statistical analysis was conducted by one-way ANOVA. (D) Western blot analysis of HIF-1a expression under hypoxic conditions (O2 concentration 1%). The absence of OS-9-mediated increment of HIF-1a expression was abrogated by ENST00000480739 overexpression. Tubulin, loading control.
To further explore our hypothesis, we incubated SW1990 cells for 24 h in hypoxic conditions following the same treatment as described above to obtain proteins. These proteins were then subjected to western blot assay with the anti-HIF-1α antibody. We found that the knockdown of OS-9 markedly abolished ENST00000480739-induced downregulation of HIF-1α (Figure 5D). These data indicate that ENST00000480739 may negatively regulate HIF-1α expression by enhancing OS-9 expression. Moreover, we have looked through the literatures and found MXI-1, PDGFC, FN1 and MMP28 as the representative target genes of HIF-1α (Zhang et al, 2007; Wan et al, 2011). Real-time PCR data showed that forced expression of the lncRNA in PDAC cells significantly decreased these genes’ mRNA levels, which were regulated by HIF-1α (Supplementary Figure 2A). The data suggest that this lncRNA may not only regulate the expression of HIF-1α but also modulate the activity of HIF-1α.
Discussion
This study emphasised lncRNA and explored potential functions of ENST00000480739, a novel lncRNA we identified, in PDAC. Long non-coding RNAs are broadly defined as RNA molecules >200 bp in length that lack an open reading frame (Wilusz et al, 2009). Although the specific functions of the large majority of lncRNAs remain unknown, recent advances in genome and transcriptome analysis have begun to shed light on the critical roles played by these molecules in a variety of cellular processes, including differentiation, development and tumorigenesis (Hung et al, 2011). The relationship between lncRNAs and tumour growth has recently attracted much attention. Altered expressions of lncRNAs have been documented in several different human cancers and lncRNAs have been suggested as biomarkers for diagnosis and prognosis, as well as potential therapeutic targets (Ponting et al, 2009; Gibb et al, 2011; Prensner and Chinnaiyan, 2011). For example, high expression of HOTAIR in breast cancer has been correlated with a poor prognosis and tumour metastasis (Gupta et al, 2010). MALAT1 is upregulated in multiple malignancies, including lung cancer, uterine endometrial stromal sarcoma, cervical cancer and hepatocellular carcinoma (HCC) (Ji et al, 2003; Lin et al, 2006; Yamada et al, 2006), and it can serve as an independent prognostic parameter for patient survival in early-stage lung adenocarcinoma (Müller-Tidow et al, 2004). MVIH was found to be overexpressed in HCC, which could promote tumour growth and intrahepatic metastasis by activating angiogenesis (Yuan et al, 2012).
Through the lncRNA expression microarray, we found a new lncRNA transcript, ENST00000480739, which was significantly downregulated in PDAC tumour tissue. In a cohort of 35 PDAC patients, we determined that the downregulation of ENST00000480739 was associated with advanced TNM stage (stage I vs stage II, P=0.0354) and lymph node metastasis (P=0.0004). Lower expression of ENST00000480739 tended to have more advanced TNM stage, suggesting that low expression of ENST00000480739 may promote cancer progression. ENST00000480739 expression was further significantly downregulated in PDAC patients with lymph node metastasis, which indicated that ENST00000480739 might have an important role in PDAC metastasis. The survival analysis data in the present study revealed that ENST00000480739 deregulation was correlated with shorter survival time of pancreatic cancer patients. In addition, the multivariate analysis indicated that low ENST00000480739 expression, advanced TNM stage and lymph node metastasis were each independent risk factors for overall patient survival rate following surgery. Our data indicate that ENST00000480739 may be a novel biomarker for risk prognostication and personal therapy screening of PDAC patients after surgery. For example, patient ENST00000480739 expression level could be used as a warning sign to clinicians that low-expression postoperative patients should be closely monitored and receive appropriate adjuvant therapies.
