Abstract
The c-Myb gene encodes the p75c-Myb isoform and less-abundant proteins generated by alternatively spliced transcripts. Among these, the best known is p89c-Mybex9b, which contains 121 additional amino acids between exon 9 and 10, in a domain involved in protein–protein interactions and negative regulation. In hematopoietic cells, expression of p89c-Mybex9b accounts for 10–15% of total c-Myb; these levels may be biologically relevant because modest changes in c-Myb expression affects proliferation and survival of leukemic cells and lineage choice and frequency of normal hematopoietic progenitors. In this study, we assessed biochemical activities of p89c-Mybex9b and the consequences of perturbing its expression in K562 and primary chronic myeloid leukemia (CML) progenitor cells. Compared with p75c-Myb, p89c-Mybex9b is more stable and more effective in transactivating Myb-regulated promoters. Ectopic expression of p89c-Mybex9b enhanced proliferation and colony formation and reduced imatinib (IM) sensitivity of K562 cells; conversely, specific downregulation of p89c-Mybex9b reduced proliferation and colony formation, enhanced IM sensitivity of K562 cells and markedly suppressed colony formation of CML CD34+ cells, without affecting the levels of p75c-Myb. Together, these studies indicate that expression of the low-abundance p89c-Mybex9b isoform has an important role for the overall biological effects of c-Myb in BCR/ABL-transformed cells.
Similar content being viewed by others
Introduction
The proto-oncogene c-myb encodes a transcription factor that is expressed predominantly in immature hematopoietic, epithelial and endothelial cells and in many tumor types1, 2 and is downregulated as cells become more differentiated.3
In hematopoietic progenitor cells, c-Myb has an important role in the control of cell proliferation, survival and differentiation;4 in vitro, downregulation of c-Myb expression leads to decreased proliferation and colony formation of myeloid progenitors,5 whereas loss of c-Myb, in vivo, is embryonically lethal due to failure of fetal hematopoiesis.6 Conditional knockout of c-Myb expression in adult hematopoietic stem cells causes loss of self-renewal due to impaired proliferation and accelerated differentiation,7 suggesting that c-Myb has an essential role also in bone marrow primitive hematopoiesis. Constitutive overexpression of c-Myb in myeloid and erythroid cell lines blocks differentiation and prevents maturation-associated growth arrest.8, 9 Aberrant c-Myb expression has been detected in several malignancies including T-cell leukemia,10 chronic myeloid leukemia (CML), acute myeloid leukemia,11, 12 colorectal cancer,13 breast cancer14 and, more recently, adenoid cystic carcinomas.15 In CML, the increased expression of c-Myb is, in part, due to enhanced protein stability via BCR/ABL-regulated activation of PI-3K/Akt/GSKIIIβ-dependent pathways;16, 17 this altered regulatory mechanism may explain the requirement of c-Myb for in vitro proliferation and survival of leukemic progenitor cells18, 19 and for leukemogenesis in mice.20
Leukemic blast cells appear to rely on c-Myb expression more than their normal counterpart,21 suggesting that this differential requirement for c-Myb may be exploited therapeutically.
The requirement of c-Myb may depend on its ability to modulate the expression of genes (that is, CD34, c-kit, c-myc, flt-3 and Bcl-2) with important roles for the proliferation and survival of hematopoietic cells.22, 23, 24, 25, 26 c-Myb also regulates the expression of cyclin B1, contributing to the control of the G2/M phase of the cell cycle, in addition to its role during G1/S-phase transition.27
More recently, we found that c-Myb activates, in a DNA-binding manner, the expression of Slug and it promotes, via Slug, the homing of K562 cells to the bone marrow.28
The main product of the c-myb gene is a 75KDa nuclear protein that contains three functional domains: (i) an N-terminal DNA-binding domain that specifically binds to the sequence PyAACG/TG; (ii) a centrally-located transcription activation domain; and (iii) a C-terminal negative regulatory domain (NRD), which includes a leucine zipper and an EVES motif that modulate the activity of c-Myb via inter-and intra-molecular interactions.29, 30
Alternatively, spliced c-myb transcripts have been detected in hematopoietic cells of several species, including humans.31, 32, 33 The best-characterized of these transcripts includes a 363 bp segment between exons 9 and 10 (designated as exon 9b in humans), which is translated into 121 additional amino acids that disrupt the NRD domain.34, 35 p89c-Mybex9b represents 10–15% of total c-Myb protein in hematopoietic cells and has, apparently, the same properties of the predominant p75c-Myb species: it localizes into the nucleus and binds to the same DNA-binding sequence.36, 37 However, the 121 amino acids added by exon 9b could, in principle, enhance and/or modify the activity of the p89c-Mybex9b isoform by disruption of the intra- and/or inter-molecular interactions that may regulate stability and transactivation activity of c-Myb.38
The function and requirement of the p89c-Mybex9b isoform is understood only in part: it appears to transactivate the expression of certain c-Myb targets more effectively than the predominant p75c-Myb isoform,39 and yet the specific knockout of p89c-Mybex9b expression has no deleterious consequences on mammalian hematopoiesis and development,40 suggesting that its loss is compensated by expression of p75c-Myb.
However, it is unknown whether expression of p89c-Mybex9b is required for the proliferation and survival of transformed hematopoietic cells independently of p75c-Myb expression.
We show here that p89c-Mybex9b is more stable and has higher transactivation activity than p75c-Myb; moreover, its specific downregulation impairs proliferation and colony formation and enhances the imatinib (IM) sensitivity of BCR/ABL-expressing cells, in spite of unperturbed expression of p75c-Myb.
Materials and methods
Plasmids
Mig-RI-c-Myb-HA and Mig-RI-Δ(358–452)c-Myb were obtained as described.17
Cyclin B1-Luc-pGL3 was a gift from the late Dr AM Gewirtz (University of Pennsylvania, Philadelphia, PA, USA). SLUG-Luc-pGL3 was recently described.28
MSCV-p89c-Mybex9b: p89c-Mybex9b was amplified from human Ph1 K562 cells by reverse-transcription PCR using exon 9b-specific primers (Fw 5′-GCCCTCGAGATGGACTACAAGGATGACGATGACAAG-3′ and Rv 5′-ACTGCTGACGTCAGCAAATATGA-3′; Fw 5′-TCATATTTGCTGACGTCAGCAGT-3′ and Rv 5′-CGGGAATTCTCACATGACCAGCGTCCGGG-3′) designed to generate 5′ and 3′ fragments. They were digested with XhoI/AatII and AatII/EcoRI, respectively, and then ligated to XhoI/EcoRI-digested MSCV.
MSCV-p89c-Mybex9bMUT was generated by mutating four nucleotides of MSCV-p89c-Mybex9b by two subsequent site-directed mutagenesis according to manufacturer's instructions (QuickChange II site-directed mutagenesis kit, Stratagene, Santa Clara, CA). Mutated primers were: 5′-CCACTGGTCATCCTCCGGAAAAAACGGGGCCA-3′ and its reverse complement (first mutagenesis); and 5′-CACAGCACAATTCCATTAGTCATCCTCCGGAAA-3′ and its reverse complement (second mutagenesis).
