Abstract
All-trans retinoic acid (ATRA) is well established as differentiation therapy for acute promyelocytic leukemia (APL) in which the PML–RARα (promyelocytic leukemia-retinoic acid receptor α) fusion protein causes blockade of the retinoic acid (RA) pathway; however, in types of acute myeloid leukemia (AML) other than APL, the mechanism of RA pathway inactivation is not fully understood. This study revealed the potential mechanism of high ATRA sensitivity of mixed-lineage leukemia (MLL)-AF9-positive AML compared with MLL-AF4/5q31-positive AML. Treatment with ATRA induced significant myeloid differentiation accompanied by upregulation of RARα, C/EBPα, C/EBPɛ and PU.1 in MLL-AF9-positive but not in MLL-AF4/5q31-positive cells. Combining ATRA with cytarabine had a synergistic antileukemic effect in MLL-AF9-positive cells in vitro. The level of dimethyl histone H3 lysine 4 (H3K4me2) in the RARα gene-promoter region, PU.1 upstream regulatory region (URE) and RUNX1+24/+25 intronic enhancer was higher in MLL-AF9-positive cells than in MLL-AF4-positive cells, and inhibiting lysine-specific demethylase 1, which acts as a histone demethylase inhibitor, reactivated ATRA sensitivity in MLL-AF4-positive cells. These findings suggest that the level of H3K4me2 in the RARα gene-promoter region, PU.1 URE and RUNX1 intronic enhancer is determined by the MLL-fusion partner. Our findings provide insight into the mechanisms of ATRA sensitivity in AML and novel treatment strategies for ATRA-resistant AML.
Similar content being viewed by others
Introduction
Differentiation therapy using all-trans retinoic acid (ATRA) is well established for the treatment of acute promyelocytic leukemia (APL)1, 2 in which the PML–RARα (promyelocytic leukemia-retinoic acid receptor α) fusion protein causes blockade of the retinoic acid (RA) pathway.3, 4 There has been significant success using ATRA to treat APL, but there has been little research into the use of ATRA in a non-APL acute myeloid leukemia (AML) setting. In addition, ATRA does not induce myeloid differentiation in non-APL AML cells. Myeloid differentiation is regulated via the activation of RARα gene by ATRA.4, 5 Studies of the molecular mechanism blocking differentiation in non-APL AML cells indicate that expression of RARα is diminished in non-APL AML cells and that its restoration can induce differentiation of human AML cells.6, 7 These observations suggest that a lack of RARα expression impairs the RA pathway in non-APL AML cells, blocking myeloid differentiation. In addition, these studies also revealed that epigenetic mechanisms, including DNA methylation, histone methylation and acetylation, are involved in the repression of RARα expression in non-APL AML cells.6, 7 Based on these findings, recent studies have focused on the development of differentiation therapies for non-APL AML using ATRA combined with epigenetic modifiers.8, 9, 10, 11, 12 Recently, it has been reported that inhibition of lysine-specific demethylase 1 (LSD1), which demethylates dimethyl histone 3 lysine 4 (H3K4me2) to silence expression of its target gene, reactivates the RA pathway in non-APL AML.13, 14
Rearrangements of the mixed-lineage leukemia (MLL) gene, which lies on chromosome 11, can involve any of the 79 potential translocation partner genes.15 MLL-rearranged leukemia is a heterogeneous group, and clinical outcome depends on the translocation partner gene involved.16, 17 AML patients with t(9;11) and t(11;19) translocations are classified as intermediate risk, while those with all other MLL-fusion partners are classified as high risk.18 In addition, patients with MLL-rearranged AML grouped according to the translocation partner gene involved can be further stratified by the gene expression signature and promoter methylation pattern of their leukemic cells.19, 20 In terms of induction of myeloid differentiation in MLL-rearranged AML, we have previously described that the demethylating agent 5-aza-2'-deoxycitidine enhanced the sensitivity of MLL-AF9-expressing AML cells to ATRA,8 and Iijima et al.9 have also reported that the combination of the histone deacetylase inhibitor trichostatin A and ATRA was effective as a growth inhibitor and differentiation enhancer in MLL-AF9-expressing leukemia cells. In addition, Niitsu et al.21 demonstrated that ATRA sensitivity of MLL-rearranged AML cells were varied and that might be related to p16 expression level. These findings suggest that chromatin remodeling, such as histone modification by methylation or acetylation, is an important factor for ATRA sensitivity in MLL-rearranged AML, and the epigenetic priming of RA pathway is necessary for the differentiation therapy in MLL-rearranged AML.
Herein, to determine whether the different partner gene translocations of MLL-rearranged AML affect the sensitivity of ATRA, we investigated the relationship between the level of H3K4me2 in the RARα gene-promoter region, PU.1 gene upstream regulatory region (URE) and RUNX1 gene intronic enhancer and sensitivity to ATRA, and whether, by inhibiting H3K4me2 demethylation in ATRA-resistant cells, sensitivity could be induced in MLL-rearranged human AML cells and murine immortalized cells expressing MLL-AF9 or MLL-AF5q31.
Materials and methods
Cell lines and culture
Human AML cell lines, THP-1 (American Type Culture Collection, Manassas, VA, USA) and MOLM-13 (kindly provided by Dr Bert A. van der Reijden, Radboud University Nijmegen Medical Centre, Nijmegen, Netherlands) bearing the MLL-AF9 gene fusion and KOCL-48 (kindly provided by Dr Kanji Sugita, Yamanashi University, Kofu, Japan) bearing MLL-AF4 were cultured in suspension in RPMI-1640 medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (10 mg/ml) at 37 °C in a 5% CO2 humidified atmosphere.
