Abstract
Background:
We examined whether silencing of IGFBP7 was associated with survival in patients with oesophageal adenocarcinoma.
Methods:
Protein expression of IGFBP7 was determined using immunohistochemistry in a tissue microarray representing tumours from 65 patients with oesophageal adenocarcinoma who had not had neoadjuvant therapy. DNA methylation of the IGFBP7 promoter was determined with the melt curve analysis in cell lines and patient tissues.
Results:
Expression of IGFBP7 was observed in the oesophageal adenocarcinoma of 34 out of 65 (52%) patients and was associated with significantly reduced median (11 vs 92 months) and 5-year survival (25% vs 52%). Multivariate analysis identified expression as an independent prognostic indicator for survival (hazard ratio=3.24, 95% confidence interval=1.58–6.67, P-value=0.0014). Hypermethylation of IGFBP7 was associated with silencing of gene expression in cell lines and patient tissues (P-value=0.0225). Methylation was observed in the squamous mucosa of 2 out of 15 (13%) patients with Barrett’s oesophagus and 3 out of 17 (18%) with oesophageal adenocarcinoma. Methylation was observed in 14 out of 18 (78%) of biopsies of Barrett’s mucosa and 23 out of 34 (68%) patients with oesophageal adenocarcinoma.
Conclusion:
Reduced IGFBP7 protein expression was associated with longer survival in patients with oesophageal adenocarcinoma. Methylation of the IGFBP7 promoter was associated with silencing of gene expression and was frequent in Barrett’s oesophagus and oesophageal adenocarcinoma.
Similar content being viewed by others
Main
Oesophageal adenocarcinoma has increased rapidly in incidence in the Western world over recent decades and continues to have a poor prognosis, with 5-year survival rates of <10% (Umar and Fleischer, 2008). Approximately 0.1–0.3% of patients with the premalignant lesion Barrett’s oesophagus will develop oesophageal adenocarcinoma each year (Bhat et al, 2011; Hvid-Jensen et al, 2011). The majority of patients present with late-stage disease that is not amenable to resection and have a poor response to non-surgical therapy.
The insulin-like growth factor (IGF) signalling pathway has a key role in mediating normal growth and development, and in the malignant progression of many cancers including oesophageal adenocarcinoma (Doyle et al, 2012). The IGF system involves the complex coordination of growth factors (IGF1 and IGF2), cell surface receptors (IGF1R, IGF2R and INSR), high- and low-affinity IGF-binding proteins (IGFBPs), as well as the IGFBP-degrading proteases. The IGFBPs serve as carrier proteins for IGFs, prolonging their half-life and modulating their binding to the IGF receptors. Some IGFBP proteases cleave IGFBPs, release IGFs, and increase signalling. IGFBP7 is one of 16 members of the IGFBP superfamily of proteins. It binds IGFs with relatively low affinity but binds insulin with very high affinity (500-fold higher than other IGFBPs) (Burger et al, 2005).
Changes in DNA methylation are frequent, early events in carcinogenesis, widely present even in premalignant lesions such as Barrett’s oesophagus (Smith et al, 2008, 2010). Hypermethylation in promoter regions is associated with gene silencing, whereas hypomethylation in other regions is associated with genomic instability. DNA methylation is of clinical interest because it may lead to biomarkers for early detection, diagnosis, prognosis, therapeutic stratification, and post-therapeutic monitoring. Downregulation of IGFBP7 with promoter DNA methylation has been demonstrated in various cancers (Komatsu et al, 2000; Ruan et al, 2006; Lin et al, 2007; Jiang et al, 2008; Lin et al, 2008; Hinoue et al, 2009; Suzuki et al, 2010; Heesch et al, 2011; Chen et al, 2011b; Sullivan et al, 2012; Dimberg et al, 2013; Suzuki et al, 2013) but has not been previously reported in Barrett’s oesophagus or oesophageal adenocarcinoma.
Here we report the correlation between IGFBP7 expression and clinicopathological parameters, including survival, in patients with oesophageal adenocarcinoma, and the association between IGFBP7 expression and DNA methylation in oesophageal adenocarcinoma and Barrett’s oesophageal cell lines and tissues.
Materials and Methods
Tissue microarrays and immunohistochemistry
Tissue microarrays (TMAs) were constructed as previously described (Thompson et al, 2011). Sections (4 μm) were mounted on polylysine-coated slides, dewaxed and rehydrated. Antigen retrieval was performed by heating the sections for 5 min in 10 mmol l−1 citrate buffer (pH 6) in a microwave pressure cooker. After cooling to room temperature, sections were immunostained using an Autostainer Plus (Dako, Glostrup, Denmark). Sections were incubated for 60 min with a 1 : 75 dilution of rabbit anti-human IGFBP7 polyclonal IgG antibody (H-102; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) and then with MACH 4 Universal Horseradish Peroxidase-Polymer (Biocare Medical, Concord, CA, USA). Liquid 3,3-diaminobenzidine (Dako) was used as the chromogen, and sections were counterstained with Meyer’s haematoxylin. The expression of IGFBP7 was scored as positive (present) or negative (absent) by an experienced gastrointestinal pathologist (ARR).