Mig-RI-p89c-Mybex9 plasmid was generated by digesting MSCV-p89c-Mybex9b with XhoI/AatII and AatII/EcoRI and ligating the two fragments into XhoI/EcoRI-digested Mig-RI.
MSCV-p89c-Mybex9b_shMUT and MSCV-p75c-Myb_shMUT, the mutant forms of p89c-Mybex9b and p75c-Myb, not inhibitable by the doxycycline (DOX)-regulated c-Myb-short hairpin RNA (shRNA)_pLVTSH lentivirus (gift of Dr TJ Gonda, Brisbane University, Australia),41 were generated using the In-Fusion HD Cloning Kit (Clontech Laboratories, Mountain View, CA, USA). The c-Myb-coding region was amplified from MSCV-p89c-Mybex9b and MSCV-p75c-Myb used as templates with primer set A (Fw 5′-CGCCGGAATTAGATCTATGGACTACAAGGATGACGATGAC-3′, Rv 5′-AATTCTAA C AG A TTCTTAACATTATCCAG-3′, which amplify a region of 1221 bp common to both p89c-Mybex9b and p75c-Myb) and primer set B (Fw 5′-C GC C GA G AC G CTCCAATTTATAGATTCT-3′, Rv 5′-ATTCGTTAACCTCGAGTCACATGACCAGCGTCC-3′, which amplify a region of 1094 and 732 bp of p89c-Mybex9b and p75c-Myb, respectively). Mutated bases in the primers are underlined, those targeted by the shRNA are in italics. Mutations maintain same amino acids as in p89c-Mybex9b and p75c-Myb. Amplification products and linearized MSCV retroviral vector were ligated and cloned according to the kit's instructions. Correct mutations were confirmed by sequencing.
Cells and culture conditions
293T cells and parental and derivative K562 cell lines were maintained as described.42 Derivative K562 cell lines (Δ(358–452)c-Myb, p89c-Mybex9b, c-Myb shRNA or c-Myb shRNA expressing p89c-Mybex9b_shMUT or p75c-Myb_shMUT) were established by retroviral infection and isolation of green fluorescent protein-positive cells or selection with puromycin.
Peripheral blood (granulocyte colony-stimulating factor (G-CSF) mobilized) normal CD34+ cells were purchased from Stem Cell Technologies (Stem Cell Technologies, Vancouver, British Columbia, Canada). CML chronic-phase fresh leukapheresis or peripheral blood samples were obtained with informed written consent from five patients. Samples were enriched for CD34+ and cultured as described.43
Transient transfection and dual-luciferase reporter assay
The Mig-RI Δ(358–452)c-Myb or p89c-Mybex9 (3 μg) was co-transfected with cyclin B1-Luc-pGL3 or SLUG-Luc-pGL3 (1 μg) and Renilla luciferase (0.02 μg) into 106 K562 cells using nucleofector kit V (Amaxa; Lonza Group, Basel, Switzerland) according to the manufacturer's protocol. After 48 h, cells were diluted 1:5 in fresh medium and cultured for an additional 6–8 h. Cells were then lysed (100 μl of passive lysis buffer; Promega, Madison, WI, USA) and Firefly and Renilla luciferase activities were measured on a luminometer using the dual-luciferase reporter assay kit (Promega). For the assay in 293T cells, cells were seeded into six-well plates at 5 × 104 cells per well. The following day, DNA (5 μg of reporter plasmid, 5 μg of effector plasmid and 0.5 μg of Renilla luciferase) was transfected using ProFection Mammalian Transfection System-Calcium Phosphate (Promega) following the manufacturer's protocol. Cells were washed 16 h after transfection and incubated in fresh medium for an additional 24 h. At 40 h post transfection, cells were lysed in 500 μl of passive lysis buffer per well (Promega) and Firefly and Renilla luciferase activities were measured as described. All the measurements were performed in triplicate.
Chromatin immunoprecipitation and co-immunoprecipitation
For chromatin immunoprecipitation experiments performed in Mig-RI-p89c-Mybex9b-FLAG K562 cells, samples were prepared as described.44 IPs were performed with anti-c-Myb antibody (30 μg; clone 1–1, Upstate Biotechnology, Millipore Corporation, Billerica, MA, USA) or anti-FLAG antibody (30 μg; clone M2, Sigma-Aldrich, St Louis, MO, USA) or anti-rabbit IgG or without antibody at 4 °C for 1 h with rotation. Immune complexes were collected with 45 μl of protein G-agarose beads at 4 °C overnight with rotation. Recovered DNA (2 μl) was used as a template for Real-Time PCR using GoTaq Real-Time PCR Mix (Promega) and primers on the 5′-flanking region of the human cyclin B127 or SLUG28 promoter. Data were analyzed with the percent input method: percentage of input was calculated by 100 × 2(CtInput−CtEnriched).
For co-immunoprecipitations, 293T cells were transfected with plasmids expressing tagged c-Myb and lysates were immunoprecipitated with Anti-FLAG M2-Affinity gel (Sigma-Aldrich) following the manufacturer's instructions. Tagged proteins were detected by anti-FLAG M2-peroxidase-HRP (Sigma-Aldrich) and anti-HA-peroxidase High Affinity 3F10 (Roche, Nutley, NJ, USA).
Measurement of c-Myb half-life
To asses the half-life of p75c-Myb and p89c-Mybex9b, cells (pre-treated or not with 2 μM IM for 16 h) were treated with cycloheximide (25 μg/ml) for 2 h to suppress protein synthesis. Samples were collected every 30 min and lysates (100 μg each) analyzed by anti-c-Myb western blotting. Later, bands corresponding to the c-Myb isoforms and to β-actin as loading control were scanned and densitometric analysis was carried out with ImageJ Software (National Institutes of Health). The half-lifes of p75c-Myb and p89c-Mybex9b were calculated using the formula: t1/2=(0.693xt)/ln(Nt/No) as described.17
Small interfering RNA (siRNA) transfection and real-time PCR
Human c-Myb siRNA pool (catalog no. L-003910-00-0005) and control siRNAs (catalog no. D-001810-10-05) were purchased from Dharmacon, Thermo Fisher Scientific (Lafayette, CO, USA).
p89c-Mybex9b-siRNA-1 and p89c-Mybex9b-siRNA-2 were customized through the Dharmacon web site and purchased from Sigma-Aldrich. A total of 106 K562 cells were resuspended in Ingenio Electroporation Solution (Mirus Bio LLC, Madison, WI, USA) and mixed with 5 μg of each siRNA. The solution was added to Ingenio Cuvettes (Mirus Bio LLC) and electroporated according to Amaxa Nucleofector II using program T-16. Then, cells were diluted in 2 ml of Iscove's modified Dulbecco's medium supplemented with 20% fetal bovine serum, penicillin/streptomycin (100 μg/ml each) and 2 mM L-glutamine. 24 h later siRNA electroporation was repeated in order to downregulate c-Myb expression completely.