Reagents
ATRA (Sigma-Aldrich, St. Louis, MO, USA), dissolved in dimethyl sulfoxide, and cytarabine (Sigma-Aldrich), dissolved in water, were stored as 1 mM stock solutions in small aliquots at −20 °C. Tranylcypromine (TCP), a nonreversible LSD1 inhibitor purchased from Sigma-Aldrich, was prepared as a 10 mM stock solution in dissolved dimethyl sulfoxide and stored at −20 °C.
Retroviral constructs and transduction of Lin− murine hematopoietic progenitors
Retroviral constructs encoding either human MLL-AF9 or MLL-AF5q31 fusion genes were used to establish murine cell lines expressing MLL-AF9 or MLL-AF5q31 fusion proteins as described previously.8
Clinical AML cell samples
Primary leukemic cells expressing the MLL-AF9 fusion were obtained from the diagnostic bone marrow samples of two pediatric AML patients following informed parental consent, in accordance with the revised Helsinki protocol. The AML cells were purified from bone marrow as mononuclear cells using Ficoll density-gradient centrifugation and stored in liquid nitrogen. On thawing, cells were plated in methylcellulose medium (MethoCult H4434, Stem Cell Technologies, Vancouver, BC, Canada) at 1.0 × 106/ml and cultured with ATRA (1 μM).
Morphological studies
Cells at 1.0 × 106/ml were cultured with ATRA (1 μM) and/or TCP (10 μM) for 3 days. Cytospin preparations were stained with May-Grünwald Giemsa.
Nitroblue tetrazolium (NBT) reduction test for detection of myeloid differentiation
After culture with ATRA (1 μM) and/or TCP (10 μM) for 3 days, cells were subjected to a NBT reduction test using the NBT Reduction Kit (Sigma-Aldrich) according to the manufacturer’s instructions. The percentage of cells containing the precipitated formazan particles was determined by light microscopy. At least 200 cells were counted per sample.
Cytotoxicity assay
ATRA, TCP and cytarabine cytotoxicity were measured by cell viability using a WST assay (Cell Count Reagent SF, Nacalai Tesque Inc., Kyoto, Japan). The concentration of each drug causing 50% growth inhibition (IC50) was determined. The effect of ATRA and cytarabine and of ATRA and TCP cytotoxic interactions were determined from the combination index (CI); CI<1, CI=1 and CI>1 indicate a synergistic, an additive and an antagonistic effect, respectively. The CI was calculated using the following equation: CI=((D)1/(Dx)1)+((D)2/(Dx)2). (Dx)1 and (Dx)2 are concentrations of each drug when each drug inhibits cell proliferation at the 50% growth level individually. (D)1 and (D)2 are concentrations of each drug when combination treatment of two drugs inhibit cell proliferation at the 50% growth level.
Flow cytometric analysis
Cells (1.0 × 106) were harvested, washed twice with PBS and incubated for 30 min with phycoerythrin-conjugated anti-human and anti-mouse Mac-1 (murine CD11b) antibody (BD Biosciences, Sparks, MD, USA) and analyzed on a FACS Calibur (BD Biosciences) with the FlowJo software (Treestar, San Carlos, CA, USA).
Cell-cycle analysis
Cells were harvested, washed twice with PBS and incubated for 30 min with propidium iodide (PI) to stain DNA. Propidium iodide fluorescence was analyzed using a FACS Calibur, and the cell-cycle phase was determined on the basis of DNA content using the ModFit LT software (Verity Software House, http://www.vsh.com/).
Real-time reverse transcriptase–PCR (RT-PCR)
Total RNA was extracted from cells using the RNeasy Mini Kit (Qiagen, Venio, The Netherlands) according to the manufacturer’s instructions. The SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) was used to synthesize cDNA according to the manufacturer’s instructions, and real-time RT-PCR was performed using the 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA) with SYBR Green 1 Master Mix (Takara Bio, Tokyo, Japan). Relative expression of target mRNA was determined using the comparative threshold (ΔCt) method. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. The primer pairs used in this study are listed in Supplementary Table S1. A standard curve analysis with stepwise sample dilution demonstrated that all primer pairs had similar efficiency (data not shown).
Immunoblot analysis
Immunoblot analysis was performed as described previously.22 The following primary antibodies were used: anti-RARα (diluted 1:200), anti-C/EBPα (diluted 1:750), anti-C/EBPɛ (diluted 1:200) and anti-PU.1 (diluted 1:500). All antibodies were purchased from Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA.
Chromatin immunoprecipitation (ChIP)
ChIP was carried out using the Simple ChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology Inc., Danvers, MA, USA) according to the manufacturer’s instructions. IP was performed with the following antibodies: anti-H3K4me2 (Abcam, Tokyo, Japan) and anti-histone H3 (Cell Signaling Technology Inc.). IP DNA in each IP sample of human cell lines was analyzed by quantitative RT-PCR using RARα gene-specific primers covering the promoter and 5′ untranslated region (5′UTR): from−1000 to+1000 bp from the transcriptional start site,23, 24 PU.1 URE25 and RUNX1+24/+25 intronic enhancer.26 Also, IP DNA in each IP sample of murine cell lines was analyzed by quantitative RT-PCR using Rarα gene-specific primers covering the promoter and 5′UTR:from−1000 to+1000 bp from the transcriptional start site, Sfpi.1 URE and Runx1+24/+25 intronic enhancer.25, 26 The list of primer sequences are shown in Supplementary Tables 2 and 3. All expression values were normalized against histone H3.