Cell lines
Human oesophageal adenocarcinoma (OE33, OE19, and JH-EsoAd1 (Alvarez et al, 2008)) and squamous cell carcinoma (OE21 and TE7 (Nishihira et al, 1993)) cell lines were maintained in RPMI-1640 supplemented with 10% foetal bovine serum, 4 mmol l−1 L-glutamine, 200 U ml−1 penicillin, and 200 μg ml−1 streptomycin at 37 °C in a humidified incubator with 5% CO2 in air. The human telomerase immortalised Barrett’s oesophagus epithelial cells, CP-A, CP-B, and CP-D were maintained as previously described (Palanca-Wessels et al, 1998). The nontumourigenic SV40T-transformed squamous oesophageal cell line HET1A (Stoner et al, 1991) was maintained as previously described (Underwood et al, 2010). All cell lines were cultured at 37 °C in a humidified incubator with 5% CO2 in air. To study the effects of demethylation, triplicate cultures were treated with either 1 μmol l−1 5-aza-2’-deoxycytidine (aza, Sigma-Aldrich, St Louis, MO, USA) or vehicle (veh; 0.0027% v/v final concentration acetic acid), as described previously (Smith et al, 2008, 2005).
Patient samples
Multiple biopsies every 2 cm from within circumferential columnar lined oesophagus (n=40) and a single biopsy of squamous mucosa proximal to the squamocolumnar junction (n=15) from 18 patients with Barrett’s oesophagus were collected into RNAlater. For a diagnosis of Barrett’s oesophagus, goblet cells had to be observed in at least one biopsy from the columnar-lined oesophagus. All patients with Barrett’s oesophagus were being treated for gastro-oesophageal reflux disease with anti-secretory therapy. Single samples of primary tumour oesophageal adenocarcinoma (n=34), adjacent high-grade dysplasia (n=7), and histologically normal squamous mucosa from the proximal resection margin (n=17) from 34 patients with oesophageal adenocarcinoma were collected into liquid nitrogen or into RNAlater (Ambion, Austin, TX, USA). The study complied with the appropriate institutional guidelines.
Isolation of RNA and DNA from cell lines and patient samples
Patient tissues were disrupted in either disposable pestles (Edwards Instruments, Narellan, NSW, Australia) or a TissueLyser with 5-mm stainless steel beads (Qiagen, Hilden, Germany). The RNA and DNA were isolated from cell lines and oesophageal adenocarcinoma patient tissues using Trizol (Invitrogen, Carlsbad, CA, USA) and from all other biopsies using either the RNA/DNA Kit or the AllPrep DNA/RNA Mini Kit (Qiagen).
Methylation analysis
Genomic DNA (500 ng) was bisulphite-modified using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA). The bisulphite-modified DNA was amplified using primer sets, which did not discriminate between methylated and unmethylated sequences. The primer sequences were (forward) 5′-GGGTTGTTTGTTGGGYGAGATT-3′ and (reverse) 5′-ACTTTACCCTTCCRCCTCTTAC-3′. The primers and PCR conditions were specific for bisulphite-modified DNA and did not amplify unmodified DNA. All methylation analysis PCRs were performed using the QuantiTect SYBR Green PCR Kit (Qiagen) in a final volume of 15 μl, containing 1 μl of bisulphite-modified DNA and a final concentration of 0.5 μmol l−1 of each forward and reverse primer. Bisulphite-modified lymphocyte DNA, CpG methyltransferase (M.SssI) (New England Biolabs Inc, Ipswich, MA, USA) treated lymphocyte DNA, and unmodified DNA were included in each PCR run and served as unmethylated, methylated, and negative controls, respectively. Reactions were run in a Rotor-Gene 3000 (RG-3000) (Corbett Life Science, Sydney, NSW, Australia) at 95 °C for 15 min, then 45 cycles of 95 °C for 30 s and 59 °C for 60 s, followed by a final extension of 72 °C for 4 min. At the end of the amplification cycle, the PCR products were melted by increasing the temperature from 60 to 95 °C, rising 0.5 °C at each step, waiting 30 s on the first step and then 5 s for each step thereafter. The raw fluorescence data were normalised as previously described (Smith et al, 2009). Methylation was also measured using an Infinium HumanMethylation450 BeadChip Kit (Illumina, San Diego, CA, USA), as previously described (Lim et al, 2013).
