ABSTRACT
The genes encoding type II DNA topoisomerases were investigated in Giardia lamblia genome, and a type IIA gene, GlTop 2 was identified. It is a single copy gene with a 4476 bp long ORF without intron. The deduced amino acid sequence shows strong homology to eukaryotic DNA Top 2. However, some distortions were found, such as six insertions in the ATPase domain and the central domain, a ∼100 aa longer central domain; a ∼200 aa shorter C-terminal domain containing rich charged residues. These features revealed by comparing with Top 2 of the host, human, might be helpful in exploiting drug selectivity for antigiardial therapy. Phylogenetic analysis of eukaryotic enzymes showed that kinetoplastids, plants, fungi, and animals were monophyletic groups, and the animal and fungi lineages shared a more recent common ancestor than either did with the plant lineage; microsporidia grouped with fungi. However, unlike many previous phylogenetic analyses, the “amitochondriate” G. lamblia was not the earliest branch but diverged after mitochondriate kinetoplastids in our trees. Both the finding of typical eukaryotic type IIA topoisomerase and the phylogenetic analysis suggest G. lamblia is not possibly as primitive as was regarded before and might diverge after the acquisition of mitochondria. This is consistent with the recent discovery of mitochondrial remnant organelles in G. lamblia.
Similar content being viewed by others
INTRODUCTION
DNA topoisomerases, found in eukaryotes and prokaryotes, are enzymes that have evolved to solve the topological problems associated with DNA metabolisms, such as transcription, replication, packing and unpacking of DNA in the cell 1. They are classified as type I topoisomerases if they create single-stranded breaks in DNA duplex or as type II topoisomerases if double-stranded breaks are created.
Type II DNA topoisomerases are a class of ubiquitous enzymes found in almost all the living cells. Eukaryotes contain at least one type II enzyme: one DNA Top 2 in most eukaryotes or two isoforms, DNA Top 2α and Top 2β, in vertebrates 2, 3. Bacteria possess two kinds, gyrase and Top IV, which are heterotetramers composed of two subunits, GyrB and GyrA, and ParE and ParC, respectively 4. Archaea also have two kinds: gyrase and Top VI, both of which are composed of two subunits: subunit B and A. Top VI is a very different form. Its sequence is distinct from those of any other DNA topoisomerases. This led to the classification of type II DNA topoisomerases into two evolutionary distinct protein families, type IIA and type IIB. Top VI belongs to type IIB family, others belong to type IIA 5.
Eukaryotic type II topoisomerases are also important for their essential role in chromosome segregation and maintenance of chromosome structure 6, as well as their involvement in the reforming of nucleolus at anaphase 7.
Giardia lamblia is one of the most widespread intestinal protozoan parasites. It was long recognized as of evolutionary importance due to its “lacking mitochondria” and many typical membrane-bound organella characteristics of eukaryotic cells 8, its primitive features of nucleus 9, and its earliest branching among extant eukaryotes in most molecular phylogenetic trees 10. Not a few authors thought that this organism was one of the most primitive eukaryotes that its divergence laid close to the transition between prokaryotes and eukaryotes, remaining in a stage of evolution before acquisition of organelle. However, this has been challenged recently by several reports such as the discovery of genes of mitochondrial origin 11, 12, 13, and the further discovery of the mitochondrial remnant organelle (mitosome) 14. Moreover, it was considered that as a parasite, the earliest branching of G. lamblia in phylogenetic trees was due to long-branch attraction artifact (LBA) resulted from fast evolution in parasitic life 15, but some people did not think so.
G. lamblia has some special features that relate to the functions of eukaryotic DNA Top 2 mentioned above. These features include: 1) although five chromosomal bands was demonstrated by PFGE, no condensed chromosomal structures have been observed so far 10, which might suggest that it has no transition between chromatin and chromosome; 2) no nucleoli have been identified and its rRNA transcription and processing are not localized to certain regions of the nuclei 10, 16, 17. Given the fact that enzymic activities of type II DNA topoisomerase were previously detected from G. lamblia, what are the characteristics of its type II DNA topoisomerase(s) and the gene(s) in the organism 18? Is its type II enzymes eukaryotic type or prokaryotic type (gyrase, and Top IV or Top VI)? Therefore, studies of type II DNA topoisomerase gene in G. lamblia may reveal some special features of the enzyme and also provide new evidence for the evolutionary position of the organism. Furthermore, topoisomerases are often considered to be targets of antiparasitic drugs 19. Thus, delineating features differentiating type II topoisomerase of the parasite from its hosts would contribute to exploiting drug selectivity for antigiardial therapy.
In the present work, we undertook identification, characteristic and phylogenetic analysis of type II topoismerase gene(s) from G. lamblia genome.
MATERIALS AND METHODS
G. lamblia and DNA extraction
G. lamblia (isolate C2), which was isolated from a giardiasis patient from Sichuan, China, was axenically cultured in modified TYI-S-33 medium at 37°C 20 and harvested at 48–72 h after inoculation. After washed three times with PBS, genomic DNA was extracted according to standard phenol-chloroform method.
