Abstract
Background:
Chemokines and chemokine receptors not only have significant roles in cancer metastasis and tumorigenesis but also act as antitumour agents. The interaction between the Crk-like adaptor protein (CrkL), which is encoded by the CRKL gene, and non-receptor tyrosine kinase c-ABL is reported to transform many cells into malignant cells. We examined the effects of CC chemokine receptor 7 (CCR7), CCR7 ligands and CrkL and c-ABL in lung adenocarcinoma.
Methods:
One hundred and twenty patients with lung adenocarcinoma were included in this historical cohort analysis. We examined CCR7 and CCR7 ligands and CrkL and c-ABL mRNA expressions in surgically resected lung adenocarcinoma specimens and evaluated their contribution to prognosis, and the relationship with epidermal growth factor receptor (EGFR) and TP53 mutations.
Results:
High CCR7 mRNA expressions indicated better prognoses than those of the groups with low CCR7 mRNA expressions (P=0.007, HR=2.00, 95% CI of ratio: 1.22 –3.31). In lung adenocarcinoma, CrkL and c-ABL mRNAs were related to CCR7 mRNA expression (P<0.0001). CrkL and c-ABL mRNA expressions were influenced by EGFR mutations. A high expression of CCL19 was a good prognostic factor of lung adenocarcinoma.
Conclusion:
We propose that CCR7 and CCL19 are clinically good prognostic factors and that CCR7 is strongly related to CrkL and c-ABL kinase mRNA expression in lung adenocarcinoma.
Similar content being viewed by others
Main
Lung cancer is one of the leading causes of malignancy-related death worldwide. Non-small-cell lung cancer accounts for 80% of lung cancer cases, with adenocarcinoma being the major subtype. The virulence of this cancer is mediated in part by its specific and aggressive metastasis to regional lymph nodes, brain, bone marrow, liver, adrenal grand and contralateral lung. Indeed, <15% of lung cancer patients survive beyond 5 years after diagnosis (Breuer et al, 2005).
Chemokines belong to a superfamily of small proteins that bind to and activate a family of chemokine receptors. In particular, the interaction between a chemokine receptor and its ligand has a very significant role in cancer metastasis and tumorigenesis (Müller et al, 2001; Richmond, 2002). CC chemokine receptor 7 (CCR7) is a G-protein-coupled receptor for the CC chemokine ligands CCL21 and CCL19. CC chemokine receptor 7 is preferentially expressed on naive T cells and mature dendritic cells, and the CCR7–CCL21 axis contributes to the acquired immune response (Itakura et al, 2001). Many reports have revealed that the expression of the chemokine receptor CCR7 on cancer cells induces metastasis to regional lymph nodes (Müller et al, 2001; Andre et al, 2006; Cabioglu et al, 2007; Koizumi et al, 2007). On the other hand, Cao et al (2007) reported an antitumour effect of CCL19. Therefore, we became interested in the function of the CCR7–ligands axis, which appears to be associated with both good and bad outcomes in cancer prognosis. None of these earlier studies have examined the expression profiles of these chemokines and their CCR7 receptor in lung adenocarcinoma patients who had undergone resection.
Many gene amplifications and fusion of genes have been reported to be the major causes of lung cancer. The CRKL gene encodes an adaptor protein (CrkL) that has functions in signal transduction, and is best known as a substrate for the BCR-ABL kinase in chronic myelogenous leukaemia (CML) (Sattler and Salgia, 1998). The CrkL Src homolog 3 domain binds constitutively to the non-receptor tyrosine kinase c-ABL in haematopoietic and non-haematopoietic cells (Sattler and Salgia, 1998). c-ABL is involved in the regulation of cell proliferation, cell survival, cell adhesion, cell migration and apoptosis (Sirvent et al, 2008). Kim et al (2010) reported that RNA-interference-mediated knockdown of CrkL in lung cancer cell lines with amplification led to significantly decreased cell proliferation, cell-cycle progression, cell survival, and cell motility and invasion. So far, no report has described an association between CCR7 chemokine receptor and c-ABL and CrkL in lung cancer; however, if a link between these entities could be established, it might lead to a better understanding of the growth mechanism and metastasis in lung cancer.
The epidermal growth factor receptor (EGFR) is a transmembrane phosphoglycoprotein that has been identified in almost all adult tissues, with the exception of haematopoietic cells, and overexpression and constitutive activation of EGFR occurs in several solid human cancers, including lung adenocarcinoma (Breuer et al, 2005). Some authors have reported on a relationship between EGFR gene mutations and CrkL and c-ABL functions (Tanos and Pendergast, 2006; Cheung et al, 2011). TP53 is a tumour suppressor gene, and its mutation in genomic DNA causes many malignant diseases, including lung cancer (Breuer et al, 2005).