Expression of p75c-Myb- and p89c-Mybex9b-specific transcripts was detected by Real-Time PCR in control or c-Myb pool or p89c-Mybex9b-siRNA-2-transfected K562 cells using the following primers: p89c-Mybex9b-specific transcript: Fw 5′-AACATCACACAGGCAAAGC-3′ and Rv 5′-TGAACTCTCACTCAACATACG-3′; p75c-Myb-specific transcript: Fw 5′-TAGATTCTTTCTTAAACACTTCC-3′ and Rv 5′-GTCTCTATGAAATGGTGTTGTAAC-3′; for normalization, expression of hypoxanthine-guanine phosphoribosyltransferase was detected with the following primers: Fw 5′-AGACTTGCTTTCCTTGGTCAGG-3′; Rv 5′-GTCTGGCTTATATCCAACACTTCG-3′. Control siRNAs, c-Myb siRNAs pool and p89c-Mybex9b-siRNA-2 (5 μg) were delivered to normal or CML CD34+ cells using nucleofector kit V, program U-08 (Amaxa; Lonza Group), according to the manufacturer's protocol.
Cell proliferation, cell-cycle analysis and colony-formation assay
Proliferation of K562 cells transfected with different siRNAs was assessed by seeding 1.2 × 105 cells/ml and counting the cells every 24 h by trypan blue exclusion.
For induction of apoptosis, parental or derivative K562 cells were seeded at 2 × 105 cells/ml and treated with 2 μM IM (1 μM added every 12 h) (LC Laboratories, Woburn, MA, USA). Cells were counted every 24 h by trypan blue exclusion and percentage of apoptosis evaluated by the hypotonic propidium iodide method45 through Coulter Epics XL-MCL (Beckman Coulter Inc., Indianapolis, IN, USA).
G2/M-phase cells were also evaluated by propidium iodide staining. Phospho-histone-H3 positivity was assessed by Phospho-(Ser10)-histone-H3 (Cell Signalling Technology, Danvers, MA, USA) staining according to the manufacturer's instructions.
For colony-formation assays, K562 cells were plated in methylcellulose (Stem Cell Technologies) with or without IM (1 μM; pre-treatment in liquid culture for 24 h and added to the plates) and with or without doxycycline (5 μg/ml; pre-tratment in liquid culture for 24 h and added to the plates); colonies were counted 6 days later. CD34+ cells were plated in methylcellulose supplemented with Cytokine Cocktail CC100 (Stem Cell Technologies) and colonies were counted 9 days later.
Statistical analyses
Data (presented as the means±s.d. of two or three experiments) were analyzed for statistical significance by the unpaired, two-tailed Student's t-test. P-values of <0.05 were considered statistically significant.
Results
Increased transactivation of c-Myb-regulated promoters by p89c-Mybex9b
Two c-Myb mutans, Δ(358–452)c-Myb and Δ(389–418)c-Myb, which lack 93 and 30 amino acids, respectively, in the leucine-zipper-NRD (LZ-NRD) are more potent than wild-type p75c-Myb in enhancing proliferation and blocking apoptosis of normal and transformed hematopoietic cells.17
p89c-Mybex9b, a naturally occurring alternatively spliced form of c-myb, which accounts for approximately 10–15% of total c-Myb levels, contains an insertion of 121 amino acids in the LZ which disrupts its structure, presumably preventing the interaction with regulatory c-Myb-interacting proteins.
A comparison (Figure 1a) of p75c-Myb, p89c-Mybex9b and the LZ-NRD mutant Δ(358–452)c-Myb shows that the LZ is altered in p89c-Mybex9b and in the c-Myb mutant, suggesting that p89c-Mybex9b may function like the artificial LZ-NRD Δ(358–452)c-Myb.
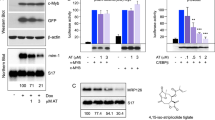
Transactivation activity of p89c-Mybex9b. (a) Schematic diagram of c-Myb proteins; levels of p75c-Myb, p89c-Mybex9b and Δ(358–452)c-Myb in transfected 293T cells (b) and K562 cells (c) used for luciferase assays. Expression of β-actin was detected as loading control; c-Myb and β-actin were detected by anti-c-Myb (clone 1-1, Upstate Biotechnology) and anti-β-actin antibody (sc-47778, Santa Cruz Biotechnology). (d) Densitometric analysis of c-Myb protein levels in transfected K562 normalized by corresponding β-actin expression. (e) Luciferase activity in 293T cells co-transfected with c-Myb plasmids and pGL3-cyclin B1-Luc (upper panel) or pGL3-Slug-Luc (lower panel). Results (from three and two experiments performed in duplicate, respectively) are expressed as fold activation relative to MSCV empty vector-transfected cells, after normalization for Renilla luciferase activity. (f, upper panel) Luciferase activity in K562 cells transfected with pGL3-cyclin B1-Luc and c-Myb plasmids. Results (means of three different experiments) are reported as fold activation relative to the pGL3 empty vector, after normalization for Renilla activity. (f, lower panel) Luciferase activity in K562 cells transfected with pGL3-Slug-Luc and c-Myb plasmids. Results (from two independent experiments, performed in duplicate) show fold activation relative to the MSCV empty vector, after normalization for Renilla activity. Error bars in e and f represent the s.d. of the means. P-values were calculated using unpaired, two-tailed Student's t-test, *denotes statistical significance of differences in transactivation activity of p75c-Myb versus p89c-Mybex9b or Δ(358–452)c-Myb and of p89c-Mybex9b versus Δ(358–452)c-Myb.
The transactivation potential of p89c-Mybex9b, Δ(358–452)c-Myb, and p75c-Myb was compared by dual-luciferase assays using reporter plasmids consisting of a fragment of the Myb-regulated human cyclin B1 or SLUG promoter driving the luciferase gene (cyclin B1-Luc or SLUG-Luc).
Following co-transfection of reporter and effector plasmids in 293T cells, expression and transactivation of each c-Myb protein were analyzed.
Expression of the three c-Myb isoforms was essentially identical in 293T cells (Figure 1b). p89c-Mybex9b and Δ(358–452)c-Myb were slightly more effective than p75c-Myb in transactivation of the cyclin B1 promoter but not of the SLUG promoter (Figure 1e). The transactivating ability of the three c-Myb proteins was also tested in Ph1 K562 cells; because K562 cells express high levels of endogenous c-Myb, the increase in expression of p75c-Myb in transfected cells (Figure 1c) was quantitated by densitometry (Figure 1d), whereas expression of p89c-Mybex9b and Δ(358–452)c-Myb was detected because of size differences and quantitated by densitometry (Figures 1c and d).
