ABSTRACT
Asip is a mammalian homologue of polarity protein Par-3 of Caenorhabditis elegans and Bazooka of Drosophila melanogaster. Asip/Par-3/Bazooka are PDZ-motif containing proteins that localize asymmetrically to the cell periphery and play a pivotal role in cell polarity and asymmetric cell division. In the present study, we have cloned human asip cDNA and its splicing variants by 5′-RACE and RT-PCR using candidate human EST clones which have a high homology to rat asip cDNA. The full-length cDNA of human asip encodes a 1,353 aa protein exhibiting 88% similarity to the rat one. Human asip is a single copy gene consisting of at least 26 exons and localizing in human chromosome 10, band p11.2, with some extraordinarily long introns. All exon/intron boundary nucleotides conform to the “gt-ag” rule. Three main transcripts were detected by Northern blot analysis, and at least five variants, from alternative splicing and polyadenylation, have been identified by RT-PCR and liver cDNA library screening. Exon 17b deleted asip mRNAs expressed ubiquitously in normal human tissues, including liver, on RT-PCR analysis. However, they were absent from most human liver cancer cell lines examined. More interestingly, the expression of exon 17b deleted variants was down regulated in 52.6% (10/19) clinic specimens of human hepatocellular carcinomas (HCCs), compared with the surrounding nontumorous liver tissues from the same patients. The presence of various splicing transcripts, the variation of their distribution among different tissues and cells, and their differential expressions in human HCCs suggest that human Asip isoforms may function in different context.
Similar content being viewed by others
INTRODUCTION
Cell polarity is the reflection of complex mechanisms that establish and maintain the functionally specialized regions in the plasma membrane and cytoplasm, and is fundamentally important for differentiation, proliferation, morphogenesis and other functions of simple and complicated organisms1.
Molecular mechanisms of cell polarity during animal development have been analyzed mainly in the nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster2. In early C. elegans embryos, Par-3 protein is required for embryonic polarity and become localized asymmetrically to the anterior periphery of the one-cell embryo. Mutations in the par-3 gene affect the asymmetric distribution of itself and other proteins involved in cell fate determination and the orientation of mitotic spindles in successive cell cycle3. As a homologue of Par-3, Drosophila Bazooka protein also controls the asymmetric distribution of other proteins and the orientation of mitotic spindles. In bazooka- deficient germ line clones that lack maternal and zygotic expression, loss of cell polarity is fully penetrant, which was shown by defective spindles orientation, mispositioning of the daughter cell after division and deforming of zonula adherens4, 5, 6. Mammalian epithelial cells also provide an experimental system to reveal essential features of cell polarity1. Asip, a rat homologue of Par-3 and Bazooka, localizes to tight junctions in epithelial cells or to adherens junctions in cells without tight junctions, and expresses in cells without cell-cell junctions7. Tight junctions are specialized structures that play an essential role in epithelial cell polarity by creating a barrier to diffusion between cells in the epithelial sheet and forming an intramembrane diffusion fence that restricts intermitting of apical and basal-lateral membrane components8.
A search of available databases revealed the presence of some human expressed sequence tag (EST) clones exhibiting high homology to rat asip cDNA, which means asip is conserved in rat and human. To explore the function of human asip and its association to any human disease, we cloned the human asip cDNA. Here, we describe the cDNA cloning and molecular characterization of the novel human gene, asip. At least five variants can be isolated from adult human liver cDNA by RT-PCR, which are resulted from alternative splicing and polyadenylation. We also report here the results of an investigation of the distribution of expressed variants in various human tissues, cell lines and clinic specimens of human hepatocellular carcinomas (HCCs).
MATERIALS AND METHODS
Sequence analysis
The acquired sequence data were aligned against the Genbank databases (nr, human EST, and htgs) at the National Center for Biotechnology Information (National Institute of Health), using Blast to search for sequence matches. Sequence editing, comparing, and primer designing were performed using Lasergene software packages (Dnastar).