In this report, we examined and evaluated the expressions of chemokine receptor CCR7 and the ligands (CCL19 and CCL21) CrkL and c-ABL in lung adenocarcinoma specimens, as well as their effect on the prognosis after surgical operation in a historical cohort study. We also analysed the effects of EGFR and TP53 gene mutations on the CrkL and c-ABL mRNA expressions. Moreover, we examined the distributions of CCR7, CrkL and c-ABL proteins by immunohistochemistry in the lung adenocarcinoma specimens.
Materials and methods
Patients and tissue samples
This study was conducted at Chiba Cancer Center according to the principles of the Declaration of Helsinki with its current amendments. The study was approved by the institutional review boards/ethics committees of Chiba Cancer Center. All patients provided written informed consent. A total of 120 consecutive patients with adenocarcinoma who were treated in the Department of Thoracic Disease, Chiba Cancer Center from 1997 to 2004 were randomly selected for this analysis. Clinical data for patients involved were obtained from patient medical records, chest X-ray films, whole-body computed tomography films, bone scanning data and operation records.
A total of 120 patients underwent complete resection of the lobe or segment in which the tumour resided, and received hilar and mediastinal lymph node dissections. All tissue samples were immediately snap-frozen in liquid nitrogen and stored at −80 °C until use.
For histological examination, samples were fixed in 10% (v v−1) formalin and then embedded in paraffin. A 4-μm section was prepared from each sample and stained with haematoxylin and eosin.
Pathological staging was determined using the current tumour node metastasis classification system (International Association for the Study of Lung Cancer). The histological type and the grade of cell differentiation in these patients were determined using the pathological diagnosis by two pathologists at the Chiba Cancer Center of Pathology, and then confirmed by one pathologist specialising in lung pathology according to the World Health Organization Classification System 2004.
RNA isolation and RT–PCR analysis
Total RNA was isolated from the resected specimens during the operation using TRIzol (Invitrogen, Carsbad, CA, USA) and in accordance with the manufacturer’s instructions.
To digest the contaminating DNA, the RNA extract was incubated with RNase-free DNase I (Amersham Pharmacia Biotech, Uppsala, Sweden) during RNA isolation. Reverse transcription (RT) of total RNA was performed using Ready-To-Go You-Prime First-Strand Beads (Amersham Biosciences, Piscataway, NJ, USA) and random hexamers (Amersham Biotech, USA). To quantify the expression of the chemokine ligand and receptor genes, real-time polymerase chain reaction (PCR) was performed using the ABI 7700 (Perkin-Elmer Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. The sense and antisense primers used for quantitative amplification of CCR7, CCL19, CCL21, CrkL and c-ABL mRNAs and those used for amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), used as an internal control, are provided in the Supplementary Table 1.
The quantification data were analysed with the Light Cycler analysis software version 5.03 (Rosche Applied Science, Penzberg, Germany). Each mRNA expression level was standardised with GAPDH mRNA expression, and then they were indicated as the ratio to the lowest expression sample in the gene.
EGFR gene and TP53 gene mutation detection
Genomic DNA was extracted from each frozen tumour sample, and each exon of the EGFR gene (exons 18–21) and TP53 gene (exons 4–8) was amplified by PCR (Shingyoji et al, 2011).
Immunohistochemical analysis
For immunohistochemical (IHC) staining of CrkL, c-ABL and CCR7, formalin-fixed and paraffin-embedded (FFPE) tissue sections were heated at 98 °C for 20 min and then rinsed with PBS. The sections were incubated with goat anti-CCR7 polyclonal antibody (8 μg ml−1; Abcam, Cambridge, MA, USA), antiphospho CrkL (Tyr207) antibody (20 μg ml−1; Cell Signaling Technology, Danvers, MA, USA), antiphospho (Y412) c-ABL antibody (20 μg ml−1; Abcam) and control goat IgG antibody (8 μg ml−1; Beckman Coulter, Brea, CA, USA) for a negative control, as recommended by the manufacturer or suppliers.
Following staining by biotin-conjugated secondary antibody, horseradish peroxidase–streptavidin were applied to the samples, counterstaining was performed and the staining pattern was evaluated by an expert pathologist. All tissue sections of IHC staining were scanned by the NanoZoomer scan system (Hamamatsu Photonics, Hamamatsu, Japan). All IHC analyses were carried out by the co-workers and medical technologists, and the analyses were supervised by expert pathologists.
Laser capture microdissection, RNA isolation and RT from FFPE sections
After toluidine blue staining, dissection was performed using a laser capture microdissection microscope (Leica AS LMD; Leica Microsystems, Wetzlar, Germany) with a pulsed 337-nm UV laser (Leica Microsystems). Tumour sites and normal lungs in FFPE tissue sections were dissected and collected into individual tubes filled with PKD buffer of the RNeasy FFPE kit (Qiagen, Venlo, Netherlands). Total RNA was purified with the RNeasy FFPE kit (Qiagen). Complementary DNA synthesis was performed with a Reverse Transcription Kit (Takara, Shiga, Japan), and then quantitative PCR was carried out as described above.