Compared with 293T cells, the effects of the c-Myb isoforms in K562 cells were markedly different: p75c-Myb induced approximately a three-fold increase in transactivation of the cyclin B1 promoter, whereas p89c-Mybex9b and Δ(358–452)c-Myb were much more potent (∼fifteen- and twelve-fold increase, respectively) (Figure 1f, upper panel); likewise, p89c-Mybex9b and Δ(358–452)c-Myb transactivated the SLUG promoter more effectively than p75c-Myb (∼five- and six- versus three-fold, respectively) (Figure 1f, lower panel).
p89c-Mybex9b binds to the human cyclin B1 and SLUG promoters
To assess whether p89c-Mybex9b interacted with the cyclin B1 and SLUG promoters, we performed chromatin immunoprecipitation assays in K562 cells overexpressing FLAG-tagged-p89c-Mybex9b using either an anti-FLAG antibody (which binds only the ectopically expressed protein) or an anti-c-Myb antibody (which binds the endogenous and the ectopically expressed protein). The ability of p89c-Mybex9b to bind the two promoters was assessed by real-time PCR amplification of a 5′-flanking region segment, which includes putative c-Myb-binding sites (Figures 2a and b, right). The −122 to +177 nucleotide segment of the cyclin B1 promoter and the −77 to +175 segment of the SLUG promoter were amplified above background in both the anti-c-Myb and anti-FLAG chromatin IPs, indicating that p89c-Mybex9b binds to these c-Myb-regulated promoters (Figures 2a and b, left).
p89c-Mybex9b binds the cyclin B1 and SLUG promoter. Chromatin immunoprecipitation assays show binding (detected by real-time PCR) of p75c-Myb and p89c-Mybex9 to a segment of the cyclin B1 promoter (a, left) and SLUG promoter (b, left). Error bars denote s.d. of the means of two experiments performed in triplicate. (a and b, right): partial sequence of the human promoter of cyclin B1 (GenBank: U22364.1) and SLUG (GenBank: AB300659.1). Putative c-Myb-binding sites located in the 5′-untranslated region (italics) are underlined; primers used for real-time PCR are in bold.
Enhanced stability of p89c-Mybex9b in BCR/ABL-positive cells
In addition to the transactivation ability, we also tested the stability of p89c-Mybex9b because the insertion of ex9b disrupts the LZ domain and c-Myb LZ-domain mutant proteins are more stable than wild-type p75c-Myb.17
Thus, parental and p89c-Mybex9b-expressing K562 cells were treated with cycloheximide for different times and cell lysates blotted with an anti-c-Myb antibody to assess half-life of endogenous p75c-Myb and ectopic p89c-Mybex9b, expressed at comparable levels. Expression of p75c-Myb was more rapidly downmodulated of p89c-Mybex9b; by densitometry analysis, the half-life of p89c-Mybex9b is ∼90 min, nearly 30 min longer than that of p75c-Myb (Figures 3a and b).
Half-life of p89c-Mybex9b in K562 cells. Representative western blot shows levels of p75c-Myb and p89c-Mybex9b in: (a) cells treated with cycloheximide only and (c) cells pre-treated for 16 h with 2 μM IM before cycloheximide treatment. Levels of β-actin were measured as loading control. (b–d) Densitometric analysis from two combined experiments assessing the half-life of c-Myb isoforms.
Because p210BCR/ABL enhances the stability of c-Myb,17 we also assessed the half-life of the two c-Myb isoforms in IM-treated cells. Inhibition of BCR/ABL tyrosine kinase activity decreased the half-life of p75c-Myb (∼35 min in IM-treated versus 60 min in untreated cells); in contrast, the half-life of p89c-Mybex9b was unaffected by treatment with IM (Figure 3c). By densitometry analysis, the half-life of p89c-Mybex9b in IM-treated cells was ∼75 min longer than that of p75c-Myb (110 versus 35 min, Figure 3d).
Inhibition of p89c-Mybex9b expression suppresses proliferation and colony formation of K562 cells.
We investigated the role of p89c-Mybex9b expression in transformed cells by assessing whether its specific knockdown has any effect on the proliferation and clonogenic potential of Ph1 K562 cells.
Two siRNAs targeting two different regions of human exon 9b c-Myb transcript (siRNA-1 and siRNA-2, Figure 4a) suppressed very effectively p89c-Mybex9b expression in K562 cells (Figure 4b, left) with no effects on levels of the p75c-Myb isoform. As control, we also used a pool of siRNAs targeting both p75c-Myb and the p89c-Mybex9b transcripts; transfection with this c-Myb siRNA pool led to the disappearance of both c-Myb isoforms (Figure 4b, right). Real-Time PCR using primers specific for p75c-Myb or p89c-Mybex9b transcripts further demonstrated that silencing of p89c-Mybex9b expression had no effect on levels of p75c-Myb transcripts (Supplementary Figure 1).
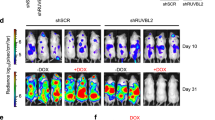
Effects of p89c-Mybex9b downregulation in K562 cells. (a) Schematic diagram representing the siRNAs target sequences of p89c-Mybex9b and the mutations in p89c-Mybex9b MUT. (b) Western blot shows levels of p75c-Myb and p89c-Mybex9b in K652 cells transfected with the c-Myb siRNA pool, c-Myb ex9b-specific siRNAs (sequence 1 and 2) or their relative controls. β-actin levels were measured as control of equal loading. (c) Cell counts and (d) colony formation of K562 cells transfected with c-Myb siRNA pool or its control (upper) or c-Myb ex9b siRNA 1 or siRNA2 or their scramble control (lower). (e) Levels of p89c-Mybex9b in MSCV-p89c-Mybex9b and MSCV-p89c-Mybex9bMUT K562 cells transfected with a control or the p89c-Mybex9b-siRNA 2. Expression of β-actin detected as loading control. (f) Methylcellulose colony formation of MSCV-p89c-Mybex9b and MSCV-p89c-Mybex9bMUT K562 cells transfected with a control or the p89c-Mybex9b-siRNA 2 (values of cell counts and number of colonies are given as mean±s.d. of three independent experiments performed in triplicate; * denotes statistical significance of the differences between scrambled- and siRNA-transfected cells). (g) Levels of p75c-Myb and p89c-Mybex9b in untreated and DOX-treated (24 h) K562 cells expressing a DOX-inducible c-Myb shRNA and transduced with p75c-Myb_shMUT or p89c-Mybex9b_shMUT. Expression of β-actin is detected as loading control. (h) Colony formation of untreated and DOX-treated cells; values are expressed as % of colonies formed by treated cells, compared with untreated cells taken as 100% (data are given as mean±s.d. of two independent experiments performed in triplicate; *denotes statistical significance of the differences between K562 cells with shMUT c-Mybs and cells with only c-Myb shRNA).
K562 cells transfected with the c-Myb siRNA pool or with p89c-Mybex9b-specific siRNAs proliferated less than control siRNA-transfected cells (Figure 4c); at 48 h, the c-Myb siRNA pool was more effective than the p89c-Mybex9b-specific siRNAs (54% versus 30% inhibition, respectively), but at 72 h, the effects were comparable.
We also assessed the effect of p89c-Mybex9b-specific downregulation on colony formation of K562 cells. Thus, cells were transfected with control or specific siRNAs and 24 h after the second transfection were plated in methylcellulose (500 cells per plate) and colonies were counted 6 days later. Cells treated with control siRNA formed 350±36 colonies (Figure 4d), whereas K562 cells transfected with the c-Myb siRNA pool formed 178±27 colonies; a significant decrease in colony formation was also noted by plating cells transfected with p89c-Mybex9b-siRNA-2 (230±14 colonies) or p89c-Mybex9b-siRNA-1 (230±11) (Figure 4d).
