Abstract
Aim:
To investigate whether the insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) rs1470579 and rs4402960 polymorphisms are associated with the development of type 2 diabetes mellitus (T2DM) and the repaglinide therapeutic efficacy in Chinese T2DM patients.
Methods:
A case-control study of a total of 350 patients with T2DM and 207 healthy volunteers was conducted to identify their genotypes for the IGF2BP2 rs1470579 and rs4402960 polymorphisms using a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay. Forty-two patients were randomly selected to undergo an 8-week repaglinide treatment (3 mg/d). Fasting plasma glucose (FPG), postprandial plasma glucose (PPG), glycated hemoglobin (HbAlc), fasting serum insulin (FINS), postprandial serum insulin (PINS), homeostasis model assessment for insulin resistance (HOMA-IR), serum triglyceride, total cholesterol (TC), low-density lipoprotein-cholesterol (LDL-c), and high-density lipoprotein-cholesterol (HDL-c) were determined before and after repaglinide treatment.
Results:
The frequencies of the IGF2BP2 rs1470579 C allele and the rs4402960 T allele were higher in T2DM patients than in healthy controls (P<0.05 and P<0.001, respectively). The effects of the repaglinide treatment on FPG (P<0.05) and PPG (P<0.05) were reduced in patients with the rs1470579 AC+CC genotypes compared with AA genotype carriers. Patients with the rs4402960 GT+TT genotypes exhibited an enhanced effect of repaglinide treatment on PINS (P<0.01) compared with GG genotype subjects.
Conclusion:
The IGF2BP2 rs1470579 and rs4402960 polymorphisms may be associated with the development of T2DM, and these polymorphisms may affect the therapeutic efficacy of repaglinide in Chinese T2DM patients.
Similar content being viewed by others
Introduction
The rapidly increasing prevalence of type 2 diabetes mellitus (T2DM) is a tremendous public health problem throughout the world. T2DM affects more than 170 million patients worldwide, and the total number of diabetes patients is estimated to increase from 20.8 million in 2000 to 42.3 million in 20301. T2DM is a polygenic disease that is characterized by impaired insulin secretion and insulin resistance. Genetic variants in combination with environmental factors are thought contributive to the development of this disease. Recent genome-wide association studies (GWASs) and meta-analyses have identified several novel diabetes susceptibility loci. These loci may play roles in insulin secretion and pancreatic β-cell function2, 3, 4, 5, 6, 7. Some variations in CDK5 regulatory subunit associated protein 1-like 1 (CDKAL1), cyclin dependent kinase inhibitor 2A-2B (CKDN2A-2B), zinc transporter member 8 (SLC30A8), and insulin-like growth factor 2 mRNA-binding protein 2 gene (IGF2BP2) have been shown to be associated with development of T2DM, and these findings have been confirmed in a Han Chinese population. SNPs in CDKAL1, SLC30A8, and IGF2BP2 are also thought to be associated with impaired β-cell function8.
IGF2BP2 belongs to an mRNA-binding protein family that plays roles in RNA localization, stability and translation9. IGF2BP2 is highly expressed in pancreatic islets and binds to insulin-like growth factor 2 (IGF-2), which is an important growth and insulin signaling molecule5. IGF2BP2 is a homolog of IGF2BP1, which binds to the 5'UTR of IGF2 mRNA and regulates IGF2 translation10. Several GWASs have found that study subjects carrying mutant alleles of SNPs rs1470579 and rs4402960 showed a moderately increased risk of T2DM. Several studies have confirmed this result in Asian populations6, 7, 8, 11. T2DM patients with different IGF2BP2 genotypes showed various levels of insulin secretion. It has been demonstrated that variants in IGF2BP2 affect first-phase insulin secretion and the disposition index detected by hyperglycemic clamps12.
Repaglinide is an insulin secretagogue agent, which acts as an effective medication for treating T2DM13, 14. Repaglinide can reduce the concentration of blood glucose by enhancing the secretion of insulin from pancreatic β-cells, inhibiting ATP-sensitive K+ channels (KATP), and activating Ca2+ channels13. Individual differences in the repaglinide therapeutic efficacy have been reported. However, the possible mechanism is still unknown. Recent studies have shown that polymorphisms in the cytochrome P450 (CYP) 2C8, 3A4, and organic anion-transporting polypeptide 1B1 (OATP1B1) gene could influence the plasma concentration of repaglinide15, 16, 17. Therefore, it is possible that polymorphisms in other genes may also affect the repaglinide efficacy. IGF2BP2 participates in the insulin signaling pathway and insulin secretion. Repaglinide also reduces glucose levels by increasing insulin secretion. Thus, the study we present here aimed to explore the correlation of IGF2BP2 genetic polymorphisms with the therapeutic efficacy of repaglinide in Chinese T2DM patients.