Statistical analysis
Statistical analysis was performed using the Student’s t-test. A P-value<0.05 was considered statistically significant.
Results
ATRA induces myeloid differentiation and growth inhibition with cell-cycle arrest in human MLL-AF9-positive AML cells
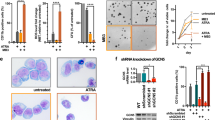
First, we evaluated whether ATRA could induce myeloid differentiation, as observed in APL, in three human AML cell lines with MLL rearrangements. ATRA induced morphological changes, such as irregularly shaped nuclei, extended cytoplasm and the appearance of fine granules, more markedly in THP-1 and MOLM-13 cell lines, which bear the MLL-AF9 fusion gene, than in KOCL-48 which bears MLL-AF4 (Figure 1a). These morphological changes corresponded with a reduction in NBT, which indicated an increase in mature myeloid cell function (for example, phagocytosis activity), and the induction of CD11b on flow cytometric analysis (Figures 1b and c). Furthermore, ATRA induced G0/G1 arrest more clearly in the MLL-AF9-positive cell lines than in the MLL-AF4-positive cells and had a much lower IC50 with the MLL-AF9-positive cell lines (THP-1 and MOLM-13) than with the MLL-AF4- (KOCL-48) positive cells (3.91±0.87 and 1.24±0.70 vs 77.2±7.37 μM; Supplementary Figure S1a and Figure 1d). Taken together, these data demonstrate that the effects of ATRA on the induction of myeloid differentiation were more apparent, and that the effects of ATRA on growth inhibition were significantly greater, on the MLL-AF9-positive AML cell line than on the MLL-AF4-positive cell line. These data were consistent with the previous studies showing MLL-AF9-positive AML cell line was sensitive to ATRA.8, 9
The effect of ATRA on human MLL-rearranged AML cell lines. (a) Photomicrographs of THP-1, MOLM-13, and KOCL-48 cell lines following incubation with ATRA (1 μM) for 72 h. Cytospin preparations were stained with May-Grünwald Giemsa. (b) Effect of ATRA on a reduction in NBT. Cells were incubated with ATRA for 72 h. Results represent the means±s.d. of three independent experiments. **P<0.05; NS, not significant. (c) CD11b expression determined by FACS analysis following incubation with ATRA (1 μM) for 72 h. Black line, untreated cells; red line, cells treated with ATRA. Representative results are shown, and the bars represent the means±s.d. of three independent experiments. **P<0.05; NS, not significant. (d) Cytotoxicity of ATRA on THP-1, MOLM-13 and KOCL-48 cell lines. The number of viable cells was assessed following incubation with titrating doses of ATRA (range, 0–10 μM) for 96 h. ATRA IC50 values were determined for each cell line. (e) Expression of RARα, C/EBPα, C/EBPɛ and PU.1 determined by immunoblot analysis after incubation with ATRA (1 μM) for 72 h. β-Actin was used as a control. Representative results from three independent experiments are shown.
The RA pathway is more profoundly impaired in human MLL-AF4-positive cells than in MLL-AF9-positive cells
The expression of RARα, C/EBPα, C/EBPɛ and PU.1 genes is modulated in myeloid differentiation by RA. Using western blotting analysis, we found that RARα and C/EBPɛ expression levels, which are directly regulated by ATRA, were increased in response to ATRA in the MLL-AF9-positive cell lines THP-1 and MOLM-13 but not in the MLL-AF4-positive cell line KOCL-48. In addition, expression of C/EBPα and PU.1, which are important transcriptional factors in myeloid differentiation, was also increased only in MLL-AF9-positive cells (Figure 1e).
The effect of ATRA on primary MLL-AF9-positive AML cells
To confirm these effects of ATRA on myeloid differentiation, we evaluated the expression levels of RARα, C/EBPα, C/EBPɛ and PU.1 in two primary AML samples bearing MLL-AF9 fusion using real-time RT-PCR analysis. In accordance with the results of the western blotting analysis of human MLL-AF9-positive AML cell lines, the expression levels of RARα, C/EBPα, C/EBPɛ and PU.1 were upregulated in both primary MLL-AF9-positive AML samples (Supplementary Figure S2). Collectively, these results showed clearly that ATRA was able to induce myeloid differentiation and activate the RA pathway in MLL-AF9-positive AML cells. In contrast, ATRA did not induce myeloid differentiation or activate the RA pathway in MLL-AF4-positive cells.
The effect of ATRA on murine MLL-rearranged immortalized cells
To confirm that the particular MLL-fusion partner (AF9 or AF4) directly determined the activity of the RA pathway, our aim was to generate immortalized murine cells expressing MLL-AF9 or MLL-AF4; however, we were unable to transform murine hematopoietic progenitor cells using MLL-AF4. Therefore AF5q31, a member of the AF4 family of genes, fused with MLL was used as an alternative.8 Similar to the results of the experiments with human AML cells, ATRA induced morphological changes only in the murine MLL-AF9-expressing cells (Figure 2a), and these changes were accompanied by a reduction in NBT and upregulation of Mac 1expression (Figures 2b and c). In addition to these effects, the percentage of cells in G0/G1 was greater among the MLL-AF9-expressing cells (Supplementary Figure S1b), and the ATRA IC50 was much lower for MLL-AF9-expressing cells than for MLL-AF5q31-expressing cells (2.01±0.39 vs 32.6±14.5 μM; Figure 2d). The expression levels of Rarα, C/ebpα, C/ebpɛ and Sfpi.1 determined by real-time RT-PCR were significantly upregulated in response to ATRA in the MLL-AF9-expressing cells but were unchanged in the MLL-AF5q31-expressing cells (Figure 2e), suggesting that murine Lin− hematopoietic progenitor cells expressing MLL-AF9, but not those expressing MLL-AF5q31, remain sensitive to ATRA. Thus, it could be concluded that particular MLL-fusion partners affect sensitivity to ATRA.