To assess the specificity of the biological effects induced by p89c-Mybex9b-siRNA-2, we generated K562-derivative lines ectopically expressing wild-type p89c-Mybex9b and a mutant, p89c-Mybex9bMUT, with four nucleotide substitutions in the sequence targeted by p89c-Mybex9b-siRNA-2; these mutations were designed to prevent interaction with siRNA-2 while preserving the amino-acid sequence of the protein (Figure 4a).
MSCV-p89c-Mybex9b and MSCV-p89c-Mybex9bMUT K562 cells were then transfected with a control or the p89c-Mybex9b-siRNA-2 and levels of p89c-Mybex9b were tested by western blotting. Levels of p89c-Mybex9b were undetectable in MSCV-p89c-Mybex9b K562 cells transfected with p89c-Mybex9b siRNA-2 (Figure 4e); in contrast, expression of p89c-Mybex9b was detected in siRNA-2-transfected MSCV-p89c-Mybex9bMUT K562 cells, reflecting downregulation of the endogenous protein but not of the p89c-Mybex9b protein derived from p89c-Mybex9bMUT, which is not targeted by p89c-Mybex9b-siRNA-2. As expected, MSCV-p89c-Mybex9b K562 cells transfected with p89c-Mybex9b-siRNA-2 formed fewer colonies (∼50% inhibition) than the scramble siRNA-transfected counterpart (Figure 4f); in contrast, there were no significant differences in colony formation of scrambled- or p89c-Mybex9b-siRNA-2-transfected MSCV-p89c-Mybex9bMUT K562 cells (Figure 4f), further confirming that the inhibition of K562 colony formation by p89c-Mybex9b-siRNA-2 is due to specific downregulation of p89c-Mybex9b expression.
The importance of p89c-Mybex9b expression for K562 colony formation was also assessed in K562 cells expressing a DOX-inducible c-Myb shRNA, which downregulates the p75c-Myb and p89c-Mybex9b isoforms, and carrying the p75c-Myb or p89c-Mybex9b expression vector non-targetable by the DOX-inducible c-Myb shRNA (p75c-Myb_shMUT and p89c-Mybex9b_shMUT, respectively) (Figure 4g). As expected, colony formation of DOX-treated K562-c-Myb shRNA cells was markedly suppressed (∼80% inhibition); in contrast, expression of p89c-Mybex9b_shMUT or p75c-Myb_shMUT rescued, in part, the inhibitory effect of the DOX-inducible c-Myb shRNA (Figure 4h), consistent with redundant and yet non-overlapping effects of the p75c-Myb and the p89c-Mybex9b-Myb isoforms.
Levels of cyclin B1 are downregulated in p89c-Mybex9b-silenced K562 cells
The reduced colony formation of c-Myb-silenced (p75c-Myb and/or p89c-Mybex9b) K562 cells may be caused by changes in the expression of c-Myb-regulated genes with a role in cell proliferation. Cyclin B1, a gene with an essential role in the G2/M transition, is one of the c-Myb targets27 whose change in expression may explain, in part, the effects of p89c-Mybex9b downregulation.
Indeed, compared with control siRNA-transfected cells, cyclin B1 expression was reduced in K562 cells transfected with the c-Myb siRNA pool (∼45% decrease of cyclin B1 levels); although expression of p75c-Myb was unaffected, the specific downregulation of p89c-Mybex9 led to a similar decrease (∼42%) of cyclin B1 expression (Figures 5a and b).
Effects of p89c-Mybex9b downregulation on cyclin B1 levels and G2/M-phase cells. (a) Western blot shows levels of c-Myb isoforms and cyclin B1 in K652 cells 16 h after treatment with the c-Myb siRNA pool or c-Mybex9b siRNA or control siRNA. Cyclin B1 expression was detected by anti-cyclin B1 antibody (sc-245, Santa Cruz Biotechnology). β-actin levels were measured as loading control. (b) Densitometric analysis of cyclin B1 levels after siRNA transfection. Densitometric values for cyclin B1 were normalized by the corresponding values for β-actin and expressed as percentage change, compared with those in control siRNA-transfected cells (taken as 100%). Results are representative of two different experiments. (c) % of G2/M-phase K562 cells 48 h after siRNA transfection (values represent the mean±s.d. of three independent experiments; *denotes statistical significance of the differences between scramble- and c-Myb siRNA-transfected cells). (d) % of phospho-H3-positivity (representative of three independent experiments) in K562 cells 48 h after siRNA transfection.
We also analyzed the fraction of G2/M-phase cells in c-Myb- or p89c-Mybex9-silenced K562 cells by evaluating DNA content of propidium iodide-stained cells and percentage of cells positive for phospho-histone-H3, which is a marker of M phase. As expected, the decrease in the cyclin B1 levels was accompanied by a reduction in the number of cells in G2/M phase (Figure 5c). Although not statistically significant, in three different experiments, the number of M-phase cells was lower in c-Myb- or p89c-Mybex9b-siRNA-2 than in control siRNA-transfected K562 cells (Figure 5d).
p89c-Mybex9b expression affects the response of K562 cells to treatment with IM
Because expression of p89c-Mybex9b appears to be important for proliferation and colony formation of K562 cells, we speculated that it may also influence the response to treatment with IM.
Parental, p89c-Mybex9b- or Δ(358–452)c-Myb-expressing K562 cells were treated with IM, and cell counts and DNA content analyses were performed every 24 h.
As expected, expression of Δ(358–452)c-Myb reduced the IM sensitivity of K562 cells (Figure 6a); expression of p89c-Mybex9b had a similar, although less potent, effect as IM-treated p89c-Mybex9b-expressing K562 cells exhibited a less pronounced decrease in cell number than parental cells (Figure 6a). Similar effects were observed upon assessing apoptosis of IM-treated K562 cells; as expected, IM-treated parental K562 cells showed a high percentage of apoptosis, which increased from 24 to 72 h. In contrast, apoptosis of IM-treated K562 cells expressing p89c-Mybex9b or Δ(358–452)c-Myb was approximately 50% lower than that of parental cells (Figure 6b). In normal growth conditions, overexpression of p89c-Mybex9b or Δ(358–452)c-Myb has no effect on the proliferation and basal apoptosis of K562 cells (Supplementary Figure 2).
Effects of p89c-Mybex9b levels on viability of IM-treated K562 cells. (a) Cell count and (b) apoptosis of IM-treated parental, p89c-Mybex9b or Δ(358–452)c-Myb ectopically expressing K562 cells. (c) Cell count and (d) apoptosis of IM-treated K562 cells transfected with the c-Myb siRNA pool or c-Mybex9b-specific siRNA or control siRNA (values indicate the increase in apoptosis at 24, 48 and 72 h over the corresponding normalized apoptosis at time 0 and represent the mean±s.d. of three independent experiments); colony formation of IM-treated: (e) parental, p89c-Mybex9b or Δ(358–452)c-Myb ectopically expressing K562 cells and (f) K562 cells transfected with c-Myb siRNA pool or c-Mybex9b-specific siRNA or control siRNA. Values are expressed as % of colonies from treated cells compared with those derived from the corresponding untreated cells taken as 100%. *denotes statistical significance of the differences between control (parental or scramble-transfected) and experimental (c-Myb ectopically expressed or siRNA-transfected) groups.