Materials and methods
Subjects
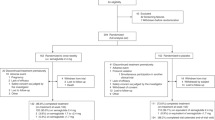
A total of 350 unrelated T2DM patients (178 male and 172 female), aged 25–70 years (mean 49.06±10.75 years), and 207 healthy controls (117 male and 90 female), aged 25–70 years (mean 47.96±10.78 years), were recruited for this study. T2DM patients were recruited from the Department of Endocrinology, the Second Xiangya Hospital and Diabetic Center of Xiangya Hospital of Central South University, and the control subjects were from the Health Screening Center of Xiangya Hospital of Central South University, Changsha, China. All subjects were evaluated through collecting medical histories and conducting physical examinations and routine clinical laboratory tests. T2DM was diagnosed according to a fasting plasma glucose (FPG ≥7.0 mmol/L) and/or postprandial plasma glucose test (PPG ≥11.1 mmol/L) (World Health Organization criteria, 1999). The criteria for enrollment were that the subjects fell within the body mass index (BMI) range from 18.5 to 30 kg/m2 and had not been treated with any insulin secretagogue, agonist or inhibitor of CYP2C8, CYP3A4, and OATP1B1 in the past 3 months. Patients with type 1 diabetes mellitus, a history of ketoacidosis, ischemic heart disease, congestive heart failure or trauma, kidney or liver diseases, patients receiving insulin treatment and pregnant or lactating women were excluded from this study. All of the healthy volunteers had normal fasting plasma glucose levels and blood pressure (data not shown). The clinical characteristics of the study groups are given in Table 1. The study protocol was approved by the Ethics Committee of Xiangya School of Medicine, Central South University and were in accordance with the Helsinki Declaration II. Written informed consent was obtained from each individual before the start of this study. We applied for clinical admission to the Chinese Clinical Trial Register (registration number: ChiCTR-CCC00000406). A total of 42 T2DM patients (23 male and 19 female) with different IGF2BP2 rs1470579 and rs4402960 genotypes and the same CYP2C8 and OATP1B1 genotypes took oral doses of 3 mg repaglinide daily (1 mg×3/per day preprandial treatment) for 8 consecutive weeks.
Genotyping analysis
Genomic DNA was isolated from peripheral blood leukocytes using an SQ Blood DNA Kit (Omega, Colorado, USA). Genotypes for the IGF2BP2 polymorphisms were analyzed using a PCR-RFLP assay. For the rs4402960 locus, the following primer pairs were used: sense primer: 5′-AGACCAGCCTTGGCAATGTAGTG-3′, antisense primer: 5′-CTAAAGCACTGAGAGAAACAGCCCT-3′. The 439-bp PCR products of rs4402960 were digested with Mbo II (Fermentas, Maryland, USA) into fragments of 282 bp and 157 bp (rs4402960 homozygosity resulted in the production of a single 439-bp fragment). To determine the genotyping success rate, Mse I (Fermentas, Maryland, USA) was used to digest DNA from rs4402960 homozygotes into fragments of 284 bp and 155 bp, while DNA from individuals who were wild type for rs4402960 resulted in production of only a single 439-bp fragment. For amplification of the rs1470579 locus, the following primer pairs were used: sense primer: 5′-CAGGGGTAGATGATGTAAGTGGT-3′, antisense primer: 5′-ACCTAATTTGATTTTGAGTTTCC-3′. The 460-bp PCR products of rs1470579 were digested with Mse I (Fermentas, Maryland, USA) into fragments of 226 bp, 157 bp, 61 bp, and 16 bp (rs1470579 homozygosity resulted in as the production of fragments of 287 bp, 157 bp, and 16 bp). For the same purpose, Fok I (Fermentas, Maryland, USA) was used to digest DNA from rs1470579 homozygotes into fragments of 396 bp and 64 bp, while DNA from individuals who were wild type for rs1470579 produced only a single 460-bp fragment. Direct sequencing of randomly selected samples confirmed the accuracy of genotyping results. The genotyping success rate was >98%, and genotyping of duplicate samples revealed no errors.