Detection of CCR7 expression area in tissue sections by IHC staining
Twenty FFPE samples were selected from each of 10 upper and lower CCR7 mRNA expression samples in the 120 samples analysed in this examination. To measure CCR7 expression areas in lung cancer, the whole tissue sections of IHC staining were scanned by the NanoZoomer scan system (Hamamatsu Photonics, Hamamatsu, Japan). Then, five images of 3.5 × 1.96 mm were selected from the scanned image data by the NDP image software (Hamamatsu Photonics). In the ImageJ analysis software, the ImageJ plugin for colour deconvolution has a built-in vector for separating haematoxylin (H) and diaminobenzidine (DAB) staining (Konsti et al, 2011). After colour deconvolution, DAB images were processed. Thresholding creates binary masks of DAB-positive areas. The number of DAB-positive pixels was calculated. The results were indicated as the percentage of DAB-positive pixels in all the pixels of the image. The mean value of five images was calculated for 20 tissue samples. The results were indicated as percentages of the mean value of 10 tissue samples of CCR7 mRNA high-expression (CCR7-hi) and low-expression (CCR7-lo) groups.
Statistical analysis
Box and whisker plots summarise the values of distributions in this study: the central box covers the interquartile range, with the median indicated by the line within the box. The whiskers extend either to the 10 and 90% percentile values or to the most extreme values within 1.5 interquartile ranges of the quartiles, in which case more extreme values are plotted individually (Figures 1C–F, 2C–F and ).
Effects of CrkL and c-ABL on the survival of the patients with surgically resected lung adenocarcinoma. (A) Overall survival in relation to high (hi) and low (lo) CrkL mRNA expression in the tumour site (P=0.42, HR=0.81, 95% CI of ratio 0.49–1.34). (B) Overall survival in relation to hi and lo c-ABL mRNA expression in the tumour site (P=0.72, HR=0.91, 95% CI of ratio 0.55–1.50). In (C – F), the effects of EGFR and TP53 gene mutations on CrkL and c-ABL mRNA expression in surgically resected lung adenocarcinoma specimens are indicated. The values of CrkL and c-ABL mRNA expression according to EGFR gene mutation status are indicated as box and whisker plots (C and D, respectively). The values of c-ABL and CrkL mRNA expression according to TP53 gene mutation status are indicated as box and whisker plots (E and F respectively). *P-value is <0.01.
Effects of CCR7 in patients with surgically resected lung adenocarcinoma. (A) Overall survival in relation to high (hi) and low (lo) CCR7 mRNA expression in the tumour site (P=0.007, HR=0.50, 95% CI of ratio 0.30–0.82). (B) Recurrence-free survival in relation to hi and lo CCR7 mRNA expression in the tumou site (P=0.007, HR=0.50, 95% CI of ratio 0.30–0.82). Two groups were formed according to the CrkL and c-ABL mRNA expression status, and then the values of CCR7, c-ABL and CrkL mRNA relative quantities were evaluated within each group (C–F). *P-value is <0.0001.
The median value of the series was used to classify each case as having high or low CCR7, CrkL and c-ABL mRNA expression, and to compare their relationships with the other biological parameters and clinical follow-up. The significance of differences in categorical data was tested using the χ2 test. Differences between continuous variables were examined using the Student’s t-test if the continuous variables were normally distributed, or using Mann–Whitney’s U-test if not normally distributed. Comparison among three continuous variables was performed using analysis of variance. Survival was defined as the time from surgery to death from any cause. Time to recurrence was defined as the time from surgery to documented disease progression or cancer-specific death. The postoperative survival rate and recurrence-free survival rate were analysed using the Kaplan–Meier (Mantel–Cox test) method with the completely resected 120 patients, and the differences between groups were assessed by the log-rank test.
Prism 5 (GraphPad Software Inc., San Diego, CA, USA) was used to perform statistical calculations. The analysis of prognostic factors was performed using the Cox proportional hazard model by Prism 5, and Spearman’s rank correlation coefficient was analysed by Prism 5. Differences were considered to be statistically significant if the P-value was <0.05.
Results
Patient status in tumour tissues obtained from lung adenocarcinoma
As shown in Table 1, mutation detection analysis of the EGFR and TP53 genes were evaluated in 120 patients’ lung specimens. Our results showed that EGFR gene mutations existed in 57 of 120 patients (47.5%). TP53 mutations were detected in 53 of 120 patients (44.2%) (Table 1). Epidermal growth factor receptor mutations had a significantly high frequency in non-smokers and women, but no significant differences were found between higher age and lower age (data not shown).
No significant difference between high and low CrkL and c-ABL mRNA expressions in lung adenocarcinoma patients related to overall survivals
To evaluate the prognostic significance of CrkL mRNA and c-ABL mRNA expression status, patients were divided into groups with higher or lower mRNA expression (CrkL-hi and CrkL-lo, c-ABL-hi and c-ABL-lo), with the median value as a cutoff. High and low CrkL and c-ABL mRNA expressions related to clinical characteristics and gene mutation status are indicated in Table 2. In the overall survival rates, there were no significant differences between patients with higher or lower CrkL mRNA (P=0.42, HR=0.81, 95% CI of ratio: 0.49–1.34), or c-ABL mRNA (P=0.72, HR=0.91, 95% CI of ratio: 0.55–1.50) expression (Figure 1A and B, respectively).