The effect of ATRA on murine MLL-rearranged immortalized cells. (a) Photomicrographs of murine immortalized cell lines expressing MLL-AF9 and MLL-AF5q31 following incubation with ATRA (1 μM) for 72 h. Cytospin preparations were stained with May-Grünwald Giemsa. Representative results from three independent experiments are shown. (b) Effect of ATRA on reduction in NBT. Murine immortalized cell lines were each cultured with ATRA (1 μM) for 72 h. Results represent the mean±s.d. of three independent experiments. **P<0.05; NS, not significant. (c) Mac 1 expression determined by FACS analysis following incubation for 72 h with ATRA (1 μM). Black line, untreated cells; red line, cells treated with ATRA. Representative results are shown, and the bars represent the means±s.d. of three independent experiments. **P<0.05; NS, not significant. (d) Cytotoxicity of ATRA on MLL-rearranged murine immortalized cell lines. The number of viable cells was assessed following incubation with titrating doses of ATRA (range, 0–10 μM) for 72 h. ATRA IC50 values were determined for each cell line. (e) Expression of Rarα, C/ebpα, C/epbɛ and Sfpi.1 determined by real-time RT-PCR. Cells were harvested after incubation with ATRA (1 μM) for 72 h. Results represent the mean±s.d. of three independent experiments. **P<0.05; NS, not significant.
A synergistic antileukemic effect between cytarabine and ATRA in MLL-AF9-positive AML cells
To explore the possibility of a synergistic antileukemic effect between ATRA and cytarabine, one of the major drugs used to treat AML, we treated human and murine MLL-AF9-positive cells (THP-1 and MOLM-13 human AML cell lines and the immortalized murine MLL-AF9-expressing cells) with a titrating dose of cytarabine in combination with 1 μM ATRA or without ATRA. We found that the presence of ATRA decreased the cytarabine IC50 in THP-1 and MOLM-13 cell lines and in the murine MLL-AF9-expressing cells (cytarabine IC50 of 3.69±1.00, 0.042±0.030 and 0.060±0.012 μM, respectively, without ATRA, vs 0.17±0.075, 0.0038±0.0031 and 0.015±0.0026 μM, respectively, with ATRA). The combined effects of cytarabine and ATRA on cell-growth inhibition were clearly synergistic in the three cell lines (Figures 3a and b). ATRA in combination with cytarabine was, therefore, more effective than cytarabine alone for ablating MLL-AF9-positive cells in vitro.
Synergistic cytotoxic effects between ATRA and cytarabine in MLL-AF9-positive cells. (a) THP-1, MOLM-13 and murine MLL-AF9-expressing immortalized cells were cultured with a range of concentrations of cytarabine (0–10 μM) with 1 μM ATRA or without. After incubation for 72 h in MLL-AF9-expressing immortalized cells and 96 h in human cell lines (THP-1 and MOLM-13), the number of viable cells was assessed by a WST assay. (b) Cytarabine IC50 with or without 1 μM ATRA values were determined for each cell line.
ChIP assays of H3K4me2 on MLL-rearranged human AML cell lines and murine MLL-rearranged immortalized cells
Next, we carried out ChIP assays on MLL-rearranged human AML cell lines to investigate whether inactivation of the RA pathway was correlated with decreased H3K4me2 in the RARα-promoter region, which contained the RA response elements.23 IP DNA was analyzed using RARα gene-specific primers mapping to the promoter and 5′UTR from−1000 to+1000 bp from the transcriptional start site. The levels of H3K4me2 normalized against histone H3 were lower in the MLL-AF4-positive cell line, KOCL-48, than in the MLL-AF9-positive cell lines, THP-1 and MOLM-13. These findings revealed that ATRA sensitivity correlates with the H3K4me2 level in the RARα gene in MLL-rearranged AML cell lines (Figure 4a). In addition, we evaluated the histone modification status at the PU.1 proximal and distal URE and RUNX1+24/+25 intronic enhancer, which were also associated with myeloid differentiation.25, 26 The levels of H3K4me2 at these regions were lower in the MLL-AF4-positive cell line, KOCL-48, than in the MLL-AF9-positive cell lines, THP-1 and MOLM-13 (Figure 4b). Collectively, MLL-AF9-positive cell lines bear higher H3K4me2 level in the RARα gene, PU.1 URE and RUNX1 intronic enhancer than MLL-AF4 positive cell line, resulting in efficient induction of myeloid differentiation in MLL-AF9-positive cell lines by ATRA.
H3K4me2 modification of the RARα, Rarαgene, PU.1/Sfpi.1 URE and RUNX1/Runx1 intronic enhancer in human AML cell lines and murine immortalized cell lines. After cells were incubated with ATRA (1 μM) for 72 h, ChIP assays were performed using antibodies to H3K4me2 and histone H3 (H3). Precipitated DNA was analyzed by real-time RT-PCR using primers mapping to within the promoter region and 5′UTR) of the RARα gene (a) and PU.1 URE or RUNX1+24/+25 intronic enhancer (b), the promoter region and 5′UTR of the Rarα gene (c) and Sfpi.1 URE or Runx1+24/+25 intronic enhancer (d). Results represent the mean±s.d. of two independent ChIP experiments and a total of four independent PCR analysis.