Measurement of gene expression by quantitative real-time reverse-transcription PCR
To measure gene expression, cDNA was synthesised using SuperScript II (Invitrogen) from 2 μg of total RNA that had been treated with the TURBO DNA-free Kit (Ambion). Quantitative real-time reverse-transcription polymerase chain reaction (QPCR) was performed using the QuantiTect SYBR Green PCR Kit in a final volume of 10 μl, containing 0.2 μl of cDNA and a final concentration of 0.5 μmol l−1 of each forward and reverse primer. The primer sequences were (forward) 5′-AGCTGTGAGGTCATCGGAAT-3′ and (reverse) 5′-ACCAGCACCCAGCCAGTTA-3′. Triplicate reactions were run in an RG-3000 at 95 °C for 15 min, then 45 cycles of 95 °C for 30 s and 57 °C for 60, followed by a final extension of 72 °C for 4 min. Data were collected and analysed using the RG-3000 Application Software. Threshold cycle Ct values were determined on auto-threshold settings with reference to a standard dilution curve. All mRNA quantitation data were normalised to hydroxymethylbilane synthase (HMBS). Following the PCR, the products were melted to confirm specificity and electrophoresed on 1.5% (w/v) agarose gels stained with ethidium bromide to confirm expected product size.
Statistical analyses
Statistics were performed using SAS Version 9.2 (SAS Institute Inc., Cary, NC, USA) and Prism version 6.0b for Macintosh (GraphPad Software, San Diego CA, USA; www.graphpad.com). The median overall survival and the 5-year survival were calculated using the Kaplan–Meier estimator. Kaplan–Meier survival curves were compared using the Log-rank (Mantel–Cox) test. Hazard ratios (HRs), 95% confidence intervals (CIs), and P-values were calculated from univariate and multivariate Cox proportional hazards models. Gene expression in cell lines comparing treatment with aza to veh was performed using multiple t-tests and significance determined using the Holm–Sidak method, with alpha of 5%. Methylation frequency in tissues and the correlation between methylation and expression was compared using Fisher’s exact test. All statistics were considered significant when the two-tailed P-value was ⩽0.05.
Results
Expression of IGFBP7 in oesophageal adenocarcinoma and Barrett’s oesophagus
The protein expression of IGFBP7 was measured using immunohistochemistry in oesophageal adenocarcinoma resections from 65 patients. None of the patients had preoperative chemotherapy or radiotherapy. Follow-up data were available from all patients, with a median follow-up time of 33 months (range 1–149). The median patient age at surgery was 63 years (range 35–80). The overall 5-year survival rate was 28%, and the overall median survival was 27 months (range 1–129).
Positive staining for IGFBP7 in tumour cells was observed in 34 of 65 (52%) patients (Figure 1). In positive tumour cells, staining was localised to the cytoplasm and was uniformly of moderate intensity. Sporadic staining of positive stromal cells, including endothelial cells, was localised to the cytoplasm and ranged in intensity from weak to intense.
Immunohistochemistry of IGFBP7 in human oesophageal adenocarcinoma. Representative images of IGFBP7 immunohistochemistry displaying (A) negative expression in tumour cells and stroma; (B) negative expression in tumour cells, and positive in stroma; (C) positive expression in tumour cells, but negative in stroma; (D) positive expression in tumour cells and stroma.
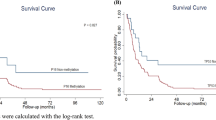
Expression of IGFBP7 protein was associated with decreased patient survival (Figure 2). With univariate analysis, IGFBP7 positivity, advanced pT stage, lymph node metastases, a positive resection margin, and vascular and perineural invasion were significantly associated with decreased survival (Table 1). Expression of IGFBP7 was associated with a significant decrease in the median overall survival (11 vs 92 months; P-value=0.0140) and decreased 5-year survival (24.5% vs 51.6%), (HR=2.16; 95% CI=1.14–4.09; P-value=0.0177).
Owing to the limited number of patients, only IGFBP7 expression, pT, pN, and positive resection margin were retained in the multivariate model (Table 1). The prognostic value of IGFBP7 expression, pT stage, and a positive resection margin were maintained even after adjustment for the other covariates, supporting the independent role of these markers in predicting survival. Survival was significantly reduced for patients who were positive compared with those who were negative for IGFBP7 expression (HR=3.25; 95% CI=1.58–6.67; P-value=0.0014).