Lymph node metastasis is an initial step in pancreatic cancer metastasis, and is a crucial factor in the determination of the clinical staging, prognosis and survival of pancreatic cancer patients. Elucidating the molecular mechanisms underlying pancreatic cancer lymph node metastasis is thus urgently needed. Research in lncRNAs may provide new insights for understanding the molecular mechanisms and hence influence the design of better therapeutic strategies to treat cancer patients with lymph node metastasis. In our study, the PDAC patients with lymph node metastasis had a much lower ENST00000480739 expression compared with those without lymph node metastasis. Therefore, ENST00000480739 may be a novel risk biomarker for judging PDAC metastasis status. Furthermore, our functional experiments demonstrated that ENST00000480739 had an important role in PDAC metastasis, and that overexpression of ENST00000480739 could suppress PDAC cells invasion in vitro and metastasis in vivo. We have also analysed the expressions of MT1-MMP, which can regulate cell invasion (Petrella et al, 2005), and the expressions of ANGPTL4 and L1CAM, which may regulate lung colonisation (Zhang et al, 2012). We found that overexpression of ENST00000480739 significantly decreased the expressions of MT1-MMP, ANGPTL4 and L1CAM in pancreatic cancer SW1990 cells (Supplementary Figure 4A), indicating that lncRNA ENST00000480739 may block cancer cell metastasis by inhibiting cancer cell invasion and lung colonisation. In general, these data indicate that ENST00000480739 may be closely related to lymph node metastasis and may act as a metastasis suppressor in PDAC.
OS-9 was originally identified as being amplified in osteosarcomas. However, further studies indicated that a dominant-negative form of OS-9 is expressed in tumours, and that developmental or physiologic alterations in OS-9 expression or activity may provide a means to alter the set point of the oxygen-sensing system, which ultimately decreases HIF-1α level, HIF-1α transcriptional activity and the expression of an endogenous HIF-1α target gene (Baek et al, 2005). Hypoxia-inducible factor-1α is a transcription factor that activates gene expression in response to hypoxia. The majority of human cancers are characterised by overexpression of HIF-1α and inhibition of HIF-1α impairs tumour growth and metastasis (Semenza, 2003). We found that OS-9 expression was associated with ENST00000480739 level in clinical samples, and OS-9 was also an independent risk factor for patient overall survival following surgery. Thus, we predicted that ENST00000480739 could upregulate OS-9 expression in PDAC. The study results verified the hypothesis that overexpression of ENST00000480739 could increase both mRNA and protein level of OS-9. ENST00000480739 could directly increase OS-9 by activating the promoter region. Previous studies have shown that one group of lncRNAs was able to bind specific proteins and then direct the localisation of the resultant complex to specific targets (Maruyama, 2012). For example, lincRNA-p21 and HOTAIR are able to alter and regulate epigenetic states and gene expression across multiple sites in trans (Gupta et al, 2010; Huarte et al, 2010). We observed that ENST00000480739 could inhibit tumour invasion in vitro. Thus, we suggest that ENST00000480739 inhibits invasion in this manner. We divided SW1990 cells into four groups, two of them were transiently co-transfected with pcDNA-ENST00000480739 and either OS-9-specific or control siRNA, and the other two groups were co-transfected with pcDNA3.1-NC and either OS-9-specific or control siRNA. We found that knockdown of OS-9 partially improved the ability of invasion in ENST00000480739-overexpressng cells, and also influenced the concentrations of MMP2/9, which were lowest in the group co-transfected with pcDNA-ENST00000480739 and control siRNA, and highest in the group co-transfected with pcDNA-NC and OS-9-specific siRNA. Overexpression of lncENST00000480739 significantly decreased the HIF-1α expression at the basal level. Knockdown of OS-9 dramatically blocked lncENST00000480739-induced HIF-1α downregulation. Taken together, results from this study suggest that ENST00000480739 may negatively regulate HIF-1α by upregulating OS-9, thereby suppressing invasion and metastasis. This discovery suggests a promising new insight with regard to the essential mechanisms of regulating HIF-1α in PDAC. Dysregulation of HIF-1α via OS-9 is potentially an important mechanism underlying cancer metastasis, and thus the lncRNA ENST00000480739 may serve as a potential treatment target for modulating this pathway in the metastatic diseases.
In summary, our results showed that ENST00000480739 is an important antimetastatic lncRNAs, which when downregulated is associated with lymph node metastasis in PDAC, as well as being an independent risk factor for patient overall survival after surgery. Moreover, in vitro experiments demonstrated that enforced expression of ENST00000480739 resulted in suppressed PDAC cell invasion and metastasis by directly targeting OS-9, and thus indirectly targeting HIF-1α. These findings suggest that the frequently downregulated ENST00000480739 in PDAC contributes to tumour metastasis and progression. ENST00000480739 may have not only a therapeutic potential to suppress metastasis but it may also be a novel biomarker for risk prognostication and personal therapy screening of PDAC patients.