To confirm the particular MLL-fusion partner (AF9 or AF4) directly modified the histone modification, such as the H3K4me2 level, we investigated the levels of H3K4me2 at Rarα gene-promoter, Sfpi.1 URE, and Runx1 intronic enhancer in immortalized murine cells expressing MLL-AF9 or MLL-AF5q31. Consistent with the result of human AML cell lines, the levels of H3K4me2 normalized against histone H3 were lower in the murine cells expressing MLL-AF5q31 than in the murine cells expressing MLL-AF9 at Rarα gene-promoter, Sfpi.1 URE, and Runx1 intronic enhancer (Figures 4c and d).
TCP restores ATRA sensitivity in the MLL-AF4-positive and ATRA-resistant cell line, KOCL-48
LSD1 demethylates H3K4me2, silencing the target gene; inhibition of LSD1 leads to an increase in H3K4me2. To determine whether the H3K4me2 status of the RARα promoter region and 5′UTR affected the RA pathway, KOCL-48 cells were treated with TCP, an inhibitor of LSD1. A combination of 1 μM ATRA and 10 μM TCP (ATRA/TCP) induced more marked morphological changes and caused a bigger reduction in NBT than either ATRA or TCP alone (Figures 5a and b). Furthermore, ATRA/TCP induced higher-intensity expression of CD11b and increased the proportion in G0/G1 cell-cycle arrest (Figure 5c and Supplementary Figure S1c). Western blotting analysis revealed that the expression of RARα, C/EBPα, C/EBPɛ and PU.1 increased in response to ATRA/TCP (Figure 5d), suggesting that the combination of ATRA and TCP reactivates the RA pathway. We therefore treated KOCL-48 cells with a titrating dose of ATRA with 10 μM TCP or without TCP. The ATRA IC50 was significantly decreased when added in combination with TCP, and the CI showed that ATRA and TCP had a synergistic effect (Figure 5e, ATRA IC50 of 72.2±7.95 μM without TCP vs 12.9±3.04 μM with TCP). Finally, we tested whether TCP could upregulate the level of H3K4me2 in the RARα gene-promoter region and 5′UTR. The levels of H3K4me2 in the presence of TCP were increased in a dose-dependent manner (Figure 5f). Similarly, the level of H3K4me2 in the PU.1 URE and RUNX1 intronic enhancer were also upregulated in the presence of TCP (Figure 5g). These findings suggest that inactivation of the RA pathway and myeloid differentiation block are induced by a decrease in the level of H3K4me2 in the RARα gene-promoter, the PU.1 URE and RUNX1 intronic enhancer.
ATRA and TCP can reactivate the RA pathway in the MLL-AF4-positive AML cell line, KOCL-48. (a) Photomicrographs of KOCL-48 cells untreated and treated with ATRA (1 μM) and/or TCP (10 μM). Cytospin preparations were stained with May-Grünwald Giemsa. Representative results from three independent experiments are shown. (b) Effect of ATRA/TCP on reduction in NBT. KOCL-48 cells were cultured with ATRA/TCP for 72 h. Results represent the mean±s.d. of three independent experiments. **P<0.05; NS, not significant. (c) CD11b expression determined by FACS analysis following incubation with ATRA/TCP for 72 h. Black line, untreated cells; red line, ATRA; blue line, TCP; green line, ATRA/TCP. Representative results are shown, and the bars indicate the means±s.d. of three independent experiments. **P<0.05; NS, not significant. (d) Expression of RARα, C/EBPα, C/EBPɛ and PU.1 determined by immunoblot analysis after incubation with ATRA/TCP. β-Actin was used as a control. Representative results from three independent experiments are shown. (e) ATRA IC50 values with or without 10 μM TCP in KOCL-48 cells. (f) After KOCL-48 cell lines were incubated with TCP (0, 10, 25 μM) for 72 h, DNA was obtained from the IP chromatin using antibodies to H3K4me2 and histone H3 (H3). Precipitated DNA was analyzed by real-time RT-PCR using primers mapping to within the promoter region and 5′UTR of the RARα gene and (g) the PU.1 URE or RUNX1+24/+25 intronic enhancer. The results shown are normalized to the level of H3. Results represent the mean±s.d. of two independent ChIP experiments and a total of four independent PCR analysis.
Discussion
In this study, we have examined the mechanisms of ATRA sensitivity in non-APL AML cells. We found that MLL-AF9-positive cells were sensitive to ATRA and that this was accompanied by the upregulation of myeloid differentiation genes, such as C/EBPα,27, 28, 29 C/EBPɛ30, 31, 32 and PU.1.33, 34 In contrast, MLL-AF4/AF5q31-positive cells were not sensitive to ATRA, suggesting that the RA pathway is more strongly inactivated in MLL-AF4/AF5q31-positive cells than in MLL-AF9-positive cells. In addition, the experiments on murine MLL-rearranged immortalized cells revealed that the response to ATRA was dependent on the MLL-fusion partner gene.
Furthermore, we demonstrated a synergistic antileukemic effect between cytarabine and ATRA in MLL-AF9-positive cells, which were sensitive to ATRA, in vitro. Previous studies have shown that ATRA induces cell-cycle arrest and apoptosis in nucleophosmin 1 (NPM1)-mutated AML cells and that the NPM1 mutation sensitizes the AML cells to ATRA and cytarabine in vitro.35, 36 However, several subsequent studies have failed to reveal any clinical benefit of combining ATRA with conventional chemotherapy.37, 38, 39 Thus it will be important to determine whether the synergistic antileukemic effect of cytarabine and ATRA seen in MLL-AF9-positive AML cells in vitro is corroborated in vivo.