Clinical laboratory tests
After an overnight fast, venous blood samples were collected both in the fasting state and 2 h after a standardized breakfast on study d 0 and on the 8th weekend after treatment administration. Concentrations of FPG, TC, and triglyceride were determined by enzymatic colorimetric assay. HDL-c concentration was measured by lipoprotein electrophoresis. LDL-c concentration was calculated according to the Friedewald formula18. Plasma insulin and HbAlc levels were measured using a radioimmunoassay kit (BNIBT, Beijing, China) and by high performance liquid chromatography (HPLC) assay, respectively. The HOMA-IR value was used to estimate the level of insulin sensitivity and calculated according to the following formula: fasting serum insulin (mU/L)×fasting blood glucose (mmol/L)/22.519.
Statistical analysis
Statistical analyses were performed with SPSS software (Version 15.0 for Windows; SPSS Inc, Chicago, IL, USA). All continuous variables were given as means±SD and confidence intervals (95% CI). Variables that were not normally distributed were log-transformed before analysis. Hardy-Weinberg equilibrium and allelic frequencies in different groups were assessed with Pearson χ2 test of goodness-of-fit in the study sample. Student's t-test was used to compare continuous variables between the T2DM and control groups. Paired Student's t-tests and ANOVA tests were used to compare the differences in the degree of reduction or increase in plasma concentrations among the different genotypic groups before and after repaglinide treatment. LD between SNPs was estimated using Haploview version 3.2. The association between each SNP and the risk of T2DM was examined using logistic regression. All association analyses assumed an additive effect of the risk allele and were adjusted for sex, age, and BMI. A two-sided test with the type error level (α) set at 5% was used in all statistical analysis. Results were regarded as significant when P<0.05. Statistical power was calculated using a power calculator software PASS (www.ncss.com).
Results
Clinical and biochemical characteristics
The clinical and biochemical characteristics of all subjects in the current study were summarized in Table 1. There were no significant differences in age, BMI, waist/hip ratio (WHR) and HDL-c, but the levels of FPG, triglycerides, TC, and LDL-c were higher in T2DM cases than in healthy controls (P<0.01 and P<0.001, respectively).
Genotyping analysis and allelic frequencies
A total of 350 T2DM patients (178 male and 172 female) and 207 (117 male and 90 female) healthy volunteers were genotyped unambiguously for IGF2BP2 rs1470579 and rs4402960 polymorphisms. The genotypic distributions of rs1470579 and rs4402960 SNPs were in agreement with Hardy-Weinberg equilibrium (P>0.05). The frequency of the rs1470579 C allele was higher in the T2DM group than in the control group (30.29% vs 24.64%, P<0.05). The frequency of the T allele at the rs4402960 locus was 27.14% in T2DM patients, which was higher than in healthy controls (27.14 vs 21.26%, P<0.001). We found significant linkage disequilibrium between the rs1470579 locus and the rs4402960 locus (D′=0.642, r2=0.358, P<0.05, Table 2). There was no significant difference with respect to clinical parameters among the different haplotype groups (data not shown).
Comparison of baseline characteristics of T2DM patients with different rs1470579 and rs4402960 genotypes
The baseline clinical characteristics of 350 T2DM patients with different rs1470579 and rs4402960 genotypes were summarized in Table 3. There were no significant differences in sex, age, BMI, or WHR between different genotype groups. In patients with the IGF2BP2 rs1470579 genotype, there were significant differences in FPG (mmol/L) (8.66±3.87 vs 9.96±3.82, P<0.01), PINS (mU/L) (44.95±40.04 vs 60.60±47.92, P<0.001), TC (mmol/L) (4.52±2.40 vs 4.86±1.64, P<0.05) and LDL-c (mmol/L) (2.91±1.09 vs 3.17±1.27, P<0.01) between subjects with the AA and AC+CC genotype (Table 3, Figure 1). We also found that there were significant differences in FPG (mmol/L) (8.26±3.23 vs 10.45±4.23, P<0.001), PPG (mmol/L) (15.21±5.36 vs 17.06±6.61, P<0.05), PINS (mU/L) (47.74±38.01 vs 60.90±49.29, P<0.05) and TC (mmol/L) (4.43±2.43 vs 4.94±1.44, P<0.001) between individuals with the rs4402960 GG and GT+TT genotypes (Table 3, Figure 2).