Change history
25 November 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Baek JH, Mahon PC, Oh J, Kelly B, Krishnamachary B, Pearson M, Chan DA, Giaccia AJ, Semenza GL (2005) OS-9 interacts with hypoxia-inducible factor 1α and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1α. Mol Cell 17 (4): 503–512.
Brooks PC, Strömblad S, Sanders LC, von Schalscha TL, Aimes RT, Stetler-Stevenson WG, Quigley JP, Cheresh DA (1996) Localization of matrix metalloproteinase MMP-2 to the surface of invasive cells by interaction with integrin αvβ3. Cell 85 (5): 683–693.
Cai Z, Zhao J-S, Li J-J, Peng D-N, Wang X-Y, Chen T-L, Qiu Y-P, Chen P-P, Li W-J, Xu L-Y (2010) A combined proteomics and metabolomics profiling of gastric cardia cancer reveals characteristic dysregulations in glucose metabolism. Mol Cell Proteom 9 (12): 2617–2628.
Fujiwara S, Nakagawa K, Harada H, Nagato S, Furukawa K, Teraoka M, Seno T, Oka K, Iwata S, Ohnishi T (2007) Silencing hypoxia-inducible factor-1α inhibits cell migration and invasion under hypoxic environment in malignant gliomas. Int J Oncol 30 (4): 793–802.
Gibb EA, Brown CJ, Lam WL (2011) The functional role of long non-coding RNA in human carcinomas. Mol Cancer 10 (1): 38–55.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464 (7291): 1071–1076.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458 (7235): 223–227.
Hashimoto O, Shimizu K, Semba S, Chiba S, Ku Y, Yokozaki H, Hori Y (2011) Hypoxia induces tumor aggressiveness and the expansion of CD133-positive cells in a hypoxia-inducible factor-1α-dependent manner in pancreatic cancer cells. Pathobiology 78 (4): 181–192.
Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA (2006) Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 20 (10): 1218–1249.
Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142 (3): 409–419.
Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P (2011) Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43 (7): 621–629.
Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, Vilardell F, Wang Z, Keller JW, Banerjee P, Herman JM, Cameron JL, Yeo CJ, Halushka MK, Eshleman JR, Raben M, Klein AP, Hruban RH, Hidalgo M, Laheru D (2009) DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol 27 (11): 1806–1813.
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ (2009) Cancer statistics, 2009. Cancer J Clin 59 (4): 225–249.
Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E (2003) MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22 (39): 8031–8041.
Jones S, Zhang X, Parsons DW, JC-H Lin, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321 (5897): 1801–1806.
Koong AC, Mehta VK, Le QT, Fisher GA, Terris DJ, Brown JM, Bastidas AJ, Vierra M (2000) Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys 48 (4): 919–922.
Lin MF, Jungreis I, Kellis M (2011) PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27 (13): i275–i282.
Lin R, Maeda S, Liu C, Karin M, Edgington T (2006) A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26 (6): 851–858.
Luebeck EG (2010) Cancer: Genomic evolution of metastasis. Nature 467 (7319): 1053–1055.
Müller-Tidow C, Diederichs S, Thomas M, Serve H (2004) Genome-wide screening for prognosis-predicting genes in early-stage non-small-cell lung cancer. Lung Cancer 45: S145–S150.
Maruyama R (2012) Long noncoding RNA involvement in cancer. Biochem Mol Biol Rep 45 (11): 604–611.
Mazur PK, Siveke JT (2012) Genetically engineered mouse models of pancreatic cancer: unravelling tumour biology and progressing translational oncology. Gut 61 (10): 1488–1500.
Muders MH, Dutta SK, Wang L, Lau JS, Bhattacharya R, Smyrk TC, Chari ST, Datta K, Mukhopadhyay D (2006) Expression and regulatory role of GAIP-interacting protein GIPC in pancreatic adenocarcinoma. Cancer Res 66 (21): 10264–10268.
Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, Cromer A, Brugge JS, Sansom OJ, Norman JC, Vousden KH (2009) Mutant p53 drives invasion by promoting integrin recycling. Cell 139 (7): 1327–1341.