PCR, cloning and sequencing
Known Top 2 sequences of eukaryotes ranging from unicellular protists to mammalians were selected from GenBank and DDBJ datebase, and aligned using Clustal W of DNAStar software and on-line Multalin (http://prodes.toulouse.inra.fr/multalin/cgi-bin/multalin.pl). Degenerate primers were designed corresponding to three highly conserved motifs showed by the alignment. The forward primer (P1), 5′-TIAT(TCA)ITIACIGA(AG)GGI(GCT)(AT)I(TA)(CG)IGC-3′, was constructed to a sequence coding for the conserved protein sequence LILTEGDSA. The reverse primer (P2), 5′-GTICCIA(AG)ICC(TC)TT(GA)TA(GA)TA(CT)TT-3′, was designed to a degenerate olignucleotide corresponding to the protein sequence KYYK-GLGT. The intermediate primer (P3), 5′-GA(TC)GGI(AT)(GC)ICA(TC)AT(TAC)AA(AG)GGI(CT)T-3′, was designed to a degenerate olignucleotide corresponding to the protein sequence DGSHIKGL, which could also make a pair with P2.
PCR was carried out on a Biometra Tgradient Thermocycler in 50 μl reaction volume containing about 100 ng of gDNA of G. lamblia, 1× PCR buffer, 0.8 mM dNTPs, 2.7 mM MgCl2, 2.5 mM each of primer P1 and P2, and 1.25 U of Ex Taq polymerase (TaKaRa). PCR cycling parameters were as follows: denaturation at 94°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 1 min, for a total of 35 cycle. An additional extension step was finally performed at 72°C for 10 min. Then, with 1/2000 of mixture of the PCR products as template and P1 and P3 as primer set, the secondary PCR was carried out under the same conditions.
The secondary PCR products (about 220 bp) were isolated using Gel Extraction Mini Kits (Watson Biotechnologies, Inc) and cloned into the PMD18-T vector (TaKaRa). JM109 Competent Cells (TaKaRa) were transformed with the ligated DNA. Positive clones were picked up and the recombinant plasmids were prepared according to Uniq-10 Column Plasmid Minipreps Kit (Sangon) protocol and evaluated by sequencing on an ABI PRISMR3100 Genetic Analyzer (Applied Biosytems).
Screening database, assembling the overlapping reads and verifying the obtained single sequence
The PCR-derived sequence above was used as a probe to screen shotgun sequences in G. lamblia single-pass read database from Giardia Genome Project in the Josephine Bay Paul Centre Web site at the Marine Biological Laboratory (http://jbpc.mbl.edu/Giardia-HTML/). Overlapping reads were obtained and assembled into a single sequence using BLASTN 21. Based on this sequence, a much longer single sequence of about 5316 bp, which was estimated to have included the entire gene, was constructed by contig assembly of single-pass reads from the same database. To verify the correct assembling of the sequence and to make sure some uncertain sites in the assembled sequence, we designed several pairs of primer to amplify and sequence some concerned regions.
Identification of GlTop 2 and properties analysis
To find ORF, the Finder program of GenBank (http://www.ncbi.nlm.nih.gov/) was applied. The deduced amino acid sequence was translated by Primer Primier Version 5.0. Then the sequence was analyzed with several database search tools to identify what kind of protein the sequence was. These tools included FingerPRINTScan (http://www.bioinf.man.ac.uk/dbbrowser/fingerPRINTScan/), Blocks/ PRINTS (http://www.block.fhcrc.org/block_search.html), PROSITE (http://www.ebi.ac.uk/ppsearch/), ProDom (http://protein.toulouse.inra.fr/prodom/blast_form.html), SBASE (http://www3.icgeb.trieste.it/~sbasesrv/), Pfam (http://www.sanger.ac.uk/Software/Pfam/search.shtml), eMotif (http://motif.stanford.edu/emotif), SMART (http://smart.embl-heidelberg.de/) and PSORT (http://psort.ims.u-tokyo.ac.jp/). The homology of the sequence with other known type II topoisomerase was also searched with BLASTP of GenBank. Sequence properties of GlTop 2 were analysed by comparing with other eukaryotic type II topoisomerase sequences.
Reverse transcription (RT)-PCR and PCR with the same primer pair
Total RNA was extracted from about 107 cells and the isolated total RNA was treated with RNase-free DNase I (Promega) at 37°C for 10 min. To perform RT-PCR, the following primer pair was designed: a non-degenerate sense primer, 5′-CACGCAATTCTACGAAAC-3′, and an antisense primer, 5′-TGACTCTGTAAAGGACGACT-3′, corresponding to nucleotide position 786–803 and 4352–4371 of the GlTop 2 ORF, respectively. The RT-PCR was carried out using the BcaBEST™ RNA PCR kit Ver.1.1 (TaKaRa). PCR with the same primer set was performed using G. lamblia gDNA as templates. At the same time, RNase-free DNase-treated total RNA was used as templates to serve as a control. All of the products of RT-PCR and PCR were analysed on 1.2% agarose gel.