We also assessed the effect of c-Myb downregulation on IM-treated K562 cells. Parental cells transfected with the c-Myb siRNA pool, p89c-Mybex9b isoform-specific or control siRNAs were counted and analyzed by flow cytometry 24, 48 and 72 h after IM treatment. Downregulation of c-Myb or p89c-Mybex9b enhanced the proliferation inhibitory effects of IM, compared with cells transfected with the control siRNA (at 72 h, ∼60% fewer cells than in control siRNA-transfected K562 cells) (Figure 6c). Evaluation of apoptosis by DNA content analysis showed a similar pattern: c-Myb-silenced cells exhibited increased apoptosis compared with cells transfected with control siRNAs. Apoptotic cells were slightly more numerous after transfection with c-Myb siRNAs than with the p89c-Mybex9b-specific siRNA but the differences were not significant (Figure 6d).
We also assessed the relationship between levels of p89c-Mybex9b and colony formation after IM treatment. Thus, parental or ectopically expressing p89c-Mybex9b or Δ(358–452)c-Myb K562 cells were treated for 24 h with IM (1 μM) and plated in methylcellulose (500 cells per plate, in the presence of 1 μM IM); colonies were counted after 6 days.
Consistent with the effects on cell proliferation, expression of the p89c-Mybex9b isoform enhanced the number of clonogenic cells after IM treatment (13.8% and 6.5% of residual clonogenic cells in p89c-Mybex9b-expressing versus parental K562 cells). Residual colonies formed by p89c-Mybex9b or Δ(358–452)c-Myb-expressing K562 cells were nearly identical (Figure 6e).
Colony assays were also performed after c-Myb or p89c-Mybex9b-specific siRNA tranfection and IM treatment of K562 cells. As expected, the combination of Myb downregulation and IM treatment caused a marked decrease in the clonogenic potential of K562 cells. Residual colony formation of cells transfected with the c-Myb siRNA pool or p89c-Mybex9b-siRNA was similar (1.1% and 1.9%, respectively) and lower of that from cells transfected with control siRNAs (3.1%) (Figure 6f). Together, these data suggest that expression of the p89c-Mybex9b isoform can modulate the effects of IM in K562 cells.
Inhibition of p89c-Mybex9b suppresses colony formation of normal and CML CD34+ progenitors
The role of p89c-Mybex9b expression was also assessed in colony-formation assays of CD34+ cells from healthy donors and CML patients.
Normal human CD34+ cells (n=3) were transfected with the c-Myb siRNA pool or a p89c-Mybex9b-specific siRNA or the control siRNA. 24 h after transfection, levels of c-Myb proteins were tested and cells were plated for colony-formation assays. A representative western blot shows that levels of p75c-Myb and p89c-Mybex9b were completely downregulated in cells transfected with the c-Myb siRNA pool, whereas transfection with p89 c-Mybex9b-siRNA reduced only the levels of the p89c-Mybex9b isoform (Figure 7a). Normal CD34+ cells transfected with the c-Myb siRNA pool or the p89c-Mybex9b-specific siRNA formed fewer colonies than control siRNA-transfected cells (∼33% and 28% inhibition, respectively; range: 23–43% and 22–37%, respectively) (Figure 7b).
Colony formation of c-Myb or p89c-Mybex9b siRNA-transfected normal or CML CD34+ progenitors. (a and c) Western blots show levels of p75c-Myb and p89c-Mybex9b in a representative sample of normal (a) or CML (c) CD34+ cells after transfection with control or c-Myb siRNAs; β-actin was detected as loading control. (b and d) Methylcellulose colony formation of normal (n=3) (b) or CML (n=5) (d) CD34+ cells transfected with control or c-Myb siRNAs. Values of colony formation are mean±s.d.; all the experiments were performed in triplicate. *denotes statistical significance of the differences in colony formation between cells transfected with control siRNA, the c-Myb siRNA pool, or the c-Mybex9b-specific siRNA.
Peripheral blood CD34+ cells from CML chronic-phase patients (n=5) were also transfected with c-Myb or control siRNAs. Like in normal CD34+ cells, expression of p75c-Myb and p89c-Mybex9b was markedly downregulated in CML CD34+ cells transfected with the c-Myb siRNA pool, whereas only p89c-Mybex9b levels decreased in cells transfected with p89c-Mybex9b-siRNA (Figure 7c). Transfected CML CD34+ cells were seeded in cytokine-supplemented methylcellulose and colonies were counted 9 days later. Colonies arising from cells transfected with the c-Myb siRNA pool or p89c-Mybex9b-siRNA were fewer (63% and 41% decrease, respectively; range: 50–90% and 28–58% , respectively) than those from the scramble siRNA-transfected counterpart (Figure 7d).
Together, these results suggest that expression of p89c-Mybex9b is more important for colony formation of CML than normal CD34+ progenitors.
Discussion
The c-Myb gene generates multiple transcripts that encode the predominant p75c-Myb isoform and, potentially, several less-abundant species.46 However, only the p89c-Mybex9b isoform is readily detectable in hematopoietic cells, suggesting that other alternatively spliced transcripts are too rare to generate detectable proteins or that the encoded gene products are unstable and rapidly degraded. p89c-Mybex9b accounts for 10–15% of total c-Myb protein, an amount that could be biologically relevant because small changes in the levels of c-Myb can affect lineage choice and progenitor cell frequencies in normal hematopoiesis.47, 48 p89c-Mybex9b isoform-specific knockout mice have no apparent defect in the number of steady-state mature hematopoietic cells,40 but the long-term proliferative potential, survival and differentiation of specific progenitor subsets was not investigated in detail. Although this study suggests that, in normal cells, loss of p89c-Mybex9b expression is compensated by expression of the more abundant p75c-Myb isoform, it is conceivable that expression of p89c-Mybex9b is biologically more relevant in leukemic cells, which typically rely on c-Myb expression more than their normal counterparts.18, 21
We show here that perturbation of p89c-Mybex9b levels by ectopic expression or by specific downregulation modulates the proliferation, survival and IM sensitivity of BCR/ABL-transformed cells. The increased proliferation, survival and IM resistance of p89c-Mybex9b ectopically expressing K562 cells is reminiscent of similar effects observed in experiments carried out with artificial degradation-resistant mutants of the c-Myb LZ-NRD.17 The similarity of the effects is likely to reflect the comparable increase in protein stability and transactivation ability of the artificial c-Myb mutants and the p89c-Mybex9b isoform. Of greater interest, downregulation of the p89c-Mybex9b isoform was associated with reduced proliferation and colony formation and with enhanced IM sensitivity of BCR/ABL-transformed cells comparable to that observed after transfection with the c-Myb siRNA pool, which caused the downregulation of both the predominant p75c-Myb isoform and the less-abundant p89c-Mybex9b species.
An explanation for these surprising findings could be that downregulation of p89c-Mybex9b expression suppresses an autoregulatory loop, which may maintain elevated levels of the p75c-Myb isoform.49 However, this putative mechanism does not appear to be involved because p75c-Myb levels were not reduced in p89c-Mybex9b-silenced cells.