Correlation between EGFR gene mutations and CrkL and c-ABL mRNA expression
The influence of EGFR and TP53 mutations on CrkL and c-ABL mRNA expressions in lung cancer specimens was examined. The EGFR mutation-positive group (n=57) showed significantly higher CrkL and c-ABL mRNA expressions (Figure 1C and D) than did the EGFR mutation-negative group (n=63). On the other hand, there were no significant differences in CrkL and c-ABL mRNA expressions between TP53 mutation-positive (n=53) and -negative (n=67) groups (Figure 1E and F).
CCR7-hi mRNA expression is a good prognostic factor in lung adenocarcinoma patients for overall and recurrence-free survival
To evaluate the prognostic significance of CCR7 mRNA expression status, patients were divided into two groups with higher or lower mRNA expression, respectively, with the median value as a cutoff. The clinical characteristics and gene mutation status of each group of CCR7-hi mRNA expression and CCR7-lo mRNA expression are indicated in Table 2. There were no significant differences in the nodal status and lymphatic invasion between the CCR7-hi and CCR7-lo samples (Supplementary Table 2). In the overall survival rate, there was a significantly better prognosis for patients with higher CCR7 mRNA expression compared with that for patients with lower CCR7 mRNA expression (P=0.007, HR=0.50, 95% CI of ratio: 0.30–0.82) (Figure 2A). Similar results were seen in recurrence-free survival rates for higher and lower CCR7 mRNA expression (P=0.007, HR=0.50, 95% CI of ratio: 0.30–0.82) (Figure 2B).
CCR7 mRNA expression was related to CrkL and c-ABL mRNA expressions in lung adenocarcinoma specimens
The correlation between CrkL and c-ABL mRNA expression with the level of chemokine receptor CCR7 mRNA expression was examined. A total of 120 patients were divided into two groups CrkL-hi mRNA expression (n=60) and CrkL-lo mRNA expression (n=60), respectively. As shown in Figure 2C, the mean value of CCR7 mRNA in the CrkL-hi mRNA expression group was significantly higher than that of the CrkL-lo mRNA expression group (P<0.0001). Furthermore, CCR7 mRNA of the c-ABL-hi group was also significantly higher than that of the c-ABL-lo group (Figure 2D; P<0.0001).
A statistical correlation analysis indicated significant correlation between CCR7 mRNA and CrkL mRNA expressions by Spearman’s rank correlation coefficient (Table 3). In addition, a significant correlation was seen between expressions of the mRNAs for CCR7 and c-ABL, and between CrkL and c-ABL (Table 3).
Combination of CCR7-hi mRNA expression and CrkL-lo or c-ABL-lo mRNA expression was a good prognostic factor in lung adenocarcinoma specimens
Next, we evaluated the influence of the combination of CrkL and c-ABL with CCR7 mRNA expression on the lung cancer prognosis. A total of 120 patients were divided into two groups, one with CrkL-hi mRNA expression and the other with CrkL-lo mRNA expression. In addition, each group was divided into two subgroups according to CCR7 mRNA expression in their lung adenocarcinoma specimens. The same division and analysis were carried out for c-ABL and CCR7. The CrkL-lo and CCR7-hi mRNA expression groups showed a significantly higher survival rate than the CCR7-lo and CrkL-lo mRNA expression group (P=0.0006, hazard Ratio=0.29, 95% CI of ratio: 0.14–0.59) (Figure 3A). In addition, the combination of c-ABL-lo mRNA and CCR7-hi mRNA expression resulted in a higher survival rate than those of the CCR7-lo and c-ABL-lo mRNA expression group (P=0.014, hazard ratio=0.41, 95% CI of ratio: 0.20–0.84) (Figure 3B). However, we could not find significant differences of survival rates between the other combination groups in Figure 3A and B.
Effect of combinations between CCR7 and CrkL and c-ABL mRNA expression on the survival of patients with surgically resected lung adenocarcinoma. (A) Overall survival of patients with various combinations of CCR7 and CrkL mRNA expression status. The CrkL-lo and CCR7-hi mRNA expression groups showed a significantly higher survival rate than the CCR7-lo and CrkL-lo mRNA expression groups (P=0.0006, hazard Ratio (HR)=0.29, 95% CI of ratio: 0.14–0.59) (CCR7-hi/CrkL-hi, n=42; CCR7-hi/CrkL-lo, n=18; CCR7-lo/CrkL–hi, n=18; CCR7-lo/CrkL–lo, n=42). (B) Overall survival in relation to the combination of CCR7 and c-ABL mRNA expression status. The combination of c-ABL-lo mRNA and CCR7-hi mRNA expression resulted in a significantly higher survival rate than that of the CCR7-lo and c-ABL-lo mRNA expression groups (P=0.014, HR=0.41, 95% CI of ratio: 0.20–0.84) (CCR7-hi/c-ABL–hi, n=39; CCR7-hi/c-ABL–lo, n=21; CCR7-lo/c-ABL–hi, n=21; CCR7-lo/c-ABL–lo, n=39).