Phylogenetic analyses
A BLASTP search with the identified GlTop 2 sequence hit a number of type II topoisomeras sequences of eukaryotes, eubacteria and archaebacteria, with the highest scores for eukaryotic DNA topoisomerase II (E-value <1e-40). After elimination of duplicates, 33 eukaryotic sequences were included in the analysis, including: Homo sapiens Top 2β (Q02880), Homo sapiens Top 2α (CAA09762), Cricetulus longicaudatus Top 2β (CAA76313), Cricetulus longicaudatus Top 2α (Q64399), Mus musculus Top 2β (NP_033435), Mus musculus Top 2α (XP_126710), Rattus norvegicus Top 2α (NP_071519), Sus scrofa Top 2α (O46374), Gallus gallus Top 2α (O42130), Gallus gallus Top 2β (BAA22540), Drosophila melanogaster Top 2 (AAF53802), Bombyx mori Top 2 (O16140), Caenorhabditis elegans Top 2 (NP_496536), Arabidopsis thaliana Top 2 (NP_189031), Pisum sativm Top 2 (O24308), Schizosaccharomyces pombe Top 2 (P08096), Saccharomyces cerevisiae Top 2 (AAM00578), Aspergillus niger Top 2 (BAB84102), Aspergillus terreus Top 2 (BAA82356), Emericella nidulans Top 2 (T30516), Talaromyces flavus Top 2 (BAB84106), Penicillius chxysogenum Top 2 (Q9Y8G8), Candida glabrate Top 2 (O93794), Candida albicans Top 2 (P87078), Encephalitozoon cuniculi Top 2 (NP_584718), Crithidia fasciculate Top 2 (A45648), Leishmania major Top 2 (CAB72310), Dictyostelium discoideum Top 2 (P90520), Trypanosoma brucei Top 2 (P12531), Trypanosoma cruzi Top 2 (P30190), Plasmodium falciparum Top 2 (T10466), Leishmania donovani Top 2 (AAD34021), Leishmania infantum Top 2 (AAF86355). Taking the combined sequences of gyrase A (B91208) and B (B91018) from E. coli and gyrase A (NP_279849) and B (NP_279848) from archaebacterium Halobacterium salinarum NRC-1 as outgroup, after exclusion of gaps and several areas where the alignment was uncertain, a dataset of 1165 aa were analysed with protein distance (NEIGHBOR, FITCH, KITSCH), maximum parsimony (PROTPAR) methods from the PHYLIP package, version 3.6a3, and maximum-likelihood method (PROTML) from MORPHY version 2.3. A 3000 bootstrap resampled dataset was generated by the program SEQBOOT. 1000 ml heuristic searches (-j-q-n 1000) were done to search for the optimal topology using PROTML. Consensus trees were produced with CONSENSE. Branch length was calculated with the JTT aa substitution model incorporating among-site rate variation (JTT +r model) (discrete distribution, eight categories) using TREE-PUZZLE. Treefile produced by CONSENSE was viewed by TREEVIEW.
RESULTS and DISCUSSION
Identification of GlTop 2
After PCR with degenerate primers designed on the basis of conserved amino acid motifs of known DNA Top 2 sequences, a 224 bp fragment was obtained after two rounds of PCR amplification. Its sequence displayed high homologous with other known Top 2. After screening shotgun sequences in G. lamblia single-pass read database from G. lamblia Genome Project, nine shotgun sequences were obtained. The nine overlapping reads could be assembled into a single sequence. At this basis, a much longer sequence of about 5316 bp was constructed by contig assembly of 13 single-pass reads from the same database. Due to the conservatism of all (eukaryotic or prokaryotic) type II topoismerase sequences, especially in our PCR-amplified region which contains two of the most conserved motifs, the screen could find all the type II topoisomerase genes in Giardia genome, if any. But no other homologous sequences could be assembled. Thus, we got only one single complete sequence.
But there were two uncertain sites in the assembled sequence due to the nucleotide discrepancy existing between the overlapping regions of shotgun sequences. Additionally, due to that some features of the G. lamblia genome can lead to important errors in assembled contigs (see http://jbpc.mbl.edu/Giardia-HTML/index2.html), we also amplified and sequenced some regions concerned by ourselves. According to the sequencing results, it was sure that there was not a G between 491G and 492C and that there existed a C between 2854C and 2856T. At the same time, the almost identity between our sequenced regions and the assembled sequence proved that our overlapping shotgun sequences were assembled correctly.
Using the on-line GENSCAN (http://genes.mit.edu/GENESCAN.html) and GeneFinder (http://argon.cshl.org/genefinder) programs, an ORF of 4476 bp was found in the 5316 bp sequence without any spliceosomal introns.