We also demonstrated that the level of H3K4me2 in the RARα gene-promoter region was closely associated with ATRA sensitivity. Our results showed that loss of H3K4me2 levels in RARα resulted in the inactivation of the RA pathway and that inhibition of LSD1 could reactivate the RA pathway in ATRA-resistant MLL-AF4-positive cells. In addition, we revealed that the levels of H3K4me2 in the PU.1 URE and RUNX1 intronic enhancer were also associated with the degree of ATRA-mediated myeloid differentiation, which was restored by the inhibition of LSD1.
Recent studies demonstrated that MLL was directly bound to RUNX1 physically and functionally.40, 41 Also, RUNX1 bound to the PU.1 URE, which has enhancer and repressor activity to regulate PU.1 expression.24, 42 Furthermore, RUNX1+24/+25 intronic enhancer was identified as a cis-regulatory element of RUNX1 gene expression in hematopoietic stem cells.25 Collectively, MLL, RUNX1 and PU.1 were closely interacted, and these interactions were modified by epigenetic regulation at the gene regulatory region, such as PU.1 URE or RUNX1 intronic enhancer, suggesting MLL fusions might interfere the myeloid differentiation caused by RUNX1 and PU.1 in different manner. Epigenetic research in leukemia is a rapidly developing field43 and in APL the PML–RARα fusion gene is now known to recruit various histone-modifying enzymes. Binding of the large PML–RARα fusion-protein complex to the promoter of the RARα target gene ‘closes’ the chromatin structure, resulting in the repression of gene expression, blocking the RA pathway, and thereby allowing oncogenic programs to progress.44, 45 Similarly, the AML1–ETO fusion protein (produced by the t(8;21) translocation) also recruits histone deacetylase and DNA methyltransferase, which block the RA pathway and promote leukomogenesis.46, 47 In addition, the demethylating agent or histone deacetylase inhibitor enhance the ATRA-mediated myeloid differentiation in MLL-AF9-expressing AML cells.8, 9 These observations suggest that epigenetic modification is an important factor for leukomogenesis in some AML subtypes, including MLL-rearranged AML.
LSD1, which demethylates histone H3 at lysine 4 (H3K4) and lysine 9 (H3K9), was the first histone demethylase to be identified.48 LSD1 is overexpressed in various cancers, and recent researches showed that this epigenetic modifier is associated with the development of drug resistance.49, 50 Schenk et al.13 reported that the LSD1 inhibitor TCP can reactivate ATRA sensitivity in the ATRA-resistant TEX cell line in vitro and diminish the engraftment of primary AML samples in vivo. In addition, they demonstrated that the combination of ATRA and TCP led to upregulation of gene expression associated with the myeloid differentiation program and apoptosis, accompanied by increased amounts of H3K4me2 near the transcriptional start sites.13 Similarly, in the current study we found that the amount of H3K4me2 in the region from−1000 to+1000 from the transcriptional start site of RARα was correlated with ATRA sensitivity and that TCP could reactivate ATRA sensitivity in an ATRA-resistant cell line expressing the MLL-AF4 fusion protein. We also revealed that TCP restored the levels of H3K4me2 in the PU.1 URE and RUNX1 intronic enhancer in the ATRA-resistant cell line. The IC50 of ATRA in combination with TCP was, however, much higher than the pharmacological concentration of ATRA for clinical use, suggesting that a more potent inhibitor of histone demethylase is required for reactivation of ATRA sensitivity in an ATRA-resistant cell.51, 52, 53
In conclusion, we found that MLL-AF9-positive AML cells were sensitive to ATRA and that a high level of H3K4me2 in the promoter region of the RARα gene, PU.1 URE and RUNX1 intronic enhancer is associated with ATRA sensitivity in this subtype. Addition of ATRA to cytarabine had a synergistic antileukemic effect on MLL-AF9-positive AML cells in vitro, suggesting that in cases of AML with high levels of H3K4me2 in the RARα promoter region, PU.1 URE and RUNX1 intronic enhancer, ATRA would be able to sensitize leukemic cells to cytarabine; an in vivo study is now required to confirm this. Furthermore, our findings indicate that epigenetic modifiers, such as LSD1 inhibitors, are potentially useful for treating ATRA-resistant AML, including MLL-rearranged AML.
References
Breitman TR, Selonick SE, Collins SJ . Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc Natl Acad Sci USA 1980; 77: 2936–2940.
Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L et al. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood 1988; 72: 567–572.
Zelent A, Guidez F, Melnick A, Waxman S, Licht JD . Translocations of the RARalpha gene in acute promyelocytic leukemia. Oncogene 2001; 20: 7186–7203.
Nowak D, Stewart D, Koeffler HP . Differentiation therapy of leukemia: 3 decades of development. Blood 2009; 113: 3655–3665.
Brown G, Hughes P . Retinoid differentiation therapy for common types of acute myeloid leukemia. Leuk Res Treatment 2012; 2012: 939021.
Glasow A, Barrett A, Petrie K, Gupta R, Boix-Chornet M, Zhou DC et al. DNA methylation-independent loss of RARA gene expression in acute myeloid leukemia. Blood 2008; 111: 2374–2377.
Glasow A, Prodromou N, Xu K, von Lindern M, Zelent A . Retinoids and myelomonocytic growth factors cooperatively activate RARA and induce human myeloid leukemia cell differentiation via MAP kinase pathways. Blood 2005; 105: 341–349.