CCL19-hi mRNA expression is a good prognostic factor in lung adenocarcinoma patients for overall survival and recurrence-free survival
Because a CCR7-hi expression in the tumours was associated with antitumour activity, we analysed the status of the CCR7 ligands’, CCL19 and CCL21, expression in the tumour samples. A total of 120 patients were divided into two groups, one with a CCL19-hi mRNA expression (n=60) and the other with a CCL19-lo mRNA expression (n=60), with the median value as a cutoff. There was a significantly better prognosis for patients with higher CCL19 mRNA expression compared with that for patients with lower CCL19 mRNA expression, as indicated in the overall survival rates (P=0.040, HR=0.59, 95% CI of ratio 0.36–0.97) (Figure 4A). Similar results were seen in higher recurrence-free survival rates for patients with higher CCL19 mRNA expression (P=0.035, HR=0.58, 95% CI of ratio 0.35–0.96) (Figure 4B).
Effects of chemokine ligands 19 and 21 (CCL19, CCL21) mRNA expression on survival of patients with surgically resected lung adenocarcinoma. (A) Overall survival in relation to high (hi) and low (lo) CCL19 mRNA expression in the tumour site (P=0.040, HR=0.59, 95% CI of ratio 0.36–0.97). (B) Recurrence-free survival in relation to hi and lo CCL19 mRNA expression in the tumour site (P=0.035, HR=0.58, 95% CI of ratio 0.35–0.96). (C) Overall survival in relation to hi and lo CCL21 mRNA expression in the tumour site (P=0.31, HR=0.77, 95% CI of ratio 0.47–1.27). (D) Recurrence-free survival in relation to hi and lo CCL21 mRNA expression in the tumour site (P=0.15, HR=0.69, 95% CI of ratio 0.42–1.14). Effect of combinations of CCR7 and CCL19 or CCL21 mRNA expression on the survival of patients with surgically resected lung adenocarcinoma. (E) Overall survival in relation to combinations of CCR7 and CCL19 mRNA expression status. The overall survival rate of the CCR7-lo/CCL19-lo group was significantly lower than those of the CCR7-hi/CCL19-hi group and CCR7-hi/CCL19-lo group (P=0.0043, HR=0.45, 95% CI of ratio 0.26–0.77; P=0.041, HR=0.46, 95% CI of ratio 0.22–0.97, respectively). We cannot find significant differences of survival rates between the other combination groups (CCR7-hi/CCL19-hi, n=47; CCR7-hi/CCL19-lo, n=13; CCR7-lo/CCL19-hi, n=14; CCR7-lo/CCL19-lo, n=46). (F) Overall survival in relation to combinations of CCR7 and CCL21 mRNA expression status. We did not find significant differences in the survival rates (CCR7-hi/CCL21-hi, n=20; CCR7-hi/CCL21-lo, n=40; CCR7-lo/CCL21-hi, n=40; CCR7-lo/CCL21-lo, n=20).
The survival rates for different combinations of CCL19 and CCL21 mRNA expression with CCR7 mRNA expression in the lung cancer specimens were examined. A total of 120 patients were divided into four groups according to the median values of CCR7 and CCL19 mRNA expression status in the tumour (CCR7-hi/CCL19–hi, n=47; CCR7-hi/CCL19–lo, n=13, CCR7-lo/CCL19–hi, n=14; CCR7-lo/CCL19–lo, n=46). The overall survival rate of the CCR7-lo/CCL19-lo group was significantly lower than that of the CCR7-hi/CCL19-hi group and the CCR7-hi/CCL19-lo group (Figure 4E; P=0.0043, HR=0.45, 95% CI of ratio: 0.26–0.77; P=0.041, HR=0.46, 95% CI of ratio: 0.22 to 0.97, respectively). We did not find significant differences in the survival rates of the CCL21-hi mRNA and CCL21-lo mRNA expression groups, or of the groups with combination expressions of CCL21 mRNA and CCR7 mRNA (Figure 4C, D and F).
IHC analysis of CCR7, CrkL and c-ABL in lung adenocarcinoma
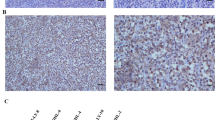
To gain a better insight into the distribution of CCR7, CrkL and c-ABL expressions, the protein expressions of each molecule were confirmed with IHC analysis of the specimens. A strong staining of CCR7 was seen in the cytoplasm of epithelial cancer cells in large fields of the tumour area (indicated by arrow, Figure 5A), and CCR7 was regionally stained in cells infiltrating the cancer stroma (indicated by arrowhead, Figure 5A).