When using BLAST of NCBI to blast protein sequences in GenBank, the deduced sequence hit more than a thousand of type II topoisomerases, all of which were the three kinds of type II topoisomerases: gyrase, Top IV and eukaryotic Top 2 (0< E-value = 4e-05) without any other protein sequences. The BLAST results also indicated that the deduced sequence had 21%–47% identity and 38%–69% positive with other known type II topoisomerases.
Using RPS-BLAST to search Conserved Domain Database of GenBank, the deduced sequence hit the following 4 CDD: Top 2C, Top 4C, DNA gyrase or Top IV sununit A, and DNA gyrase B, all the E-values were very low (3e-97, 7e-69, 2e-48 and 2e-15, respectively).
Besides the above analyses, another eight protein-sequence-searching programs in different databases were applied to identify the deduced sequence. Seven of them suggested that the deduced sequence belong to DNA Top 2 (Tab. 1). The eighth program PROSITE revealed that the deduced sequence had a consistent sequence (511-L-T-E-G-D-S-A-K-A-520) with DNA Top 2 signature ([LIVMA]-X-E-G-[DN]-S-A-X-[STAG]).
All the analyses above strongly suggested that the deduced sequence was a type II DNA topoisomerase, a homolog of eukaryotic DNA Top 2. Thus, we named the gene GlTop 2 (GenBank accession No.AY278365).
When we searched the G. lamblia genomic database with the complete sequence of GlTop 2, no other homologous sequence was found.
We also blasted Giardia genomic database for type IIB topoisomerase gene with archaeal Top VI as a query. Except a Spo11 gene, which had been reported in GenBank, no type IIB topoisomerase homologs was found.
Taken together, the above data indicate that G. lamblia genome has only one type IIA DNA topoisomerase gene GlTop 2, which is a single-copy type II DNA topoisomerase gene and encodes a eukaryotic DNA Top 2, GlTop 2.
Properties of GlTop 2 and its deduced protein sequence
Probable regulatory motifs were identified through an analysis of upstream and downstream sequences. The upstream sequence is A + T rich as has been reported for several other giardial genes 22, 23, 24. Although no TATA box element and GTTAAA, a putative TFIID/TBP binding element 22, 25, were found, there exists a 10 bp AT-rich sequence, TAAAAATTAA, at position −10 to −1 with respect to the start codon ATG. Previous researches have reported that sequence alignment of several promoter regions of giardial genes failed to reveal any highly conserved sequence 11, 23, 24, 26, suggesting that G. lamblia promoter sequences are highly degenerate 22, 23. Thus, this AT-rich region might function as a TATA box. Another upstream motif, AAATTT, spanning position −40 to −35, resembles the 6-base consensus motif CAATTT present in upstream of other gene coding regions of G. lamblia 11. Also, a putative poly(A) signal, AGTAAA, which matches the consensus for G. lamblia 22, 27, is located 54 bp downstream of the TAA stop codon.
To verify the transcription of GlTop 2, RT-PCR and gDNA-PCR with the same primer pair were carried out simultaneously. Both of their products showed the same expected size amplicons of about 3600 bp (the estimated length is 3586 bp). To avoid the contamination of gDNA in RT-PCR, the isolated RNA was treated with RNase-free DNase before it was used as templates. The negative result of the control PCR using RNase-free DNase-treated total RNA directly as templates proved that the gDNA contamination was removed. The results of RT-PCR and gDNA-PCR imply that GlTop 2 was transcribed in G. lamblia without any introns.
The deduced protein sequence contains 1491 amino acids with an estimated Mr of 168400.2 Da. It is longer than several reported type II DNA topoisomerases from other parasitic protozoan (1221–1397 aa) and is nearly the same size of those of S. pombe (1485 aa) and of human (1530 aa). Its theoretical pI is 8.39 (calculated out by ProtParam tool, http://expasy.chl/tools/protparam.html).
Analysis using the on-line PSORT program showed GlTop 2 possessed several putative nuclear locating signals (NLS), and was located in nucleus with 78.3% possibility. But which of the several putative NLS is/are really involved in the nuclear localisation is uncertain. It needs more experiments to verify.
Inspection of the alignment of GlTop 2 sequence with other known eukaryotic Top 2 sequences (of from protists to mammals) using Clustal W reveals that GlTop 2 has an average of 31.8% identity, being most identity to E. cuniculi (41.3%) and least identity to L. major (29.2%). With its human counterparts, GlTop 2 shares 31.7% and 32% identity with Top 2α and Top 2β, respectively. The degree of conservation is generally greater in the N-terminal two-thirds of the coding sequence and falls off markedly towards the C-terminus.
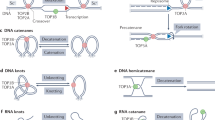
The conserved amino acid sequences that are found in all type II DNA topoisomerases are scattered throughout GlTop 2. By analogy with other eukaryotic Top 2, the putative catalytic active site tyrosine of GlTop 2 was identified as Y847 corresponding to Y805 of human Top 2α and Y771 of T. Cruzi Top 2 (Fig. 1). Like others, the active site is located within the highly conserved motif RY. Usually, when DNA is cleaved by type II DNA topoiso-merases, the tyrosine residue attacks the phosphodiester bond of DNA strands to form a covalent DNA-protein transient intermediate 1. In this process, the conserved G-loop motif, GXXGXGXK, of type II DNA topoisomerases interacts with the phosphates of ATP. GlTop 2 also has such a motive, 140-GRNGYGAK-147.