Nagano T, Fraser P (2011) No-nonsense functions for long noncoding RNAs. Cell 145 (2): 178–181.
Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143 (1): 46–58.
Petrella BL, Lohi J, Brinckerhoff CE (2005) Identification of membrane type-1 matrix metalloproteinase as a target of hypoxia-inducible factor-2 alpha in von Hippel–Lindau renal cell carcinoma. Oncogene 24 (6): 1043–1052.
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136 (4): 629–641.
Prensner JR, Chinnaiyan AM (2011) The emergence of lncRNAs in cancer biology. Cancer Discov 1 (5): 391–407.
Qiu W, Sahin F, Iacobuzio-Donahue CA, Garcia-Carracedo D, Wang WM, Kuo CY, Chen D, Arking DE, Lowy AM, Hruban RH, Remotti HE, Su GH (2011) Disruption of p16 and activation of Kras in pancreas increase ductal adenocarcinoma formation and metastasis in vivo. Oncotarget 2 (11): 862–873.
Ramos-DeSimone N, Hahn-Dantona E, Sipley J, Nagase H, French DL, Quigley JP (1999) Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. J Biol Chem 274 (19): 13066–13076.
Semenza GL (2003) Targeting HIF-1 for cancer therapy. Nat Rev Cancer 3 (10): 721–732.
Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62 (1): 10–29.
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y (2013) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 41 (17): e166.
Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM (2011) Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer 10: 141.
Tian D, Sun S, Lee JT (2010) The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143 (3): 390–403.
Wan J, Chai H, Yu Z, Ge W, Kang N, Xia W, Che Y (2011) HIF-1alpha effects on angiogenic potential in human small cell lung carcinoma. J Exp Clin Cancer Res 30: 77.
Wilusz JE, Sunwoo H, Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23 (13): 1494–1504.
Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T, Noguchi M (2006) Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci 97 (2): 106–112.
Yokoyama Y, Nimura Y, Nagino M (2009) Advances in the treatment of pancreatic cancer: limitations of surgery and evaluation of new therapeutic strategies. Surg Today 39 (6): 466–475.
Yuan S, Yang F, Yang Y, Tao Q, Zhang J, Huang G, Wang R, Yang S, Huo X, Zhang L (2012) Long non-coding RNA-MVIH promotes angiogenesis and serves as a predictor for HCC patients’ poor recurrence-free survival after hepatectomy. Hepatology (Baltimore, MD) 56: 2231–2241.
Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL (2007) HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11 (5): 407–420.
Zhang H, Morisaki T, Matsunaga H, Sato N, Uchiyama A, Hashizume K, Nagumo F, Tadano J, Katano M (2000) Protein-bound polysaccharide PSK inhibits tumor invasiveness by down-regulation of TGF-beta1 and MMPs. Clin Exp Metast 18 (4): 343–352.
Zhang H, Wong CC, Wei H, Gilkes DM, Korangath P, Chaturvedi P, Schito L, Chen J, Krishnamachary B, Winnard PT Jr., Raman V, Zhen L, Mitzner WA, Sukumar S, Semenza GL (2012) HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene 31 (14): 1757–1770.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Foundation: Young Physician Training and Subsidizing Project of Shanghai Municipal Commission of Health and Family Plan.
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Sun, YW., Chen, YF., Li, J. et al. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1α in pancreatic ductal adenocarcinoma. Br J Cancer 111, 2131–2141 (2014). https://doi.org/10.1038/bjc.2014.520
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.520
Keywords
This article is cited by
-
The strict regulation of HIF-1α by non-coding RNAs: new insight towards proliferation, metastasis, and therapeutic resistance strategies
Cancer and Metastasis Reviews (2023)
-
LncRNAs as epigenetic regulators of epithelial to mesenchymal transition in pancreatic cancer
Discover Oncology (2022)
-
The impact of LncRNA dysregulation on clinicopathology and survival of pancreatic cancer: a systematic review and meta-analysis (PRISMA compliant)
Cancer Cell International (2021)
-
The multifaceted roles of long noncoding RNAs in pancreatic cancer: an update on what we know
Cancer Cell International (2020)
-
Signaling in and out: long-noncoding RNAs in tumor hypoxia
Journal of Biomedical Science (2020)