Effects of the rs1470579 and rs4402960 polymorphisms on therapeutic efficacy of repaglinide treatment in patients with T2DM
A total of 42 T2DM patients (23 male and 19 female) were treated with 3 mg of repaglinide daily for 8 weeks. Repaglinide significantly decreased the concentrations of FPG (P<0.001), PPG (P<0.001), HbA1c (P<0.001), TC (P<0.05), and LDL-c (P<0.001), while treatment increased the levels of FINS (P<0.01), PINS (P<0.001), and HDL-c (P<0.01) in T2DM patients (Table 4). As shown in Table 5 and Figure 3, there were significantly lower repaglinide effects in patients with the IGF2BP2 rs1470579 AC+CC genotype on FPG (mmol/L) (from 8.27±2.02 to 7.08±1.51, DV −1.19±1.64) and PPG (mmol/L) (from 14.80±4.09 to 11.96±3.43, DV −2.84±3.91) compared with rs1470579 AA genotype carriers (from 9.14±1.84 to 6.59±1.57, DV -2.56±1.65, P<0.05; from 16.49±3.38 to 11.23±2.61, DV −5.25±3.23, P<0.05, respectively). However, patients with the GT+TT genotypes of rs4402960 showed an enhanced effect of repaglinide treatment on PINS (mU/L) (from 31.76±27.59 to 64.13±29.39, DV 32.37±25.42) compared with individuals with the GG genotype (from 40.99±27.59 to 55.04±28.71, DV 13.16±13.64, P<0.01) (Table 5).
Comparison of differential values (post-administration minus pre-administration) of FPG (A) and PPG (B) between the AA genotype and the AC+CC genotypes of the IGF2BP2 rs1470579 polymorphism in T2DM patients after repaglinide treatment, respectively. Comparison of differential values of PINS (C) between the GG genotype and the GT+TT genotypes of the IGF2BP2 rs4402960 polymorphism in T2DM patients after repaglinide treatment. Data are expressed as means±SD. bP<0.05, cP<0.01. n=42.
Discussion
In this study, we examined the effects of IGF2BP2 variations on the therapeutic efficacy of repaglinide treatment in Chinese T2DM patients. Our results suggested that the IGF2BP2 gene may represent a susceptibility gene for T2DM. We observed that the two variants in IGF2BP2 fell within one LD block (r2=0.358 for rs1470579 and rs4402960) and were significantly associated with T2DM [odds ratios 1.284 (1.175, 1.346), P=0.036 for rs1470579 and odds ratios 1.499 (1.021, 2.199), P=0.039 for rs4402960]. This study also found that patients with the rs1470579 AC+CC genotypes had poor responses to repaglinide treatment with respect to FPG and PPG compared with individuals with the AA genotype (P<0.05). Patients with the GT+TT genotypes of rs4402960 also showed a better repaglinide therapeutic effect on PINS compared with individuals with the GG genotype (P<0.01).
IGFs and IGF binding proteins (IGFBPs) regulate cellular growth and proliferation. Targeting mRNA binds to the 5′UTR, 3′UTR or coding region of IGFBPs9. IGF2 belongs to the insulin family of polypeptide growth factors, which is involved in the development and stimulation of insulin action3. IGF2BP2, also named IMP2, is an mRNA-binding protein that post-translationally regulates IGF2, which is a fetal growth factor that is involved in several developmental stages10. IGF2BP2 is involved in binding IGF-2 transcripts and regulating their translation10. The IGFBP homolog is necessary for pancreas development in Xenopus20, and IGF2BP3 transgenic mice exhibit acinar-ductal pancreatic metaplasia21. IGF2BP2 also acts in the regulation of mRNA stability. Interactions between genetic variation in IGF2BP2 and T2DM may be exerted through this IGF2 pathway and through the insulin pathway. The IGF2BP2 gene is located at chromosome 3q27.2. Intron 2 is the longest intron in the IGF2BP2 gene among mammalian species. SNPs rs1470579 and rs4402960 are located in a 50-kb region of this intron. Diabetes-predisposing variants may, therefore, affect regulation of IGF2BP2 expression3.
In the present study, we found that the distributions of both the rs1470579 and the rs4402960 alleles were in accordance with Hardy-Weinberg equilibrium. The frequency of the C allelic of rs1470579 and the T allele of rs4402960 were higher in T2DM subjects than in healthy controls (P<0.05 and P<0.001, respectively). Our data showed that the allelic frequency of IGF2BP2 SNPs in the Chinese population is different from their frequencies in Japanese6, Finns Swedish22, English23, and French Caucasian populations24. IGF2BP2 rs1470579 SNPs have dramatically different allele frequencies in white and black populations25.