RNA extraction, Northern blot analysis, and RT-PCR
Total RNAs of various human tissues, cultured human cells and human HCCs were isolated using guanidium isothiocyanate according to Chomczynski and Sacchi's description9. Poly (A)+ RNA was prepared using Quickprep Micro mRNA Purification kit (Amersham-Pharmacia Biotech) according to manufacture's instruction. Northern blot analysis was carried out as described by Sambrook et al10 using 5 μg mRNA. The RNA was electrophoresed in a 1.0% agarose gel containing formaldehyde, transferred to a nylon membrane (Hybon-N, Amersham-Pharmacia Biotech) by capillary diffusion, and then hybridized with a 32p-labelled human asip cDNA probe (2284-3387 in AF196185). Hybridized membrane was autoradiographed at −70°C using X-ray film (Kodak) with intensifying screen.
cDNA was synthesized from 3 μg total RNAs by random hexamer priming using M-MLV reverse transcriptase (Gibco BRL), normalized and subjected to PCR amplification with primers specific for human asip. The primers were 5′-CTT GAT GAA TCG CCC AGC AGA AAT G-3′(sense) and 5′-CGG CCG TGG ACG ATG GAA AGG AAT A (antisense), which amplify 567 and/or 477 bp products (due to the presence or absence of exon 17b). The conditions for amplification were an initial denaturation at 94°C for 2 min, followed by 28 cycles at 94°C for 30 sec, 66°C for 30 sec, and 72°C for 45 sec. The PCR products were analyzed by 1.5% agarose gel and ethidium bromide staining.
RESULTS
Molecular cloning of human asip cDNAs
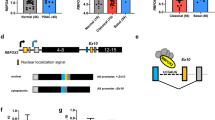
A Blast search of the human EST database using the sequence of rat asip cDNA (Accession No. AB005549) as query revealed the presence of some human expressed sequence tag (EST) clones exhibiting high homology to rat asip cDNA, which means asip is conserved in rat and human. With the help of in silico cDNA cloning, 5′-RACE and cDNA library screening techniques, we finally cloned the full-length cDNA of human asip and also found the presence of various isoforms. The complete nucleotides sequence of the human asip cDNA (Accession No. AF196185) is 5,958 bp long and contains a continuous ORF of 4,062 bp predicted to encode a polypeptide of 1,353 aa with a calculated molecular mass of 150 kDa. The flanking nucleotides of the first ATG codon partially match with Kozak's criteria of an optional eukaryotic translation start site11, although a preceded in-frame stop codon is not detected. Three polyadenylation signals, AATAAA, are identified in the 1,609 bp long 3′UTR at 4,381-4,386, 4,992-4,997, and 5,934-5,939. The nucleotides of human asip ORF and the deduced amino acids show 81% and 88% similarity to rat asip, respectively. Like rat Asip, its human homologue also contains three PDZ motifs and one aPKC-binding domain (Fig 1).
Alternative splicing and polyadenylation events of human asip pre-mRNA (A) and the five protein isoforms of human Asip (B). Note that exon 17 has two 5′ splicing donor sites, and that exons 5, 12, 21 and 22 may or may not be included, as shown in (A). Nine alternative splicing events, from (a) to (i), are shown as dashed lines. Splicing event (d) is always linked to splicing event (e). One or three polyadenylation signals (AATAAA) can be found in exon 21 or exon 26, respectively. The five protein products of alternatively spliced mRNAs are diagrammatically shown in (B). Typical conserved regions-CR1, CR2 and CR3, are shown.
Exon/intron structure of human asip gene
To clarify the structure of human asip gene, htgs database was searched by Blast program using the full-length human asip cDNA sequences as query. We found that, as part of human genome project, DNA sequencing of chromosome 10 has provided the DNA sequence of almost the entirety of the human asip gene, except for the 5′ -end sequence and gaps between different genomic DNA clones. Eight contigs can be withdrawn from chromosome 10p11.2 genomic sequences AL138768, AL012241, AL360233, AL392123, AL356462 and AL160409, which contain stretches of human asip cDNA sequence from 408 bp to 5′UTR. For the absence of 5′ -end DNA sequence, we tentatively assigned the sequence ≤407 to exon 1, thus, human asip gene has at least 26 exons and localizes in chromosome 10p11.2. The nucleotides at the exon/intron junction are presented in Tab 1; all the exon/intron boundary sequences conform to the “gt-ag” rule. A distinguished feature of human asip gene is that some of its introns are extraordinarily long.