Next, we analysed IGFBP7 mRNA expression in normal oesophagus, Barrett’s oesophagus, and oesophageal adenocarcinoma in published transcription microarray data sets. There was a significant increase in IGFBP7 mRNA in adenocarcinoma (four of four data sets) and Barrett’s oesophagus (three of four), compared with normal oesophageal tissue (Figure 3). Despite the significant increase in expression in oesophageal adenocarcinoma, it was clear that there was a wide range of expression from low to high, consistent with our finding that IGFBP7 was only expressed in a subset of patients. Some clinical data were available for one of the public data sets (Kim et al, 2010), and the expression of IGFBP7 was significantly increased in stage 3 tumours compared with stage 0; however, no other significant differences were observed.
Expression of IGFBP7 in oesophageal tissues. Expression of IGFBP7 in histologically normal squamous mucosa (Normal), Barrett’s oesophagus (Barrett’s), and oesophageal adenocarcinoma (OAC) tissues was determined using transcriptional microarrays in four different data sets as named and published on Oncomine.
DNA methylation and expression of IGFBP7 in oesophageal cell lines
The melt curve analysis was used to measure the extent of DNA methylation, and QPCR to measure expression, of IGFBP7 in three oesophageal cancer cell lines. OE21 and TE7 were unmethylated and expressed IGFBP7, whereas OE33 was methylated and did not express the gene (Figure 4). When OE33 was grown in the presence of aza, the extent of DNA methylation decreased, and the expression of IGFBP7 significantly increased (P-value <0.0001). In contrast, aza did not significantly alter IGFBP7 DNA methylation or expression in either OE21 and TE7. Methylation of IGFBP7 in OE33 was also assessed on an Infinium HumanMethylation450 BeadChip Kit (Figure 4C). All CpG probes assessed within the promoter region (−500 to +1000 nt from the transcription start site) were heavily methylated, with beta-values >0.8. Together, these results are consistent with methylation of the region of IGFBP7 assessed with the melt curve analysis being associated with silencing of its expression in oesophageal cell lines.
Expression and DNA methylation of IGFBP7 in oesophageal cell lines. The Barrett’s associated oesophageal adenocarcinoma (OE33) and oesophageal squamous cell carcinoma (OE21 and TE7) cell lines were treated with either vehicle (veh) or 1 μmol l−1 5-aza-2’-deoxycytidine (aza) and analysed for (A) expression of IGFBP7, measured by QPCR (mean±s.e.m. of three replicates), and (B) DNA methylation, measured with the melt curve analysis (mean±s.e.m. of two to three replicates). (C) Methylation of individual CpGs in the IGFBP7 promoter was measured in OE33 using an Infinium HumanMethylation450 BeadChip Kit. The position of individual CpGs was determined relative to the transcription start site (TSS) of IGFBP7 (assembly GRCh37/hg19, chr4:57898404-57976551). The black horizontal bar denotes the region analysed with the melt curve analysis. (D) Methylation, measured with the melt curve analysis, in oesophageal adenocarcinoma (OE19 and JH-EsoAd1), Barrett’s oesophagus (CP-A, CP-B, and CP-D) and squamous epithelial cell lines (mean±s.e.m. of two to three replicates).
Methylation was also assessed in the following oesophageal cell lines: oesophageal adenocarcinoma (OE19, JH-EsoAd1), telomerase immortalised non-dysplastic (CP-A), and high-grade dysplastic (CP-B, CP-D) Barrett’s oesophagus, and immortalised squamous epithelium (HET1A). Methylation was detected in OE19, JH-EsoAd1, and CP-A, but not in CP-B, CP-D nor HET1A (Figure 4D). We found that methylation of IGFBP7 is a common feature of Barrett’s and oesophageal adenocarcinoma cell lines.
DNA methylation of IGFBP7 in Barrett’s oesophagus and oesophageal adenocarcinoma tissues
Methylation was determined using the melt curve analysis in biopsies of squamous mucosa and columnar lined oesophagus from 18 patients with Barrett’s oesophagus. Methylation was detected in significantly more biopsies of columnar lined oesophagus (27 of 40; 68%) than squamous mucosa (2 of 15; 13%) (P-value=0.0006). Methylation was detected in at least one biopsy of columnar lined oesophagus in 14 of 18 (78%) patients with Barrett’s oesophagus.
Methylation was determined in biopsies of tumour, and when available, adjacent high-grade dysplasia, and histologically normal squamous proximal resection margin from 34 patients with oesophageal adenocarcinoma. Compared with squamous mucosa (3 of 17; 18%), methylation was detected in significantly more biopsies of high-grade dysplasia (5 of 7; 71%) (P-value=0.0207) and oesophageal adenocarcinoma (23 of 34; 68%) (P-value=0.0010). Together, these results demonstrate that IGFBP7 methylation is infrequent in squamous mucosa but is common in Barrett’s oesophagus and oesophageal adenocarcinoma tissue.