Schematic comparison module of Giardia Top 2 protein sequence with those of Trypanosoma Top 2 and Homo Top 2a. The conserved motifs in sequences are indicated by medium gray frame. Insertions are showed by boxes with diagonal lines. “M” is methionine which is coded by start codon. The number reflects the position of amino acid. Active site tyrosines Tyr, 847, 771, and 805 are positioned by vertical arrows. Vertical dashed lines delineate the ATPase, DNA breakage-rejoining, and C-terminal domains of Giardia conserved in all Top 2.
Several highly conserved motifs such as TEGDSA, DGSHIKGL and YYKGLG (they are located at the regions corresponding to our PCR primers), and other conserved motifs (eg. RP, KIXDEI, PLRGK, NVR, MIMTDQ) were also found in GlTop 2. But a conserved motif, GXGXP (eg. 103-GQGIP-107 in enzyme of D. melanogaster) was not found in GlTop 2.
Like other eukaryotic DNA Top 2, GlTop 2 can be divided into three functional domains, a N-terminal ATPase domain (1–482 aa), a central DNA breakage-rejoining domain (483–1334 aa) and a C-terminal domain (1335–1491 aa) (Fig. 1). However, compared with the other eukaryotic enzymes, its central domain is longer, and its C-terminal domain is shorter. There are three insertions in its ATPase domain: 252–263 aa, 275–313 aa and 434–441 aa (Fig.1). Among them the 252–263 aa insertion is unique, and although the 275–313 aa insertion also exists in P. falciparum Top 2 with rich Asn (65%), the GlTop 2 275–313 aa insertion is a rich charged cluster and includes three repeats of DX, KX, TXX (4 copies respectively). The central domain of GlTop 2 also has three distinct insertions at least: 1001–1047 aa, 1077–1090 aa and 1244–1258 aa (Fig. 1). The 1001–1047 aa insertion contains rich Lys and four repeats of KX. Like the enzymes in other known protozoan parasites, T. brucei, T. cruzi, C. fasciculate, L. donovani, L. Infantum and P. falciparum, and in a saprobe D. discoideum, the C-terminal domain of GlTop 2 is obviously (about 200 aa) shorter than those of other higher eukaryotic enzymes. But, uniquely, the C-terminal domain of GlTop 2 contains rich charged residues (residue D, E, K and R together are 43% of all the residues).
Thus, unlike typical eukaryotic Top 2, GlTop 2 possesses some distortions in its sequence. For example, having no conserved GXGXP motif; having six insertions in the central domain and C-terminal domain (some of them have obvious characters); having a ∼100 aa longer central domain; having a ∼200 aa shorter C-terminal domain containing rich charged residues. It is unknown whether these distortions is related to the special features of the organism, such as that no transition between chromatin and chromosome has been observed, and no nucleoli have been identified so far in both of which DNA Top 2 is involved in other typical eukaryotes 6, 7. While, the short C-terminal domain is a common feature of most of the unicellular eukaryote Top 2 sequences. This could have two different explanations: 1) DNA Top 2 in all these lower unicellular eukaryotes are at the primitive stage, and the higher eukaryotes have got the longer C-terminal domain later in the evolutionary process; 2) The C-terminal domain has become shorter due to the parasitic or saprobic life. Considering that the enzyme is involved in DNA metabolisms and has no direct relationship with the parasitic or saprobic life, the first explanation might be more reasonable. To confirm this, the inclusion of additional sequences from a wide variety of lower eukaryotes, especially from more free-living unicellular protists are needed.
These revealed features differentiating the enzymes of the parasite Giardia from its counterpart in higher eukaryotes, especially in the host human (Fig. 1), may help to exploit drug selectivity between the host and parasite replication apparatus for antigiardial therapy. On the base of the present work, further overexpression and functional characterisation of GlTop 2 should facilitate the screening and designing of drugs against this parasite.
PROSITE analysis showed a number of potential protein kinase C phosphorylation sites and CKII phosphorylation sites in GlTop 2. So many phosphorylation sites may imply that phosphorylation has a great impact on catalytic activities of the enzyme. This is consistent with other enzymes previously reported 28, 29, 30. These suggest that like other eukaryotic Top 2, GlTop 2 is also a substrate for protein kinase C (PKC) and casein kinase II (CKII), and phosphorylation might increase its catalytic activity and might be involved in the regulation of its functions.
Finally, GlTop 2 has rich N-myristoylation sites and N-glucosylation sites. Thus myristoylation and glucosylation may also make a notable impact on the functions of the enzyme.