Our data also demonstrated that patients with T2DM who carried the C allele of rs1470579 had higher levels of FPG, PINS, TC, and LDL-c compared with individuals with the AA genotype. The IGF2BP2 variant (rs4402960) was associated with insulin sensitivity, FPG, glucose AUC, and FPG24. rs4402960 was also associated with reductions in first-phase insulin secretion and in the disposition index, which reflects the failing adaptive capacity of pancreatic β-cells12 resulting in hyperglycemia including FPG and PPG. SNP rs4402960 has also been shown to be associated with the disposition index in Hispanic Americans26, HOMA-β in non-diabetic Japanese individuals and lower acute insulin release and tolerance27. SNP rs4402960 is strongly associated with an increased risk of T2DM and increased AUC of glucose in individuals of Dutch descent28. Our findings were, thus, in accord with several previous reports.
We also found that patients with T2DM who carried the T allele of rs4402960 had higher levels of FPG, PPG, PINS, and TC compared with subjects with the GG genotype. The results of our research agree with the observations of Wu et al, who observed a significant association of SNPs (rs1470579 and rs4402960) in IGF2BP2 [1.17 (1.03–1.32); P=0.014] with combined IFG (impaired fasting glycemia)/T2DM group. The association of these SNPs with HOMA-β reduction suggested that IGF2BP2 confers T2DM risk through a reduction of β-cell function8.
The underlying pathophysiological mechanisms that are affected by IGF2BP2 variations could be linked to nearby SNPs that have an effect on microRNAs, larger non-coding transcripts, or even antisense mRNA transcribed from intron 29. Doria et al pointed out that protein phosphatase 1, regulatory subunit 2 (PPP1R2) and insulin-sensitizing adipokine adiponectin (ADIPOQ), which are implicated in metabolism and regulation of insulin activity, are located in proximity to IGF2BP229. Replication of this research has indicated that IGF2BP2 variants were more likely to be associated with reduced β-cell function12, 27, 30. Because IGF2BP2 was shown to affect insulin secretion in a previous study, we hypothesized that patients with deteriorative β-cell function would have a poorer response to repaglinide treatment.
This study explored the influences of the rs1470579 and rs4402960 polymorphisms of IGF2BP2 on the therapeutic efficacy of repaglinide treatment in T2DM patients. In this study, to avoid the influence of pharmacokinetic changes in the action of repaglinide on its therapeutic efficacy, we selected patients with the same CYP2C8 (*3) and OATP1B1 (*1B, *5 and *15) genotypes. After T2DM patients were treated with 3 mg repaglinide daily for 8 consecutive weeks, there were significantly augmented repaglinide effects in patients with the rs1470579 AC+CC genotypes on FPG and PINS compared to subjects with the rs1470579 AA genotype (P<0.05, P<0.05, respectively). Moreover, patients with GT+TT genotypes of rs4402960 showed lower effects of repaglinide treatment on PINS compared with individuals with the GG genotype (P<0.01). Repaglinide acts to inhibit ATP-sensitive K+ (KATP) currents in pancreatic β-cells and to stimulate an increase in [Ca2+]i to promote insulin secretion31. Repaglinide is metabolized in the liver by the cytochrome P450 (CYP) 2C8 and 3A4 enzymes15. Hepatic uptake by OATP1B1 is an important step preceding the metabolism of this drug, and genetic polymorphisms in these enzymes affect the pharmacokinetics of repaglinide metabolism16, 17. In this study, to avoid the influence of pharmacokinetic changes in repaglinide action on its therapeutic efficacy, we selected patients with the same CYP2C8 (*3) and OATP1B1 (521T>C) genotypes to participate in this study (data not show).
One of the limitations of our study was its relatively small sample size. Another limitation was that the mechanisms through which IGF2BP2 contributes to the development of T2DM are not fully understood. The power values for detecting the two polymorphisms we investigated were 80%–98%. The influence of IGF2BP2 polymorphisms on the therapeutic efficacy of treatments for T2DM in large samples of T2DM patients is worthy of continued investigation.