Another possibility is that disruption of the LZ-NRD in the p89c-Mybex9b isoform prevents protein–protein interactions with p75c-Myb or p75c-Myb-interacting proteins, allowing p89c-Mybex9b to escape the negative regulation imposed by p75c-Myb and/or its interacting proteins.50, 51 Consistent with this interpretation, co-immunoprecipitation assays showed that p75c-Myb interacted with p89c-Mybex9b less efficiently than it did with itself (Supplementary Figure 3).
Thus, the biological effects of c-Myb in p210BCR/ABL-transformed cells might be exerted, to a large extent, by the less-abundant but more potent p89c-Mybex9b isoform.
The fact that p89c-Mybex9b is a more potent transactivator of p75c-Myb and downregulation of p89c-Mybex9b is almost as effective as the combined downregulation of p75c-Myb and p89c-Mybex9b in inducing a decrease in the expression of the c-Myb-target cyclin B1 supports this interpretation, although the entire catalogue of c-Myb-target genes should be examined.
Lastly, the preferential requirement of the p89c-Mybex9b isoform by BCR/ABL-transformed cells may depend on the regulation of a specific gene subset, which may be distinct from that regulated by p75c-Myb.39 This possibility is supported by the observation that restoring the expression of either the p75c-Myb or p89c-Mybex9b isoform rescued only in part the inhibition of colony formation induced by downregulation of both, consistent with non-overlapping effects of the two isoforms.
Regardless of the mechanisms involved, the observation that the effects of c-Myb in leukemic cells are, in a significant part, due to minute amounts of an alternatively spliced isoform raises new questions regarding the elusive oncogenic potential of c-Myb.
References
Kastan MB, Slamon DJ, Civin CI . Expression of protooncogene c-myb in normal human hematopoietic cells. Blood 1989; 73: 1444–1451.
Ramsay RG, Gonda TJ . MYB function in normal and cancer cells. Nat Rev Cancer 2008; 8: 523–534.
Bjerregaard MD, Jurlander J, Klausen P, Borregaard N, Cowland JB . The in vivo profile of transcription factors during neutrophil differentiation in human bone marrow. Blood 2003; 101: 4322–4332.
Oh IH, Reddy EP . The myb gene family in cell growth, differentiation and apoptosis. Oncogene 1999; 18: 3017–3033.
Gewirtz AM, Calabretta BA . c-myb antisense oligodeoxynucleotide inhibits normal human hematopoiesis in vitro. Science 1988; 242: 1303–1306.
Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 1991; 65: 677–689.
Lieu YK, Reddy EP . Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of selfrenewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci USA 2009; 106: 21689–21694.
Bies J, Mukhopadhyaya R, Pierce J, Wolff L . Only late, nonmitotic stages of granulocyte differentiation in 32Dcl3 cells are blocked by ectopic expression of murine c-myb and its truncated forms. Cell Growth Differ 1995; 6: 59–68.
Selvakumaran M, Liebermann DA, Hoffman-Liebermann B . Deregulated c-myb disrupts interleukin-6- or leukemia inhibitory factor induced myeloid differentiation prior to c-myc: role in leukemogenesis. Mol Cell Biol 1992; 12: 2493–2500.
Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (TALL), the translocation defining a new T-ALL subtype in very young children. Blood 2007; 110: 1251–1261.
Slamon DJ, Boone TC, Murdock DC, Keith DE, Press MF, Larson RA et al. Studies of the human c-myb gene and its product in human acute leukemias. Science 1986; 233: 347–351.
Ganter B, Lipsick JS . Myb and oncogenesis. Adv Cancer Res 1999; 76: 21–60.
Biroccio A, Benassi B, D'Agnano I, D'Angelo C, Buglioni S, Mottolese M et al. c-Myb and Bcl-x overexpression predicts poor prognosis in colorectal cancer: clinical and experimental findings. Am J Pathol 2001; 158: 1289–1299.
Persson M, Andrén Y, Mark J, Horlings HM, Persson F, Stenman G . Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci 2009; 106: 18740–18744.
Stenman G, Andersson MK, Andrén Y . New tricks from an old oncogene: gene fusion and copy number alterations of MYB in human cancer. Cell Cycle 2010; 9: 2986–2995.
Gonda TJ, MacMillan EM, Towsend PV, Hapel AJ . Differentiation state and responses to hematopoietic growth factors of murine myeloid cells transformed by myb. Blood 1993; 82: 2813–2822.
Corradini F, Cesi V, Bartella V, Pani E, Bussolari R, Candini O et al. Enhanced proliferative potential of hematopoietic cells expressing degradation-resistant c-Myb mutants. J Biol Chem 2005; 280: 30254–30262.
Ratajczak MZ, Hijiya N, Catani L, DeRiel K, Luger SM, McGlave P et al. Acute- and chronic-phase chronic myelogenous leukemia colony-forming units are highly sensitive to the growth inhibitory effects of c-myb antisense oligodeoxy-nucleotides. Blood 1992; 79: 1956–1961.
Luger S, O'Brien SG, Ratajczak J, Ratajczak MZ, Mick R, Stadtmaner EA et al. Oligodeoxynucleotide-mediated inhibition of c-myb gene expression in autografted bone marrow: a pilot study. Blood 2002; 99: 1150–1158.
Lidonnici MR, Corradini F, Waldron T, Bender TP, Calabretta B . Requirement of c-Myb for p210(BCR/ABL)-dependent transformation of hematopoietic progenitors and leukemogenesis. Blood 2008; 111: 4771–4779.
Calabretta B, Sims RB, Valtieri M, Caracciolo D, Szczylik C, Venturelli D et al. Normal and leukemic hematopoietic cells manifest differential sensitivity to inhibitory effects of c-myb antisense oligodeoxynucleotides: an in vitro study relevant to bone marrow purging. Proc Natl Acad Sci USA 1991; 88: 2351–2355.
Melotti P, Ku DH, Calabretta B . Regulation of the expression of the hematopoietic stem cell antigen CD34: role of c-myb. J Exp Med 1994; 179: 1023–1028.
Salomoni P, Perrotti D, Martinez R, Franceschi C, Calabretta B . Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl-2 promoter activity. Proc Natl Acad Sci USA 1997; 94: 3296–3301.
Ness SA, Marknell A, Graf T . The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 1989; 59: 1115–1125.
Schmidt M, Nazarov V, Stevens L, Watson R, Wolff L . Regulation of the resident chromosomal copy of c-myc by c-Myb is involved in myeloid leukemogenesis. Mol Cell Biol 2000; 20: 1970–1981.
Lang G, White JR, Argent-Katwala MJ, Allinson CG, Weston K . Myb proteins regulate the expression of diverse target genes. Oncogene 2005; 24: 1375–1384.
Nakata Y, Shetzline S, Sakashita C, Kalota A, Rallapalli R, Rudnick SI et al. c-Myb contributes to G2/M cell cycle transition in human hematopoietic cells by direct regulation of cyclin B1 expression. Mol Cell Biol 2007; 27: 2048–2058.