Alternative splicing and polyadenylation of human asip gene
Different transcripts were detected by the cloning of various human asip cDNAs. Analyses of these different cDNAs obtained by cDNA library screening, 5′-RACE and RT-PCR suggest that the presence of splicing variants with regard to exons 5, 12, 17, 21 and 22. The alternative splicing events of human asip gene is shown in Fig 1A. Exons 5, 12 or 22 can be spliced out in different variants. Splicing can also take place within exon 17 to delete its 3′ -end 90 bp sequence that we named exon 17b. Exon 20 can be spliced to exons 21, 22 or 23 alternatively. Because exon 21 contains polyadenylation signal, splicing to it produces short transcripts (hasips) that end with this exon, whereas variants that end with exon 26 are long transcripts (hasipl), with or without exon 22. As a result of these alternative splicing and polyadenylation events, up to five distinct variants can be expressed. They are hasipla (full-length), hasiplb ( Δ exon 12, Δexon 17b, and Δexon 22), hasipsa (Δexons 22-26), and hasipsb (Δexon 12, Δexon 17b, and Δexons 22-26). hpar-3 (Accession No. AF252293), which is deleted at exons 5, 12 and 17b, is another variant of human asip mRNA cloned recently by other authors12. The structure of protein products of the alternative splicing events is shown diagrammatically in Fig 1B. The presence of some alternative splicing forms has also been reported recently for the mouse homologue of asip13.
Expression of human asip mRNA in human adult liver tissue was further examined by Northern blot analysis. Three main bands, approximately 6.0, 4.5 and 3.6 kb in size, were observed (Fig 2). The 6.0 kb band corresponds to long transcripts (hasipla, hasiplb and hpar-3) and 3.6 kb corresponds to the short ones (hasipsa and hasipsb), whereas the 4.5 kb transcript has not been identified so far.
Northern blot analysis of the expression of human asip transcripts in adult human liver tissue. Five micrograms of mRNAs was separated in 1.0 % agarose gel containing formaldehyde. Note that three main bands were detected by the probe (2284-3387 in AF196185). The 6.0 kb and 3.6 kb bands correspond to long transcripts and short transcripts respectively, while the nature of the 4.5 kb transcript remains to be characterized.
Expression profiles of human asip variants in various human tissues, cultured cells and HCCs
In all splicing events, the deletions of exon 12 and 17b were linked. We emphasize the deletion of exon 17b in this paper because it contains the core sequence of aPKC-binding domain. Primer pair (in exon 15 and exon 19 respectively) were designed to detect its expression in various human tissues. It can be seen from Fig 3A that exon17b deleted variants are expressed ubiquitously in normal human tissues including brain, colon, heart, kidney, liver, lung, skeletal muscle, placenta, small intestine, spleen, stomach and testis. However, exon 17b containing variants are hardly detected in skeletal muscle, spleen and testis. Since HCC is among the most common malignancies in China14, we therefore investigated the expression profiles of asip splicing isoforms in HCC cells and clinic specimens to see if the alternations of asip is related to this disease. Interestingly, exon 17b containing variants are expressed in all cultured human liver cell lines examined (immortalized human liver cells L-02, hepatocellular carcinoma cells BEL-7721, 7402 and 7404, and hepatoblastoma cells Hep G2), while exon 17b deleted variants are only weakly detectable in Hep G2 cells (Fig 3B). The expression of exon 17b deleted variants was further investigated in clinic specimens of human HCCs. Among the 19 cases of human HCCs we examined, the expression of exon 17b deleted variants was down-regulated, more or less, in 10 cases of human HCCs, compared with the surrounding nontumorous liver tissues from the same patients (Fig 3C).
Expression of human asip mRNAs in various human tissues, cell lines and HCCs. Human asip mRNAs were analyzed by RT-PCR using normalized, first-strand cDNA preparations derived from different human tissues (A), cell lines (B) and HCCs (C). The amplified fragments are 567 bp for exon 17b containing mRNAs (a) and 477 bp for exon 17b deleted mRNAs (b). The human tissues used in (A) are brain (lane 1), colon (lane 2), heart (lane 3), kidney (lane 4), liver (lane 5), lung (lane 6), skeletal muscle (lane 7), placenta (lane 8), small intestine (lane 9), spleen (lane 10), stomach (lane 11), and testis (lane 12). And the human cell lines used in (B) are hepatoblastoma cells Hep G2 (lane 1), hepatocellular carcinoma cells BEL-7404 (lane 2), BEL-7402 (lane 3) and BEL-7721 (lane 4), and immortalized human liver cells L-02 (lane 5). (C) Expression of human asip mRNAs in 19 cases of clinic specimens of human HCCs (T), compared with the surrounding nontumorous liver tissues (N) from the same patients. The HCCs with down-regulated expression of the exon 17b deleted variants were marked with “*”.