Fujiki A, Imamura T, Sakamoto K, Kawashima S, Yoshida H, Hirashima Y et al. All-trans retinoic acid combined with 5-aza-2'-deoxycitidine induces C/EBPα expression and growth inhibition in MLL-AF9-positive leukemic cells. Biochem Biophys Res Commun 2012; 428: 216–223.
Iijima K, Honma Y, Niitsu N . Granulocytic differentiation of leukemic cells with t(9;11)(p22;q23) induced by all-trans-retinoic acid. Leuk Lymphoma 2004; 45: 1017–1024.
Gu ZM, Wu YL, Zhou MY, Liu CX, Xu HZ, Yan H et al. Pharicin B stabilizes retinoic acid receptor-α and presents synergistic differentiation induction with ATRA in myeloid leukemic cells. Blood 2010; 116: 5289–5297.
Fang Y, Zhou X, Lin M, Ying M, Luo P, Zhu D et al. Inhibition of all-trans-retinoic acid-induced proteasome activation potentiates the differentiating effect of retinoid in acute myeloid leukemia cells. Mol Carcinog 2011; 50: 24–35.
Cassinat B, Zassadowski F, Ferry C, Llopis L, Bruck N, Lainey E et al. New role for granulocyte colony-stimulating factor-induced extracellular signal-regulated kinase 1/2 in histone modification and retinoic acid receptor α recruitment to gene promoters: relevance to acute promyelocytic leukemia cell differentiation. Mol Cell Biol 2011; 31: 1409–1418.
Schenk T, Chen WC, Göllner S, Howell L, Jin L, Hebestreit K et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med 2012; 18: 605–611.
Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012; 21: 473–487.
Meyer C, Hofmann J, Burmeister T, Gröger D, Park TS, Emerenciano M et al. The MLL recombinome of acute leukemias in 2013. Leukemia 2013; 27: 2165–2176.
Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood 2009; 114: 2489–2496.
Balgobind BV, Zwaan CM, Pieters R, Van den Heuvel-Eibrink MM . The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia 2011; 25: 1239–1248.
von Neuhoff C, Reinhardt D, Sander A, Zimmermann M, Bradtke J, Betts DR et al. Prognostic impact of specific chromosomal aberrations in a large group of pediatric patients with acute myeloid leukemia treated uniformly according to trial AML-BFM 98. J Clin Oncol 2010; 28: 2682–2689.
Stam RW, Schneider P, Hagelstein JA, van der Linden MH, Stumpel DJ, de Menezes RX et al. Gene expression profiling-based dissection of MLL translocated and MLL germline acute lymphoblastic leukemia in infants. Blood 2010; 115: 2835–2844.
Stumpel DJ, Schneider P, van Roon EH, Boer JM, de Lorenzo P, Valsecchi MG et al. Specific promoter methylation identifies different subgroups of MLL-rearranged infant acute lymphoblastic leukemia, influences clinical outcome, and provides therapeutic options. Blood 2009; 114: 5490–5498.
Niitsu N, Hayashi Y, Sugita K, Honma Y . Sensitization by 5-aza-2'-deoxycytidine of leukaemia cells with MLL abnormalities to induction of differentiation by all-trans retinoic acid and 1alpha,25-dihydroxyvitamin D3. Br J Haematol 2001; 112: 315–326.
Yoshida H, Imamura T, Fujiki A, Hirashima Y, Miyachi M, Inukai T et al. Post-transcriptional modulation of C/EBPα prompts monocytic differentiation and apoptosis in acute myelomonocytic leukemia cells. Leuk Res 2012; 36: 735–741.
Leroy P, Nakshatri H, Chambon P . Mouse retinoic acid receptor alpha 2 isoform is transcribed from a promoter that contains a retinoic acid response element. Proc Natl Acad Sci USA. 1991; 88: 10138–10142.
Chim CS, Wong SY, Pang A, Chu P, Lau JS, Wong KF et al. Aberrant promoter methylation of the retinoic acid receptor alpha gene in acute promyelocytic leukemia. Leukemia 2005; 19: 2241–2246.
Okuno Y, Huang G, Rosenbauer F, Evans EK, Radomska HS, Iwasaki H et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol Cell Biol 2005; 25: 2832–2845.
Ng CE, Yokomizo T, Yamashita N, Cirovic B, Jin H, Wen Z et al. A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem Cells 2010; 28: 1869–1881.
Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG . Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci USA 1997; 94: 569–574.
Schepers H, Wierenga AT, van Gosliga D, Eggen BJ, Vellenga E, Schuringa JJ . Reintroduction of C/EBPalpha in leukemic CD34+ stem/progenitor cells impairs self-renewal and partially restores myelopoiesis. Blood 2007; 110: 1317–1325.
Kummalue T, Friedman AD . Cross-talk between regulators of myeloid development: C/EBPalpha binds and activates the promoter of the PU.1 gene. J Leukoc Biol 2003; 74: 464–470.
Matsushita H, Nakajima H, Nakamura Y, Tsukamoto H, Tanaka Y, Jin G et al. C/EBPalpha and C/EBPvarepsilon induce the monocytic differentiation of myelomonocytic cells with the MLL-chimeric fusion gene. Oncogene 2008; 27: 6749–6760.
Gombart AF, Kwok SH, Anderson KL, Yamaguchi Y, Torbett BE, Koeffler HP . Regulation of neutrophil and eosinophil secondary granule gene expression by transcription factors C/EBP epsilon and PU.1. Blood 2003; 101: 3265–3273.
Park DJ, Chumakov AM, Vuong PT, Chih DY, Gombart AF, Miller WH Jr et al. CCAAT/enhancer binding protein epsilon is a potential retinoid target gene in acute promyelocytic leukemia treatment. J Clin Invest 1999; 103: 1399–1408.