Phylogenetic analyses of eukaryotic enzymes
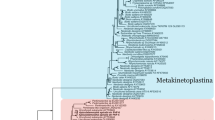
Considering that prokaryotic gyrases are homologs of eukaryotic Top 2, and their two subunits, subunit B and A, correspond to the N-terminal half and the C-terminal half of the eukaryotic enzymes respectively, we combined gyrase B and A from eubacterium E. coli and archaebacterium Halobacterium salinarum NRC-1 respectively as the outgroup. Five phylogenetic trees were constructed using five programs of the three methods (protein distance, maximum parsimony and maximum-likelihood). All the trees have an almost identical topology and the PROML tree was chosen as the representative tree here (Fig. 2). They are congruent with the SS rRNA tree, actin tree, and combined protein data tree 31, 32 in that they show that kinetoplastid protozoans, plants, fungi, and animals are monophyletic groups; the animal and fungi lineages share a more recent common ancestor than either does with the plant lineage. Moreover, in our trees, the microsporidia E. cuniculi is not related to protozoans but groups with fungi and is at the base of fungi clade, which is consistent with recently accumulating evidence that microsporidias belong to fungi 33, 34. There are two isoenzymes (Top 2α and Top 2β) in vertebrates. In our trees, they form two isolated subgroups within the vertebrate clade. This is agreement with the common presumable consideration that by gene duplication the two isoforms evolved from a common precursor to fulfill different cellular function as suggested by their different patterns of expression in the cell cycle. All these suggest that type II DNA topoisomerase is an effective and dependable promising marker for phylogenetic analysis.
Phylogeny of eukaryotic DNA Top 2. The phylogeny presented is derived by the maximum-likelihood method based on 1165 amino acid residue positions using combined gyase B and A from eubacterium E. coli and archaebacterium Halobacterium salinarum NRC-1 as outgroup. Branch lengths are proportional to the estimated number of amino acid substitutions; the scale bar indicates amino acid substitutions per site. Bootstrap support value above 50% and importance are given at branch nodes and derived from PROTML (the first), NEIGHBOR (the second), PROTPAR (the third), FITCH (the fourth), KITSCH (the fifth), separated by slash marks. “-”s are denoted no support for those nodes.
To our surprising, however, the protozoan P. falciparum falls into the plant (A. thaliana and P. sativm) clade; the kinetoplastid L. major does not group with other kinetoplastids but form a unique clade branching after slim mould D. discoideum clade and before the plant clade. For the former, if the sequence data are reliable, a possible explanation is that an unknown novel gene transfer had once occurred between P. falciparum and plants. As for the latter, the sequence alignment has showed that the used sequence of L. major is very different from those of other protozans and is much (∼260 aa) longer than those of other kinetoplastids; furthermore, our recent search of T. brucei and T. cruzi genomic databases (though uncompleted) (http://www.ncbi.nlm.nih.gov/sutils/genom_tree.cgi) indicated that there must exist two kinds of type II DNA topoisomerase genes (though some of their sequences are uncompleted and can not be involved in our phylogenetic analysis) in them, and the L. major sequence and other kinetoplastid sequences we used in the phylogenetic analysis belong to different kinds. Thus, L. major did not group with other kinetoplastids in our tree.
G. lamblia and kinetoplastid protozoans (excluding L. major) are close to the base of the tree, but they do not form a common clade and G. lamblia diverged after the kinetoplastid protozoans. The position of G. lamblia is strongly supported by high bootstrap values (99.8%, 100%, 99.5%, 93% and 97% in NEIGHBOR, FITCH, KITSCH, PROTPAR, PROTML trees, respectively).
A number of previous phylognetic trees based on various molecules, including small subunit ribosomal RNA 35, 36), elongation factors 1a 33 and 2 37, 38, the largest subunit of RNA polymerase II 39, α-tubulin 40, and V-type ATPase catalytic subunit and the front and back halves of the proteolipid subunit 41, showed that G. lamblia branched earliest among extant eukaryotes. These molecular phylogenetic analyses were taken as one of the main evidences to support that G. lamblia was a living relic of most primitive eukaryotes before the acquisition of mitochondria. However, recently, the earliest branching of G. lamblia has been recognized as an artifact of long-branch attraction (LBA) due to its fast evolution in parasitic life 15, though some authors did not agree. But, in our trees, it was showed that the “amitochondriate” eukaryote G. lamblia did not branch first but diverged after a group of mitochondriate protists, kinetoplatids. It seems that our trees were not affected by LBA. The reason for this might be as following: DNA Top 2 is involved in the basic intranuclear metabolic activities–DNA meta-bolisms, and has no direct relationship with the parasitic life, thus, it might not evolved so fast as those directly related to the parasitic life. In fact, that microsporidia E. cuniculi, a more obligatory parasite than Giardia, did not branch very early but branched much later than G. lamblia and kinetoplastids did in our trees is evidence for this.