Matched DNA methylation and expression data were available for 22 of the 65 patients. Methylation was detected in seven out of seven (100%) patients who did not express IGFBP7, compared with 47% (7 of 15) patients who did (P-value=0.0225). Survival was reduced in patients with unmethylated IGFBP7 (9 vs 56 months; Figure 5); however, this difference was not significant (log-rank test, P-value=0.071; Cox proportional hazards model, HR=0.46, 95% CI=0.19–1.10).
Discussion
We have demonstrated that, in patients with oesophageal adenocarcinoma, protein expression of IGFBP7 was associated with significantly reduced survival, independent of other clinicopathological features. In both oesophageal adenocarcinoma and Barrett’s cell lines, DNA methylation within the IGFBP7 promoter was associated with transcriptional silencing. In patient samples, DNA methylation of IGFBP7 was infrequent in squamous mucosa, but a common feature in Barrett’s and oesophageal adenocarcinoma, and it correlated with silencing of protein expression.
The levels of expression of IGFBP7 differ among different cancer types. Increased expression has been observed in glioblastoma multiforme (Jiang et al, 2008), oesophageal squamous cell (Kashyap et al, 2012), and colorectal carcinoma (Umeda et al, 1998). In Barrett’s and oesophageal adenocarcinoma samples we found, from an analysis of four independent expression array data sets in public repositories, that IGFBP7 transcript expression was increased in a proportion of patient samples compared with their squamous mucosa, consistent with our immunohistochemical findings. In contrast, decreased expression has been reported in cancers of the breast (Burger et al, 1998), lung (Chen et al, 2007), prostate (Hwa et al, 1998), pancreas (An et al, 2012), liver (Chen et al, 2011a), and thyroid (Vizioli et al, 2010), raising the possibility that, at least in some types of cancer, IGFBP7 may function as a tumour suppressor. A limitation of our correlative study is that the data do not allow any conclusions to be reached regarding the possible biological role of IGFBP7 in oesophageal adenocarcinoma.
Decreased IGFBP7 expression has been associated with DNA methylation in a range of cancers including T-lymphoblastic leukaemia (Heesch et al, 2011), liver (Komatsu et al, 2000), lung (Chen et al, 2011b; Suzuki et al, 2013), prostate (Sullivan et al, 2012), and colorectal (Ruan et al, 2006; Lin et al, 2007, 2008; Hinoue et al, 2009; Suzuki et al, 2010; Dimberg et al, 2013), as well as premalignant colorectal adenomas (Kaji et al, 2012). In our study, methylation was detected in oesophageal adenocarcinoma and Barrett’s oesophagus cell lines. The methylation was associated with decreased transcript expression, and demethylation with 5-aza-2′-deoxycytidine resulted in IGFBP7 re-expression. Methylation was more frequently observed in primary Barrett’s and oesophageal adenocarcinoma tissues than in adjacent histologically normal squamous mucosa. Finally, methylation in adenocarcinoma was more likely to be associated with a lack of protein expression. Taken together, these data demonstrate that DNA methylation of the IGFBP7 promoter is associated with the silencing of its expression and is common in Barrett’s and adenocarcinoma of the oesophagus.
Reports on the prognostic significance of IGFBP7 expression in cancer are conflicting. In accordance with our findings, increased expression of IGFPB7 was associated with decreased patient survival in colorectal (Adachi et al, 2001), T-acute lymphoblastic leukaemia (Heesch et al, 2010), and head and neck squamous cell carcinoma (Sepiashvili et al, 2012). In contrast, increased expression correlated with increased survival in pancreatic ductal (An et al, 2012), breast (Landberg et al, 2001), hepatocellular (Tomimaru et al, 2012), and colorectal carcinoma (Ruan et al, 2007). These conflicting observations may be because of heterogeneous expression within the cancer. For example, Adachi et al (2001) reported that expression was greatest in the cells located at the invasive front of tumour nests, and that it was the expression in these cells that correlated with poor patient prognosis, independent of other clinicopathological covariates. In our study, we could not be confident that the increased expression that we observed was restricted to the cells at the invasive front because we stained TMAs that were constructed using 1 mm cores taken from representative regions of the tumour, which did not always include the invasive front.
In conclusion, expression of IGFBP7 was an independent prognostic indicator for decreased patient survival in oesophageal adenocarcinoma. DNA methylation within the IGFBP7 promoter was associated with transcriptional silencing in oesophageal adenocarcinoma and Barrett’s cell lines, was a common feature in Barrett’s and oesophageal adenocarcinoma tissues, and correlated with silencing of protein expression.
References
Adachi Y, Itoh F, Yamamoto H, Arimura Y, Kikkawa-Okabe Y, Miyazaki K, Carbone DP, Imai K (2001) Expression of angiomodulin (tumor-derived adhesion factor/mac25) in invading tumor cells correlates with poor prognosis in human colorectal cancer. Int J Cancer 95 (4): 216–222.