In conclusion, we showed that variants in the IGF2BP2 gene are associated with T2DM, and also associated with reduced therapeutic efficacy of repaglinide treatment. However, understanding the biological mechanism by which variants in IGF2BP2 could mediate these effects on the biphasic pattern of insulin secretion will require further investigation.
Author contribution
Zhao-qian LIU had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis; Zhao-qian LIU and Qiong HUANG designed the research; Zhao-qian LIU, Qiong HUANG, Ji-ye YIN, Xing-ping DAI, Min DONG, Zhi-guang ZHOU, Xi HUANG, Hong-hao ZHOU, and Min YU performed the research; Zhao-qian LIU, Qiong HUANG and Qi PEI analyzed the data; Zhao-qian LIU and Qiong HUANG wrote the paper.
Abbreviations
- GWASs:
-
genome-wide association studies
- SNPs:
-
single nucleotide polymorphisms
- T2DM:
-
type 2 diabetes mellitus
- PCR-RFLP:
-
polymerase chain reaction-restriction fragment length polymorphism
- IGF2:
-
insulin-like growth factor 2
- IGF2BP2:
-
insulin-like growth factor 2 mRNA-binding protein 2 gene
- BMI:
-
body mass index
- WHR:
-
waist to hip ratio
- FPG:
-
fasting plasma glucose
- PPG:
-
postprandial plasma glucose
- HbAlc:
-
glycated hemoglobin
- FINS:
-
fasting serum insulin
- PINS:
-
postprandial serum insulin
- HOMA-IR:
-
homeostasis model assessment for insulin resistance
- TC:
-
total cholesterol
- LDL-c:
-
low-density lipoprotein-cholesterol
- HDL-c:
-
high-density lipoprotein-cholesterol
- DV:
-
differential value (post-administration minus pre-administration)
- Pre-:
-
pre-administration
- Post-:
-
post-administration
References
Wild S, Roglic G, Green A, Sicree R, King H . Global prevalence of diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care 2004; 27: 1047–53.
Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007; 445: 881–5.
Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in finns detects multiple susceptibility variants. Science 2007; 316: 1341–5.
Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008; 40: 638–45.
Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007; 316: 1336–41.
Takeuchi F, Serizawa M, Yamamoto K, Fujisawa T, Nakashima E, Ohnaka K, et al. Confirmation of multiple risk loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes 2009; 58: 1690–9.
Hinohara K, Nakajima T, Sasaoka T, Sawabe M, Lee BS, Ban J, et al. Replication studies for the association of psma6 polymorphism with coronary artery disease in east asian populations. J Hum Genet 2009; 54: 248–51.
Wu Y, Li H, Loos RJ, Yu Z, Ye X, Chen L, et al. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes 2008; 57: 2834–42.
Christiansen J, Kolte AM, Hansen TO, Nielsen FC . Igf2 mrna-binding protein 2: Biological function and putative role in type 2 diabetes. J Mol Endocrinol 2009; 43: 187–95.
Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC . A family of insulin-like growth factor ii mrna-binding proteins represses translation in late development. Mol Cell Biol 1999; 19: 1262–70.
Ng MC, Park KS, Oh B, Tam CH, Cho YM, Shin HD, et al. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes 2008; 57: 2226–33.
Groenewoud MJ, Dekker JM, Fritsche A, Reiling E, Nijpels G, Heine RJ, et al. Variants of cdkal1 and IGF2BP2 affect first-phase insulin secretion during hyperglycaemic clamps. Diabetologia 2008; 51: 1659–63.
Hatorp V . Clinical pharmacokinetics and pharmacodynamics of repaglinide. Clin Pharmacokinet 2002; 41: 471–83.
Papa G, Fedele V, Rizzo MR, Fioravanti M, Leotta C, Solerte SB, et al. Safety of type 2 diabetes treatment with repaglinide compared with glibenclamide in elderly people: A randomized, open-label, two-period, cross-over trial. Diabetes Care 2006; 29: 1918–20.
Bidstrup TB, Bjornsdottir I, Sidelmann UG, Thomsen MS, Hansen KT . CYP2C8 and CYP3A4 are the principal enzymes involved in the human in vitro biotransformation of the insulin secretagogue repaglinide. Br J Clin Pharmacol 2003; 56: 305–14.
Niemi M, Backman JT, Kajosaari LI, Leathart JB, Neuvonen M, Daly AK, et al. Polymorphic organic anion transporting polypeptide 1b1 is a major determinant of repaglinide pharmacokinetics. Clin Pharmacol Ther 2005; 77: 468–78.