DeKoter RP, Walsh JC, Singh H . PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J 1998; 17: 4456–4468.
Anderson KL, Smith KA, Perkin H, Hermanson G, Anderson CG, Jolly DJ et al. PU.1 and the granulocyte- and macrophage colony-stimulating factor receptors play distinct roles in late-stage myeloid cell differentiation. Blood 1999; 94: 2310–2318.
Martelli MP, Pettirossi V, Manes N, Liso A, Mezzasoma M, Cecchetti F et al. Selective silencing of the NPM1 mutant protein and apoptosis induction upon ATRA in vitro treatment of AML cells carrying NPM1 mutations. Blood 2007; 110: 868.
Balusu R, Fiskus W, Rao R, Chong DG, Nalluri S, Mudunuru U et al. Targeting levels or oligomerization of nucleophosmin 1 induces differentiation and loss of survival of human AML cells with mutant NPM1. Blood 2011; 118: 3096–3106.
Schlenk RF, Döhner K, Kneba M, Götze K, Hartmann F, Del Valle F et al. Gene mutations and response to treatment with all-trans retinoic acid in elderly patients with acute myeloid leukemia. Results from the AMLSG Trial AML HD98B. Haematologica 2009; 94: 54–60.
Burnett AK, Hills RK, Green C, Jenkinson S, Koo K, Patel Y et al. The impact on outcome of the addition of all-trans retinoic acid to intensive chemotherapy in younger patients with nonacute promyelocytic acute myeloid leukemia: overall results and results in genotypic subgroups defined by mutations in NPM1, FLT3, and CEBPA. Blood 2010; 115: 948–956.
Nazha A, Bueso-Ramos C, Estey E, Faderl S, O'Brien S, Fernandez MH et al. The addition of all-trans retinoic acid to chemotherapy may not improve the outcome of patient with NPM1 mutated acute myeloid leukemia. Front Oncol 2013; 3: 218.
Koh CP, Wang CQ, Ng CE, Ito Y, Araki M, Tergaonkar V et al. RUNX1 meets MLL: epigenetic regulation of hematopoiesis by two leukemia genes. Leukemia 2013; 27: 1793–1802.
Huang G, Zhao X, Wang L, Elf S, Xu H, Zhao X et al. The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations. Blood 2011; 118: 6544–6552.
Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet 2008; 40: 51–60.
Rodríguez-Paredes M, Esteller M . Cancer epigenetics reaches mainstream oncology. Nat Med 2011; 17: 330–339.
Martens JH, Brinkman AB, Simmer F, Francoijs KJ, Nebbioso A, Ferrara F et al. PML-RARalpha/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell 2010; 17: 173–185.
Hoemme C, Peerzada A, Behre G, Wang Y, McClelland M, Nieselt K et al. Chromatin modifications induced by PML-RARalpha repress critical targets in leukemogenesis as analyzed by ChIP-Chip. Blood 2008; 111: 2887–2895.
Ferrara FF, Fazi F, Bianchini A, Padula F, Gelmetti V, Minucci S et al. Histone deacetylase-targeted treatment restores retinoic acid signaling and differentiation in acute myeloid leukemia. Cancer Res 2001; 61: 2–7.
Fazi F, Zardo G, Gelmetti V, Travaglini L, Ciolfi A, Di Croce L et al. Heterochromatic gene repression of the retinoic acid pathway in acute myeloid leukemia. Blood 2007; 109: 4432–4440.
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119: 941–953.
Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010; 141: 69–80.
Hou J, Wu J, Dombkowski A, Zhang K, Holowatyj A, Boerner JL et al. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am J Transl Res 2012; 4: 247–256.
Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M et al. Identification of cell-active lysine specific demethylase 1-selective inhibitors. J Am Chem Soc 2009; 131: 17536–17537.
Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res 2011; 71: 7238–7249.
Ogasawara D, Itoh Y, Tsumoto H, Kakizawa T, Mino K, Fukuhara K et al. Lysine-specific demethylase 1-selective inactivators: protein-targeted drug delivery mechanism. Angew Chem Int Ed Engl 2013; 52: 8620–8624.
Acknowledgements
This work was supported by grants for Clinical Cancer Research and Research on Measures for Intractable Diseases from the Japanese Ministry of Health, Labor and Welfare and by grants-in-aid for scientific research from the Japanese Ministry of Education, Culture, Sports, Science and Technology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Blood Cancer Journal website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Sakamoto, K., Imamura, T., Yano, M. et al. Sensitivity of MLL-rearranged AML cells to all-trans retinoic acid is associated with the level of H3K4me2 in the RARα promoter region. Blood Cancer Journal 4, e205 (2014). https://doi.org/10.1038/bcj.2014.25
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bcj.2014.25
This article is cited by
-
Dynamic regulation and functions of mRNA m6A modification
Cancer Cell International (2022)
-
All-trans retinoic acid induces differentiation in primary acute myeloid leukemia blasts carrying an inversion of chromosome 16
International Journal of Hematology (2022)
-
Co-existence of a novel translocation t(11;22)(q23;q12.1) with PML-RARA in acute promyelocytic leukemia: a case report
Annals of Hematology (2022)
-
Diptoindonesin G promotes ERK-mediated nuclear translocation of p-STAT1 (Ser727) and cell differentiation in AML cells
Cell Death & Disease (2017)
-
The Retinoid Agonist Tazarotene Promotes Angiogenesis and Wound Healing
Molecular Therapy (2016)