Alvarez H, Koorstra JB, Hong SM, Boonstra JJ, Dinjens WN, Wu TT, Montgomery E, Eshleman JR, Maitra A (2008) Establishment and characterization of a bona fide barrett esophagus-associated adenocarcinoma cell line. Cancer Biol Ther 7 (11): 1753–1755.
An W, Ben QW, Chen HT, Zheng JM, Huang L, Li GX, Li ZS (2012) Low expression of IGFBP7 is associated with poor outcome of pancreatic ductal adenocarcinoma. Ann Surg Oncol 19 (12): 3971–3978.
Bhat S, Coleman HG, Yousef F, Johnston BT, McManus DT, Gavin AT, Murray LJ (2011) Risk of malignant progression in Barrett's esophagus patients: results from a large population-based study. J Natl Cancer Inst 103 (13): 1049–1057.
Burger AM, Leyland-Jones B, Banerjee K, Spyropoulos DD, Seth AK (2005) Essential roles of IGFBP-3 and IGFBP-rP1 in breast cancer. Eur J Cancer 41 (11): 1515–1527.
Burger AM, Zhang X, Li H, Ostrowski JL, Beatty B, Venanzoni M, Papas T, Seth A (1998) Down-regulation of T1A12/mac25, a novel insulin-like growth factor binding protein related gene, is associated with disease progression in breast carcinomas. Oncogene 16 (19): 2459–2467.
Chen D, Yoo BK, Santhekadur PK, Gredler R, Bhutia SK, Das SK, Fuller C, Su ZZ, Fisher PB, Sarkar D (2011a) Insulin-like growth factor-binding protein-7 functions as a potential tumor suppressor in hepatocellular carcinoma. Clin Cancer Res 17 (21): 6693–6701.
Chen Y, Cui T, Knosel T, Yang L, Zoller K, Petersen I (2011b) IGFBP7 is a p53 target gene inactivated in human lung cancer by DNA hypermethylation. Lung Cancer 73 (1): 38–44.
Chen Y, Pacyna-Gengelbach M, Ye F, Knosel T, Lund P, Deutschmann N, Schluns K, Kotb WF, Sers C, Yasumoto H, Usui T, Petersen I (2007) Insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1) has potential tumour-suppressive activity in human lung cancer. J Pathol 211 (4): 431–438.
Dimberg J, Hong TT, Skarstedt M, Lofgren S, Zar N, Matussek A (2013) Analysis of APC and IGFBP7 promoter gene methylation in Swedish and Vietnamese colorectal cancer patients. Oncol Lett 5 (1): 25–30.
Doyle SL, Donohoe CL, Finn SP, Howard JM, Lithander FE, Reynolds JV, Pidgeon GP, Lysaght J (2012) IGF-1 and its receptor in esophageal cancer: association with adenocarcinoma and visceral obesity. Am J Gastroenterol 107 (2): 196–204.
Heesch S, Bartram I, Neumann M, Reins J, Mossner M, Schlee C, Stroux A, Haferlach T, Goekbuget N, Hoelzer D, Hofmann WK, Thiel E, Baldus CD (2011) Expression of IGFBP7 in acute leukemia is regulated by DNA methylation. Cancer Sci 102 (1): 253–259.
Heesch S, Schlee C, Neumann M, Stroux A, Kuhnl A, Schwartz S, Haferlach T, Goekbuget N, Hoelzer D, Thiel E, Hofmann WK, Baldus CD (2010) BAALC-associated gene expression profiles define IGFBP7 as a novel molecular marker in acute leukemia. Leukemia 24 (8): 1429–1436.
Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, Whitehall VL, Leggett BA, Laird PW (2009) Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One 4 (12): e8357.
Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P (2011) Incidence of adenocarcinoma among patients with Barrett's esophagus. N Engl J Med 365 (15): 1375–1383.
Hwa V, Tomasini-Sprenger C, Bermejo AL, Rosenfeld RG, Plymate SR (1998) Characterization of insulin-like growth factor-binding protein-related protein-1 in prostate cells. J Clin Endocrinol Metab 83 (12): 4355–4362.
Jiang W, Xiang C, Cazacu S, Brodie C, Mikkelsen T (2008) Insulin-like growth factor binding protein 7 mediates glioma cell growth and migration. Neoplasia 10 (12): 1335–1342.
Kaji E, Uraoka T, Kato J, Hiraoka S, Suzuki H, Akita M, Saito S, Tanaka T, Ohara N, Yamamoto K (2012) Externalization of saw-tooth architecture in small serrated polyps implies the presence of methylation of IGFBP7. Dig Dis Sci 57 (5): 1261–1270.