Immunohistochemical detection of CCR7, CrkL and c-ABL in human lung adenocarcinoma. Lungs were removed from lung adenocarcinoma patients. Immunohistochemical analysis was performed using anti-CCR7 antibodies (Abs) for tumour epithelial cells (arrow) and infiltrated cells (arrowhead) in cancer stroma (A), antiphospho CrkL, c-ABL Abs for tumour epithelial (arrow) cells and tumour stroma (arrowhead) (B and C, respectively) and control immunoglobulin G (IgG) Abs (D), as described in Materials and Methods. Scale bars indicate 400 μm in panels A–D. Panel E shows two areas of relative CCR7 expression (%) in IHC analysis corresponding to patient groups with CCR7 mRNA high (CCR7 mRNA-hi, n=10) and low (CCR7 mRNA-lo, n=10) expression. Significant differences were detected between the two groups (panel E, P<0.05).
On the other hand, the expression of CrkL and c-ABL proteins was seen in the cytoplasm of epithelial cancer cells in parts of the tumour (indicated by arrow, Figure 5B and C, respectively) and cancer stroma lesion (indicated by arrowhead, Figure 5B and C, respectively). We detected no immunostaining in cancer epithelial cells or stroma cells in the lung cancer specimens by control goat IgG (Figure 5D).
To confirm the results of the CCR7 mRNA expression status, we measured the CCR7 IHC staining area (see the slide images in Supplementary Figure 1). The CCR7 IHC expression areas of the upper and lower CCR7 mRNA expression groups were analysed in tissue samples, as described in Materials and Methods. The mean values of percent CCR7 protein expression areas are indicated in Figure 5E. The percent CCR7 protein expression area of the CCR7-hi mRNA expression group is significantly higher than that of the CCR7-lo mRNA expression group (Figure 5E, P=0.0284).
mRNA expressions by LCM isolation and RT from FFPE sections
We dissected specimens with laser capture microdissection from the tumour site in PPFE samples (n=8) of randomly selected lung adenocarcinoma specimens that were surgically resected and performed relative quantitative RT–PCR analyses. Taken together with this process, we also dissected normal lung tissue of the PPFE samples (n=8) and examined mRNA expression. Expression of the mRNAs for chemokine receptor CCR7, CrkL and c-ABL in the tumour site was confirmed by laser capture microdissection (Figure 6A–C). On the other hand, the expression of each of the mRNAs was very low in normal lung tissue (Figure 6A–C).
Quantitative RT–PCR expressions of CCR7, CrkL and c-ABL mRNA in specimens (n=8) from resected lung adenocarcinoma by laser capture microdissection. In each panel, the relative quantities of CCR7, CrkL and c-ABL mRNA in tumour and normal lung sites are indicated (A–C, respectively). Box and whisker plots summarise these distributions: the central box covers the interquartile range, with the median indicated by the line within the box. The whiskers indicate the minimum and maximum values, and all values are plotted individually.
Discussion
The results of our study showed that a high expression of CCR7 in lung adenocarcinoma was a good prognostic factor (Figure 2A and B). This result was confirmed by CCR7 protein expression in IHC of lung adenocarcinoma specimens (Figure 5E). A high expression of CCL19, a ligand of CCR7, was a good prognostic factor in lung adenocarcinoma (Figure 4A and B). This CCL19 function in lung adenocarcinoma corroborates the report that CCL19 promotes IFN-γ-dependent antitumour responses in a lung cancer model (Hillinger et al, 2003). In addition, a double loss of CCR7 and CCL19 brought about very poor prognosis (Figure 4E). These results suggest that CCR7 cooperates with CCL19 and may have antimetastatic and antitumorigenic activity in lung adenocarcinoma. The CCR7 expression did not affect the nodal status and lymphatic invasion (Supplementary Table 2); however, we anticipate that the antitumorigenic activity of CCR7/CCL19 uses the activation of antitumour immune responses.
In our study, we detected CrkL and c-ABL mRNA in all specimens of surgically resected lung adenocarcinoma. CrkL and c-ABL are considered important factors in the pathogenesis of lung adenocarcinoma themselves. Thus, lower and higher expressions of CrkL and c-ABL mRNA did not influence the survival (Figure 1A and B). Our results showed that CrkL and c-ABL mRNA expressions are related to CCR7 mRNA expression in lung adenocarcinoma specimens (Figure 2C and D and Table 3). As Sekine et al (2012) reported with regard toCML, these results indicate the possibility that c-ABL signals may alter chemokine receptor expression, leading to the upregulation of CCR7 of lung cancer cells and cancer-related cells. In addition, high expressions of CrkL mRNA in surgically resected specimens are associated with decreased survival rates in patients with high expressions of CCR7 mRNA (Figure 3A).