Therefore, our phylogenetic analysis result suggests that G. lamblia might diverge after the acquisition of mitochondria, and implies that G. lamblia once probably possessed and secondarily lost mitochondria, or might still have a mitochondrion-like organelle not found yet. This is consistent with the recent discovery of genes of mitochondrial origin 11, 12, 13, and the later discovery of the mitochondrial remnant organelle (mitosome) in G. lamblia 14. Actually, accumulating evidence, such as the discoveries of intron and Golgi in Giardia 42, 43, 44, has also proved some previously so-called 'primitive features' to be no longer tenable. Moreover, our recent studies imply that the 'lack of nucleolus' is probably not a primitive feature of G. lamblia yet but might arise secondarily (to be published). All these suggest that G. lamblia is not possibly as primitive as was regarded before.
Accession codes
Abbreviations
- GlTop 2:
-
Giardia lamblia type IIA DNA topoisomerase
- PBS:
-
phosphate-buffer saline
- gDNA:
-
genome DNA
- cDNA:
-
DNA complementary to RNA
- RT-PCR:
-
reverse transcription-polymerase chain reaction
- ORF:
-
open reading frame
- C-terminal:
-
carboxyl terminal
- N-terminal:
-
amino terminal
- NLS:
-
nuclear localization signal
References
Wang JC . DNA Topoisomerases. Annu Rev Biochem 1996; 65:635–92.
Watt PM, Hickson ID . Structure and function of type II DNA topoisomerases. Biochem J 1994; 303:681–95
Gimenez-Abian JF, Clarke DJ, Mullinger AM, Downes CS, Johnson RT . A postprophase topoisomerase II-dependent chromatid core separation step in the formation of metaphase chromosomes. J Cell Biol 1995; 131:7–17.
Bergerat A, de Massy B, Gadelle D, et al. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 1997; 386:414–17.
Buhler C, Gadelle D, Forterre P, Wang JC, Bergerat A . Reconstitution of DNA topoisomerase VI of the thermophilic archaeon Sulfolobus shibatae from subunits separately overexpressed in Escherichia coli. Nucleic Acads Res 1998; 26:5157–62.
Wang JC . Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 2002; 3:430–40.
Tavormina PA, Come MG, Hudson JR, et al. Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J Cell Biol 2002; 158:23–9.
Gillin FD, Reiner DS, McCaffery JM . Cell biology of the primitive eukaryote Giardia lamblia. Annu Rev Microbiol 1996; 50:679–705.
Li JY . Characterization of Giardia cell nucleus: its implication on the nature and the primitive cell nucleus. Cell Res 1995; 5:115–24.
Adam RD . Biology of Giardia lambia. Clin Microbiol Rev 2001; 14:447–75.
Roger AJ, Svard SG, Tovar J, et al. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: Evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc Natl Acad Sci USA 1998; 95:229–340.
Tachezy J, Sanchez LB, Müller M . Mitochondrial type iron-sulfur cluster assembly in the amitochondriate eukaryotes Tri Chomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol Biol Evol 2001; 18:1919–28.
Arisue N, Sanchez LB, Weiss LM, Muller M, Hashimoto T . Mitochondrial-type hsp70 genes of the amitochondriate protists, Giardia intestinalis, Entamoeba histolytica and two micro-sporidians. Parasitol Int 2002; 51:9–16.
Tovar J, Leon-Avila G, Sanchez LB, Sutak R, Tachezy J, van der Giezen M, Hernandez M, Müller M, Lucocq JM . Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature 2003; 426:172–6.
Philippe H, Lopez P, Brinkmann H, et al. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc R Soc Lond B 2000; 267:1213–21.
Li JY, He YJ, Chen SF . The primitive cell nucleus, in which the nucleolus has not emerged yet. Endocyto & Cell Res 1997; 12:65–70.
Narcisi EM, Glover CV, Fechheimer M . Fibrillarin, a conserved pre-ribosomal RNA processing protein of Giardia. J Eukaryot Microbiol. 1998; 45:105–11.
Bell CA, Dykstra CC, Naiman NA, et al. Structure-activity studies of dicationically substituted bis-benzimidazoles against Giardia lamblia: correlation of antigiardial activity with DNA binding affinity and giardial topoisomerase II inhibition. Antimicrob Agents Chemother 1993; 37:2668–73.
Shapiro TA . Inhition of topoisomerases in African trypano-somes. Acta Trop 1993; 54:251–60.
Keister DB . Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg 1983; 77:487–8.
Altschul SF, Madden T L, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389–402.
Minotto L, Tutticci EA, Bagnara AS, Schofield PJ, Edwards MR . Characterisation and expression of the carbamate kinase gene from Giardia intestinalis. Mol Biochem Parasitol 1999; 98:43–51.
Holberton DV, Marshall J . Analysis of consensus sequence patterns in Giardia cytoskeleton gene promoters. Nucleic Acids Res 1995; 23:2945–53.
Sun CH, Tai JH . Identification and characterization of a ran gene promoter in the protozoan pathogen Giardia lamblia. J Biol Chem 2000; 274:19699–706.