Kashyap MK, Pawar HA, Keerthikumar S, Sharma J, Goel R, Mahmood R, Kumar MV, Kumar KV, Pandey A, Kumar RV, Prasad TS, Harsha HC (2012) Evaluation of protein expression pattern of stanniocalcin 2, insulin-like growth factor-binding protein 7, inhibin beta A and four and a half LIM domains 1 in esophageal squamous cell carcinoma. Cancer biomarkers: section A of Disease markers 12 (1): 1–9.
Kim SM, Park YY, Park ES, Cho JY, Izzo JG, Zhang D, Kim SB, Lee JH, Bhutani MS, Swisher SG, Wu X, Coombes KR, Maru D, Wang KK, Buttar NS, Ajani JA, Lee JS (2010) Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome. PLoS One 5 (11): e15074.
Komatsu S, Okazaki Y, Tateno M, Kawai J, Konno H, Kusakabe M, Yoshiki A, Muramatsu M, Held WA, Hayashizaki Y (2000) Methylation and downregulated expression of mac25/insulin-like growth factor binding protein-7 is associated with liver tumorigenesis in SV40T/t antigen transgenic mice, screened by restriction landmark genomic scanning for methylation (RLGS-M). Biochem Biophys Res Commun 267 (1): 109–117.
Landberg G, Ostlund H, Nielsen NH, Roos G, Emdin S, Burger AM, Seth A (2001) Downregulation of the potential suppressor gene IGFBP-rP1 in human breast cancer is associated with inactivation of the retinoblastoma protein, cyclin E overexpression and increased proliferation in estrogen receptor negative tumors. Oncogene 20 (27): 3497–3505.
Lim YY, Wright JA, Attema JL, Gregory PA, Bert AG, Smith E, Thomas D, Lopez AF, Drew PA, Khew-Goodall Y, Goodall GJ (2013) Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J Cell Sci 126 (Pt 10): 2256–2266.
Lin J, Lai M, Huang Q, Ma Y, Cui J, Ruan W (2007) Methylation patterns of IGFBP7 in colon cancer cell lines are associated with levels of gene expression. J Pathol 212 (1): 83–90.
Lin J, Lai M, Huang Q, Ruan W, Ma Y, Cui J (2008) Reactivation of IGFBP7 by DNA demethylation inhibits human colon cancer cell growth in vitro. Cancer Biol Ther 7 (12): 1896–1900.
Nishihira T, Hashimoto Y, Katayama M, Mori S, Kuroki T (1993) Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol 119 (8): 441–449.
Palanca-Wessels MC, Barrett MT, Galipeau PC, Rohrer KL, Reid BJ, Rabinovitch PS (1998) Genetic analysis of long-term Barrett’s esophagus epithelial cultures exhibiting cytogenetic and ploidy abnormalities. Gastroenterology 114 (2): 295–304.
Ruan W, Xu E, Xu F, Ma Y, Deng H, Huang Q, Lv B, Hu H, Lin J, Cui J, Di M, Dong J, Lai M (2007) IGFBP7 plays a potential tumor suppressor role in colorectal carcinogenesis. Cancer Biol Ther 6 (3): 354–359.
Ruan WJ, Lin J, Xu EP, Xu FY, Ma Y, Deng H, Huang Q, Lv BJ, Hu H, Cui J, Di MJ, Dong JK, Lai MD (2006) IGFBP7 plays a potential tumor suppressor role against colorectal carcinogenesis with its expression associated with DNA hypomethylation of exon 1. J Zhejiang Univ Sci B 7 (11): 929–932.
Sepiashvili L, Hui A, Ignatchenko V, Shi W, Su S, Xu W, Huang SH, O'Sullivan B, Waldron J, Irish JC, Perez-Ordonez B, Liu FF, Kislinger T (2012) Potentially novel candidate biomarkers for head and neck squamous cell carcinoma identified using an integrated cell line-based discovery strategy. Mol Cell Proteomics 11 (11): 1404–1415.
Smith E, De Young NJ, Pavey SJ, Hayward NK, Nancarrow DJ, Whiteman DC, Smithers BM, Ruszkiewicz AR, Clouston AD, Gotley DC, Devitt PG, Jamieson GG, Drew PA (2008) Similarity of aberrant DNA methylation in Barrett's esophagus and esophageal adenocarcinoma. Mol Cancer 7 (1): 75.
Smith E, Drew PA, Tian ZQ, De Young NJ, Liu JF, Mayne GC, Ruszkiewicz AR, Watson DI, Jamieson GG (2005) Metallothionien 3 expression is frequently down-regulated in oesophageal squamous cell carcinoma by DNA methylation. Mol Cancer 4: 42.
Smith E, Jones ME, Drew PA (2009) Quantitation of DNA methylation by melt curve analysis. BMC Cancer 9: 123.