CrkL and c-ABL mRNA expressions were also significantly increased by EGFR gene mutations (Figure 1C and D). These results indicated that EGFR thymidine kinase signalling influenced CrkL and c-ABL mRNA expression. Our results showing a relationship between c-ABL and EGFR are reasonable in light of a previous report that showed that the activated ABL kinase phosphorylated the EGFR and inhibited EGFR internalisation, and that the c-ABL also was activated by ligand-stimulated EGFR (Tanos and Pendergast, 2006). Thus, the EGFR and c-ABL/CrkL functions have been linked to the pathogenesis of cancer. Meanwhile, TP53 mutations did not affect CrkL and c-ABL mRNA expression in the tumour sites of lung adenocarcinoma (Figure 1E and F).
In the IHC analysis, epithelial cancer cells were stained with anti-CCR7, anti-CrkL and c-ABL antibodies (Figure 5A–C). The distribution of each expressed protein varied in different cancer cell specimens (Figure 5A–C). CC chemokine receptor 7-positive cells were widely seen among the epithelial cancer cells (Figure 5A), and CCR7-positive cells were seen in the cancer stroma (Figure 5A). Dendritic cells and memory T cells express CCR7; these cells participate in acquired immune responses and antitumour effects (Croci et al, 2007). Therefore, CCR7-positive epithelial cancer cells and cells infiltrating the cancer stroma may join up to improve the overall survival rates in a postoperative patient with lung adenocarcinoma (Figures 2A and 5A). c-ABL-positive cells were seen among some epithelial cancer cells, and large mononuclear cells those were considered to be tumour-associated macrophages infiltrating cancer epithelial spaces (Figure 5C). CrkL-positive epithelial cancer cells were seen locally in parts of the cancer lesion (Figure 5B). We used antiphospho CrkL (Tyr207) antibody and antiphospho (Y412) c-ABL antibody for IHC analysis. These are antibodies against activated phosphorylated proteins (de Jong et al, 1997; Brasher and Van Etten, 2000; Pluk et al, 2002). These results indicated that active types of c-ABL and CrkL are expressed in cancer cells or cancer-related cells and contribute to maintaining the cancer system. Furthermore, we detected these mRNAs in the tumour sites using laser capture microdissection (Figure 6).
We need to conduct further analyses on the roles of CCR7, CrkL and c-ABL in lung cancer by unravelling the complicated riddle of the functions of chemokines, interaction with their receptors and their signal factors in cancer cells. This is the first report for a clinical approach to study the interaction among chemokines, CrkL and c-ABL in cancer.
By examining the expression of CCR7, c-ABL and CrkL mRNAs and proteins in lung adenocarcinoma, we conclude that they are preferentially expressed in primary tumour lesions. A CCR7-hi mRNA expression resulted in a good prognosis, and the effects were modified by CrkL and c-ABL expression status. In addition, we also found that CCL19 expression in lung adenocarcinoma was a good prognostic factor, and that EGFR gene mutations affected the CrkL and c-ABL mRNA expressions in lung adenocarcinoma.
Change history
03 September 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Andre F, Cabioglu N, Assi H, Sabourin JC, Delaloge S, Sahin A, Broglio K, Spano JP, Combadiere C, Bucana C, Soria JC, Cristofanilli M (2006) Expression of chemokine receptors predicts the site of metastatic relapse in patients with axillary node positive primary breast cancer. Ann Oncol 17 (6): 945–951.
Brasher BB, Van Etten RA (2000) c-Abl has high intrinsic tyrosine kinase activity that is stimulated by mutation of the Src homology 3 domain and by autophosphorylation at two distinct regulatory tyrosines. J Biol Chem 275 (45): 35631–35637.
Breuer RH, Postmus PE, Smit EF (2005) Molecular pathology of non-small-cell lung cancer. Respiration 72 (3): 313–330.
Cabioglu N, Gong Y, Islam R, Broglio KR, Sneige N, Sahin A, Gonzalez-Angulo AM, Morandi P, Bucana C, Hortobagyi GN, Cristofanilli M (2007) Expression of growth factor and chemokine receptors: new insights in the biology of inflammatory breast cancer. Ann Oncol 18 (6): 1021–1029.
Cao M, Deng HX, Zhao J, Fan LY, Jiang Y, Wen YJ, Li J, Lei S, Mao YQ, Ding ZY, Wei YQ (2007) Antitumour activity of cationic-liposome-conjugated adenovirus containing the CCL19 [chemokine (C–C motif) ligand 19] gene. Biotechnol Appl Biochem 48: 109–116.
Cheung HW, Du J, Boehm JS, He F, Weir BA, Wang X, Butaney M, Sequist LV, Luo B, Engelman JA, Root DE, Meyerson M, Golub TR, Jänne PA, Hahn WC (2011) Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov 1 (7): 608–625.
Croci S, Nicoletti G, Landuzzi L, Palladini A, Chiarini F, Nanni P, Lollini PL, De Giovanni C (2007) Expression of a functional CCR7 chemokine receptor inhibits the post-intravasation steps of metastasis in malignant murine mammary cancer cells. Oncol Rep 18 (2): 451–456.
de Jong R, ten Hoeve J, Heisterkamp N, Groffen J (1997) Tyrosine 207 in CRKL is the BCR/ABL phosphorylation site. Oncogene 14 (5): 507–513.