Yee J, Mowatt MR, Dennis PP, Nash TE . Transcriptional analysis of the glutamate dehydrogenase gene in the primitive eukaryote, Giardia lamblia. Identification of a primordial gene promoter. J Biol Chem 2000; 275:11432–9.
Roger AJ, Morrison HG, Sogin ML . Primary structure and phylogenetic relationships of a malate dehydrogenase gene from Giardia lamblia. J Mol Evol 1999; 48:750–5.
Elmendorf HG, Singer SM, Nash TE . The abundance of sterile transcripts in Giardia lamblia. Nucleic Acids Res 2001; 29:4674–83.
Ackerman P, Glover CVC, Osheroff N . Phosphorylation of DNA topoisomerase II by Casein kinase II: modulation of eukaryotic topoisomerase II activity in vitro. Proc Natl Acad Sci USA 1985; 82:3164–8.
Shiozaki K, Yanagida M . Functional dissection of the phosphorylated termini of fission yeast DNA topoisomerase II. J Cell Biol 1992; 119:1023–36.
Park GH, Lee YT, Bae YS . Stimulation of human DNA topoisomerase II activity by its direct association with the â subunit of protein kinase CKII. Mol Cells 2001; 11:82–8.
Drouin G, Moniz de SaM, Zuker M . The Giardia lamblia actin gene and the phylogeny of eukaryotes. J Mol Evol 1995; 41:841–9.
Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF . A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 2000; 290:972–7.
Hirt RP, Logsdon JM, Healy BH, et al. Microsporidia are related to fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci USA 1999; 96:580–5.
Keeling PJ, Fast NM . Microsporidia: biology and evolution of highly reduced intracellar parasites. Annu Rev Microbiol 2002; 56:93–116.
Sogin ML, Gunderson JH, Elwood HJ, Alonso RA, Peattie DA . Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science 1989; 243:75–7.
Simpson AG, Roger AJ, Silberman JD, et al. Evolutionary history of “early-diverging” eukaryotes: the excavate taxon Carpediemonas is a close relative of Giardia. Mol Biol Evol 2002; 19:1782–91.
Hashimoto T, Nakamura Y, Kamaishi T, et al. Phylogenetic place of mitochondrion-lacking protozoan, Giardia lamblia, inferred from amino acid sequences of elongation factor 2. Mol Biol Evol 1995; 12:782–93
Hashimoto T, Nakeamura Y, Kamaishi T, Hasegawa M . Early evolution of eukaryotes inferred from protein phylogenies of translation elongation factors 1a and 2. Arch Protistenkd 1997; 148:287–95.
Dacks JB, Marinets AM, Doolittle WF, Cavalier-Smith T, Logsdon JM . Analyses of RNA polymerase II genes from free-living protests: phylogeny, long branch attraction, and the eukaryotic big bang. Mol Boil Evol 2002; 19:830–40.
Keeling PJ, Doolittle WF . α-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family. Mol Biol Evol 1996; 13:1297–305.
Hilario E, Gogarten JP . The prokaryote-to-eukaryote transition reflected in the evolution of the V/F/A-ATPase catalytic and proteolipid subunits. J Mol Evol 1998; 46:703–15.
Dacks JB, Doolittle WF . Reconstructing/deconstructing the earliest eukaryotes: how comparative genomics can help. Cell 2001; 107:419–25.
Lujan HD, Marotta A, Mowatt MR, et al. Developmental induction of Golgi structure and function in the primitive eukaryote Giardia lamblia. J Biol Chem 1995; 270:4612–8.
Nixon JE, Wang A, Morrison HG, et al. A spliceosomal intron in Giardia lamblia. Proc Natl Acad Sci U S A 2002; 99:3701–5.
Acknowledgements
In this work, the shotgun sequences used to assembly the complete gene sequence were from the database of Giardia lamblia Genome Project (http://jbpc.mbl.edu/Giardia-HTML/), Marine Biological Laboratory at Woods Hole. We would like to thank for their outstanding work. This work was supported by Grants (30070362, 30170135, 30021004) from the National Natural Science Foundation of China and Grants (KSCX2-SW-101C, STZ-00-23) from the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
HE, D., WEN, J., CHEN, W. et al. Identification, characteristic and phylogenetic analysis of type II DNA topoisomerase gene in Giardia lamblia. Cell Res 15, 474–482 (2005). https://doi.org/10.1038/sj.cr.7290316
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290316
Keywords
This article is cited by
-
Evolutionary History of TOPIIA Topoisomerases in Animals
Journal of Molecular Evolution (2022)
-
Evaluating the evolution of G. lamblia based on the small nucleolar RNAs identified from Archaea and unicellular eukaryotes
Parasitology Research (2009)
-
Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes
BMC Genomics (2006)
-
Evidence from bioinformatics, expression and inhibition studies of phosphoinositide-3 kinase signalling in Giardia intestinalis
BMC Microbiology (2006)
-
Identification and evolutionary implication of four novel box H/ACA snoRNAs from Giardia lamblia
Chinese Science Bulletin (2006)