Smith E, Kelly JJ, Ruskiewicz AR, Sullivan T, Jamieson GG, Drew PA (2010) The effect of long-term control of reflux by fundoplication on aberrant deoxyribonucleic acid methylation in patients with Barrett esophagus. Ann Surg 252 (1): 63–69.
Stoner GD, Kaighn ME, Reddel RR, Resau JH, Bowman D, Naito Z, Matsukura N, You M, Galati AJ, Harris CC (1991) Establishment and characterization of SV40 T-antigen immortalized human esophageal epithelial cells. Cancer Res 51 (1): 365–371.
Sullivan L, Murphy TM, Barrett C, Loftus B, Thornhill J, Lawler M, Hollywood D, Lynch T, Perry AS (2012) IGFBP7 promoter methylation and gene expression analysis in prostate cancer. J Urol 188 (4): 1354–1360.
Suzuki H, Igarashi S, Nojima M, Maruyama R, Yamamoto E, Kai M, Akashi H, Watanabe Y, Yamamoto H, Sasaki Y, Itoh F, Imai K, Sugai T, Shen L, Issa JP, Shinomura Y, Tokino T, Toyota M (2010) IGFBP7 is a p53-responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis 31 (3): 342–349.
Suzuki M, Shiraishi K, Eguchi A, Ikeda K, Mori T, Yoshimoto K, Ohba Y, Yamada T, Ito T, Baba Y, Baba H (2013) Aberrant methylation of LINE-1, SLIT2, MAL and IGFBP7 in non-small cell lung cancer. Oncol Rep 29 (4): 1308–1314.
Thompson SK, Sullivan TR, Davies R, Ruszkiewicz AR (2011) Her-2/neu gene amplification in esophageal adenocarcinoma and its influence on survival. Ann Surg Oncol 18 (7): 2010–2017.
Tomimaru Y, Eguchi H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Umeshita K, Kim T, Wakasa K, Doki Y, Mori M, Nagano H (2012) IGFBP7 downregulation is associated with tumor progression and clinical outcome in hepatocellular carcinoma. Int J Cancer 130 (2): 319–327.
Umar SB, Fleischer DE (2008) Esophageal cancer: epidemiology, pathogenesis and prevention. Nat Clin Pract Gastroenterol Hepatol 5 (9): 517–526.
Umeda F, Ono Y, Sekiguchi N, Hashimoto T, Masakado M, Nakamura K, Chijiiwa Y, Nawata H (1998) Increased mRNA expression of a novel prostacyclin-stimulating factor in human colon cancer. J Gastroenterol 33 (2): 213–217.
Underwood TJ, Derouet MF, White MJ, Noble F, Moutasim KA, Smith E, Drew PA, Thomas GJ, Primrose JN, Blaydes JP (2010) A comparison of primary oesophageal squamous epithelial cells with HET-1A in organotypic culture. Biol Cell 102 (12): 635–644.
Vizioli MG, Sensi M, Miranda C, Cleris L, Formelli F, Anania MC, Pierotti MA, Greco A (2010) IGFBP7: an oncosuppressor gene in thyroid carcinogenesis. Oncogene 29 (26): 3835–3844.
Acknowledgements
This study was supported by the Flinders University School of Nursing and Midwifery Industry Partnership Research Grant. The Barrett’s associated adenocarcinoma cell line OE33 was a gift from Associate Professor Wayne A Phillips, Peter MacCallum Cancer Research Centre, Melbourne, Australia. The squamous cell carcinoma cell line TE7 was obtained from Dr Tetsuro Nishihira, Tohoku University School of Medicine, Sendai, Japan. The immortalised Barrett’s oesophagus cell lines were a gift from Professor Peter S Rabinovitch, University of Washington, Seattle, Washington, USA.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Smith, E., Ruszkiewicz, A., Jamieson, G. et al. IGFBP7 is associated with poor prognosis in oesophageal adenocarcinoma and is regulated by promoter DNA methylation. Br J Cancer 110, 775–782 (2014). https://doi.org/10.1038/bjc.2013.783
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.783
Keywords
This article is cited by
-
Angiogenesis modulated by CD93 and its natural ligands IGFBP7 and MMRN2: a new target to facilitate solid tumor therapy by vasculature normalization
Cancer Cell International (2023)
-
Association of insulin-like growth factor binding protein-7 promoter methylation with esophageal cancer in peripheral blood
Molecular Biology Reports (2022)
-
A systematic review and meta-analysis of prognostic biomarkers in resectable esophageal adenocarcinomas
Scientific Reports (2018)
-
Fibroblasts derived from oesophageal adenocarcinoma differ in DNA methylation profile from normal oesophageal fibroblasts
Scientific Reports (2017)
-
Modulated DISP3/PTCHD2 expression influences neural stem cell fate decisions
Scientific Reports (2017)