Hillinger S, Yang SC, Zhu L, Huang M, Duckett R, Atianzar K, Batra RK, Strieter RM, Dubinett SM, Sharma S (2003) EBV-induced molecule 1 ligand chemokine (ELC/CCL19) promotes IFN-gamma-dependent antitumor responses in a lung cancer model. J Immunol 171 (12): 6457–6465.
Itakura M, Tokuda A, Kimura H, Nagai S, Yoneyama H, Onai N, Ishikawa S, Kuriyama T, Matsushima K (2001) Blockade of secondary lymphoid tissue chemokine exacerbates Propionibacterium acnes-induced acute lung inflammation. J Immunol 166 (3): 2071–2079.
Kim YH, Kwei KA, Girard L, Salari K, Kao J, Pacyna-Gengelbach M, Wang P, Hernandez-Boussard T, Gazdar AF, Petersen I, Minna JD, Pollack JR (2010) Genomic and functional analysis identifies CRKL as an oncogene amplified in lung cancer. Oncogene 29 (10): 1421–1430.
Koizumi K, Hojo S, Akashi T, Yasumoto K, Saiki I (2007) Chemokine receptors in cancer metastasis and cancer cell-derived chemokines in host immune response. Cancer Sci 98 (11): 1652–1658.
Konsti J, Lundin M, Joensuu H, Lehtimäki T, Sihto H, Holli K, Turpeenniemi-Hujanen T, Kataja V, Sailas L, Isola J, Lundin J (2011) Development and evaluation of a virtual microscopy application for automated assessment of Ki-67 expression in breast cancer. BMC Clin Pathol 11: 3.
Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verástegui E, Zlotnik A (2001) Involvement of chemokine receptors in breast cancer metastasis. Nature 410 (6824): 50–56.
Pluk H, Dorey K, Superti-Furga G (2002) Autoinhibition of c-Abl. Cell 108 (2): 247–259.
Richmond A (2002) Nf-kappa B, chemokine gene transcription and tumour growth. Nat Rev Immunol 2 (9): 664–674.
Sattler M, Salgia R (1998) Role of the adapter protein CRKL in signal transduction of normal hematopoietic and BCR/ABL-transformed cells. Leukemia 12 (5): 637–644.
Sekine Y, Ikeda O, Mizushima A, Ueno Y, Muromoto R, Yoshimura A, Kanakura Y, Oritani K, Matsuda T (2012) STAP-2 interacts with and modulates BCR-ABL-mediated tumorigenesis. Oncogene 31 (40): 4384–4396.
Shingyoji M, Kageyama H, Sakaida T, Nakajima T, Matsui Y, Itakura M, Iuchi T, Yokoi S, Kimura H, Iizasa T (2011) Detection of epithelial growth factor receptor mutations in cerebrospinal fluid from patients with lung adenocarcinoma suspected of neoplastic meningitis. J Thorac Oncol 6 (7): 1215–1220.
Sirvent A, Benistant C, Roche S (2008) Cytoplasmic signalling by the c-Abl tyrosine kinase in normal and cancer cells. Biol Cell 100 (11): 617–631.
Tanos B, Pendergast AM (2006) Abl tyrosine kinase regulates endocytosis of the epidermal growth factor receptor. J Biol Chem 281 (43): 32714–33272.
Acknowledgements
We are grateful to Dr Hidetada Kawana, Dr Akinobu Araki, Mr Tetsuji Hirata, Ms Yuko Noguchi and all other members of Chiba Cancer Center, Chiba, Japan for their advice and help. We appreciate Dr Noriyuki Tanaka, Department of Molecular and Tumor Pathology, Chiba University Graduate School of Medicine, Chiba, Japan for technical advice concerning laser capture microdissection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Itakura, M., Terashima, Y., Shingyoji, M. et al. High CC chemokine receptor 7 expression improves postoperative prognosis of lung adenocarcinoma patients. Br J Cancer 109, 1100–1108 (2013). https://doi.org/10.1038/bjc.2013.440
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.440
Keywords
This article is cited by
-
LINC02126 is a potential diagnostic, prognostic and immunotherapeutic target for lung adenocarcinoma
BMC Pulmonary Medicine (2022)
-
Targeting FROUNT with disulfiram suppresses macrophage accumulation and its tumor-promoting properties
Nature Communications (2020)
-
An assessment of the relationship between the expression of CCR7/CCL19 axis and selected regulatory miRNAs in non-small cell lung cancer
Molecular Biology Reports (2019)
-
Expression of CC chemokine receptor 9 predicts poor prognosis in patients with lung adenocarcinoma
Diagnostic Pathology (2015)
-
The effect of chemokine CC motif ligand 19 on the proliferation and migration of hepatocellular carcinoma
Tumor Biology (2014)