Niemi M, Leathart JB, Neuvonen M, Backman JT, Daly AK, Neuvonen PJ . Polymorphism in CYP2C8 is associated with reduced plasma concentrations of repaglinide. Clin Pharmacol Ther 2003; 74: 380–7.
Friedewald WT, Levy RI, Fredrickson DS . Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972; 18: 499–502.
Kang ES, Yun YS, Park SW, Kim HJ, Ahn CW, Song YD, et al. Limitation of the validity of the homeostasis model assessment as an index of insulin resistance in Korea. Metabolism 2005; 54: 206–11.
Spagnoli FM, Brivanlou AH . The rna-binding protein, VG1RBP, is required for pancreatic fate specification. Dev Biol 2006; 292: 442–56.
Wagner M, Kunsch S, Duerschmied D, Beil M, Adler G, Mueller F, et al. Transgenic overexpression of the oncofetal RNA binding protein KOC leads to remodeling of the exocrine pancreas. Gastroenterology 2003; 124: 1901–14.
Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007; 316: 1331–6.
Wellcome Trust Case Control Consortium. Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature 2007; 447: 661–78.
Ruchat SM, Elks CE, Loos RJ, Vohl MC, Weisnagel SJ, Rankinen T, et al. Association between insulin secretion, insulin sensitivity and type 2 diabetes susceptibility variants identified in genome-wide association studies. Acta Diabetol 2009; 46: 217–26.
Moore AF, Jablonski KA, McAteer JB, Saxena R, Pollin TI, Franks PW, et al. Extension of type 2 diabetes genome-wide association scan results in the diabetes prevention program. Diabetes 2008; 57: 2503–10.
Palmer ND, Goodarzi MO, Langefeld CD, Ziegler J, Norris JM, Haffner SM, et al. Quantitative trait analysis of type 2 diabetes susceptibility loci identified from whole genome association studies in the insulin resistance atherosclerosis family study. Diabetes 2008; 57: 1093–100.
Horikoshi M, Hara K, Ito C, Shojima N, Nagai R, Ueki K, et al. Variations in the hhex gene are associated with increased risk of type 2 diabetes in the Japanese population. Diabetologia 2007; 50: 2461–6.
van Hoek M, Langendonk JG, de Rooij SR, Sijbrands EJ, Roseboom TJ . Genetic variant in the IGF2BP2 gene may interact with fetal malnutrition to affect glucose metabolism. Diabetes 2009; 58: 1440–4.
Doria A, Patti ME, Kahn CR . The emerging genetic architecture of type 2 diabetes. Cell Metab 2008; 8: 186–200.
Grarup N, Rose CS, Andersson EA, Andersen G, Nielsen AL, Albrechtsen A, et al. Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: Validation and extension of genome-wide association studies. Diabetes 2007; 56: 3105–11.
Ashcroft FM, Rorsman P . Electrophysiology of the pancreatic beta-cell. Prog Biophys Mol Biol 1989; 54: 87–143.
Acknowledgements
We thank all subjects who volunteered to participate in this study. This project was supported in part by National Major Project of Science and Technology of China Grant 2009ZX09304-003, National High-tech R&D Program of China (863 Program) Grant 2009AA022704, the National Natural Science Foundation of China Grants 30572230, 30873089, and by the Hunan Provincial Natural Science Foundation Grants 08JJ3058.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huang, Q., Yin, Jy., Dai, Xp. et al. IGF2BP2 variations influence repaglinide response and risk of type 2 diabetes in Chinese population. Acta Pharmacol Sin 31, 709–717 (2010). https://doi.org/10.1038/aps.2010.47
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2010.47
Keywords
This article is cited by
-
Lin28 affects the proliferation and osteogenic differentiation of human dental pulp stem cells by directly inhibiting let-7b maturation
BDJ Open (2024)
-
Circulating expression levels of CircHIPK3 and CDR1as circular-RNAs in type 2 diabetes patients
Molecular Biology Reports (2022)
-
The role of IGF2BP2, an m6A reader gene, in human metabolic diseases and cancers
Cancer Cell International (2021)
-
Pharmacogenetics and personalized treatment of type 2 diabetes mellitus
International Journal of Diabetes in Developing Countries (2016)
-
A whole genome analyses of genetic variants in two Kelantan Malay individuals
The HUGO Journal (2014)