Tanno B, Sesti F, Cesi V, Bossi G, Ferrari-Amorotti G, Bussolari R et al. Expression of Slug is regulated by c-Myb and is required for invasion and bone marrow homing of cancer cells of different origin. J Biol Chem 2010; 285: 29434–29445.
Corradini F, Bussolari R, Cerioli D, Lidonnici MR, Calabretta B . A degradation resistant c-Myb mutant cooperates with Bcl-2 in enhancing proliferative potential and survival of hematopoietic cells. Blood Cells Mol Dis 2007; 39: 292–296.
Dash AB, Orrico FC, Ness SA . The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev 1996; 10: 1858–1869.
Schuur ER, Rabinovich JM, Baluda MA . Distribution of alternatively spliced chicken c-myb exon 9A among hematopoietic tissues. Oncogene 1994; 9: 3363–3365.
Shen-Ong GL, Skurla RM, Owens JD, Mushinski JF . Alternative splicing of RNAs transcribed from the human c-myb gene. Mol Cell Biol 1990; 10: 2715–2722.
Westin EH, Gorse KM, Clarke MF . Alternative splicing of the human c-myb gene. Oncogene 1990; 5: 1117–1124.
Dasgupta P, Reddy EP . Identification of alternatively spliced transcripts for human c-myb: molecular cloning and sequence analysis of human c-myb exon 9A sequences. Oncogene 1989; 4: 1419–1423.
Dudek H, Reddy EP . Identification of two translational products for c-myb. Oncogene 1989; 4: 1061–1066.
Biedenkapp H, Borgmeyer U, Sippel AE, Klempnauer KH . Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature 1988; 335: 835–837.
Sakura H, Kanei-Ishii C, Nagase T, Nakagoshi H, Gonda TJ, Ishii S . Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci USA 1989; 86: 5758–5762.
Woo CH, Sopchak L, Lipsick JS . Overexpression of an alternatively spliced form of c-Myb results in increases in transactivation and transforms avian myelomonoblasts. J Virol 1998; 72: 6813–6821.
Kumar A, Baker SJ, Lee CM, Reddy EP . Molecular mechanisms associated with the regulation of apoptosis by the two alternatively spliced products of c-Myb. Mol Cell Biol 2003; 23: 6631–6645.
Baker SJ, Kumar A, Reddy EP . p89c-Myb is not required for fetal or adult hematopoiesis. Genesis 2010; 48: 309–316.
Drabsch Y, Hugo H, Zhang R, Dowhan DH, Miao YR, Gewirtz AM et al. Mechanisms of and requirement for estrogen-regulated MYB expression in estrogen-receptor-positive breast cancer cells. Proc Natl Acad Sci USA 2008; 105: 825–829.
Fragliasso V, Chiodo Y, Ferrari-Amorotti G, Soliera AR, Manzotti G, Cattelani S et al. Phosphorylation of serine 21 modulates the proliferation inhibitory more than the differentiation inducing effects of C/EBPα in K562 cells. J Cell Biochem 2012; 113: 1704–1713.
Soliera AR, Mariani SA, Audia A, Lidonnici MR, Addya S, Ferrari-Amorotti G et al. Gfi-1 inhibits proliferation and colony formation of p210BCR/ABL-expressing cells via transcriptional repression of STAT 5 and Mcl-1. Leukemia 2012, e-pub ahead of print 30 January 2012; doi:10.1038/leu.2012.19.
Ferrari-Amorotti G, Mariani SA, Novi C, Cattelani S, Pecorari L, Corradini F et al. The biological effects of C/EBPalpha in K562 cells depend on the potency of the N-terminal regulatory region, not on specificity of the DNA binding domain. J Biol Chem 2010; 285: 30837–30850.
Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C . A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Meth 1991; 139: 271–279.
O'Rourke JP, Ness SA . Alternative RNA splicing produces multiple forms of c-Myb with unique transcriptional activities. Mol Cell Biol 2008; 28: 2091–2101.
Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J . Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J 2003; 22: 4478–4488.
Sakamoto H, Dai G, Tsujino K, Huang X, Fujimoto T, Mucenski M et al. Proper levels of c-Myb are discretely defined at distinct steps of hematopoietic cell development. Blood 2006; 108: 896–903.
Nicolaides NC, Gualdi R, Casadevall C, Manzella L, Calabretta B . Positive autoregulation of c-myb expression via Myb binding sites in the 5′ flanking region of the human c-myb gene. Mol Cell Biol 1991; 11: 6166–6176.
Nomura T, Sakai N, Surai A, Sudo T, Kanei-Ishii C, Ramsay RG et al. Negative autoregulation of c-Myb activity by homodimer formation through the leucine zipper. J Biol Chem 1993; 268: 21914–21923.
Kaspar P, Dvorakova M, Krolova J, Pajer P, Koznick Z, Dvorak M . Myb-interacting protein, ATBF1, represses transcriptional activity of Myb oncoprotein. J Biol Chem 1999; 274: 14422–14428.
Acknowledgements
This study was supported by grants from: Fondazione Cassa di Risparmio di Modena, Programmi di ricerca di Rilevante Interesse Nazionale (PRIN2007), Fondazione Cassa di Risparmio di Vignola, Associazione ‘Amici di Lino’, Fondazione Guido Berlucchi per la Ricerca sul Cancro and NCI grant CA95111 (BC). GM, FC, GF-A, ARS and SC were supported, in part, by a fellowship of the Associazione Italiana Ricerca sul Cancro (AIRC). ARS and SC were supported in part by a fellowship of Fondazione Cassa di Risparmio di Modena. SAM was supported by an AIRC/Marie Curie international fellowship. GF-A was supported in part by a fellowship of Associazione ‘Angela Serra’ per la Ricerca sul Cancro. This paper is dedicated to the memory of Dr Alan M Gewirtz.
Author contributions
GM performed most of the work and wrote parts of the paper. SAM performed colony assays of normal umbilical cord blood and primary CML cells. FC generated some plasmid, established K562-derivative cell lines and performed some proliferation and colony assays. RB designed p89c-Mybex9b-specific siRNAs and performed initial nucleofection experiments. VC generated some plasmids. JV established some K562-derivative cell lines. GF-A and VF helped with flow cytometry analyses and luciferase assays. ARS and SC helped in chromatin immunoprecipitation and real-time PCR analyses. GR provided reagents and critically reviewed the manuscript. TLH provided primary CML samples. BC designed experiments and wrote manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Blood Cancer Journal website .
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Manzotti, G., Mariani, S., Corradini, F. et al. Expression of p89c-Mybex9b, an alternatively spliced form of c-Myb, is required for proliferation and survival of p210BCR/ABL-expressing cells. Blood Cancer Journal 2, e71 (2012). https://doi.org/10.1038/bcj.2012.16
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bcj.2012.16
Keywords
This article is cited by
-
c-Myb regulates tumorigenic potential of embryonal rhabdomyosarcoma cells
Scientific Reports (2019)
-
Targeting acute myeloid leukemia by drug-induced c-MYB degradation
Leukemia (2018)
-
c-MYB is a transcriptional regulator of ESPL1/Separase in BCR-ABL-positive chronic myeloid leukemia
Biomarker Research (2016)