DISCUSSION
Human Asip protein exhibits 88% overall sequence similarity to the rat one, 15% to C. elegans Par-3, and 19% to Drosophila Bazooka. Similar to its homologues, human Asip also contains three PDZ motifs and an aPKC-binding domain. The three PDZ motifs of human Asip protein and its homologues differ in sequence from each other, perhaps reflecting different binding specificities for various target proteins. The PDZ2 and/or PDZ3 domains of hPar-3 was found to interact with the C-terminal tail of ephrin B, a cell-associated ligand for eph receptor tyrosine kinase, and implicated in the control of organized cell movement in several tissues15. hPar-3 also binds to Par-6 through its first PDZ domain, and this interaction is necessary for tight-junction formation12. Another class of proteins that can interact with Asip/Par-3/Bazooka is aPKC7, 16, 17. The protein complex including in Cdc42/Rac1, Par-6, Par-3 and aPKC was found to be present under physiological conditions, and this complex may be involved in maintaining the apico-basolateral polarity of epithelia cells12, 13, 18. By recruiting various proteins to the apical pole of the cell, Asip/Par-3/Bazooka are likely to be the candidates for organizing multi-protein complexes for establishing cell polarity.
So far, there is no description about splicing variants of bazooka gene in flies and par-3 gene in nematodes. In this paper, up to five splicing variants of human asip mRNA were identified. The alternative splicing was also observed in rat and mouse7, 13. Mammalian epithelial cells may have more complicated molecular mechanisms of orienting the cell polarity and, hence, need more delicate regulation and control. The various protein products of human asip mRNAs, especially those of the exon 17b deleted variants which can not be phosphorylated and bonded by aPKC, may function differently in signaling cascade when cells receive an external signal to adjust cell polarity.
The expression of exon 17b deleted variants may be also under fine regulation. The exon 17b deleted variants expressed ubiquitously in normal human tissues including liver, but they were absent in most of human liver cancer cell lines examined (Fig 3). Although L-02 was originally derived from normal human liver tissue, we can not exclude the possibility, to some extent, that the cell line studied here has undergone malignant transformation. Since Asip localizes at the tight junctions of polarized epithelial cells and may play a critical role in the development of the junctional structures and apico-basal polarization of mammalian epithelial cells7, 19. The expression of exon 17b deleted variants might be related to highly organized cell-cell contact and its down-regulation or absence occurred if the contacts among cells were disrupted. Normal tissues have highly organized cell-cell contact and junctions. However, the situation changes in tumors, in which cancer cells are typically less adhesive both to other cells and non-cellular substrate for their reduced tendency to form junctions. In fact, we also have detected the down-regulated expression of exon 17b deleted variants in about 50% human HCCs compared with the cancer adjacent liver tissues (Fig 3C). The exon 17b deleted variants were also found to be absent in non-adherent cells such as leukemia cells K562, HL-60 and Jurkat (data not shown). In conclusion, at least five splicing variants of human asip are expressed; some of which may not bind directly to aPKC for the absence of aPKC-binding domain. The down-regulation or the absence of exon 17b deleted variants in human liver cancer cell lines and HCCs indicated that they might be involved in cell growth control.
Abbreviations
- Asip:
-
atypical PKC isotype-specific interacting protein
- HCC:
-
hepatocellular carcinoma
- ORF:
-
open reading frame
- par:
-
partitioning defective
- RACE:
-
rapid amplification of cDNA ends
- RT-PCR:
-
reverse transcription-polymerase chain reaction
- UTR:
-
untranslated region(s)
- Delta; :
-
deletion
References
Drubin DG, Nelson WJ . Origins of cell polarity. Cell 1996; 84:335–44.
Knoblich JA . Mechanisms of asymmetric cell division during animal development. Curr Opin Cell Biol 1997; 9:833–41.
Guo S, Kempheues KJ . Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegans embryo. Curr Opin Genet Dev 1996; 6:408–15.
Schober M, Schaefer M, Knoblich JA . Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 1999; 402:548–51.
Wodarz A, Ramrath A, Kuchinke U, Knust E . Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblast. Nature 1999; 402:544–7.
Muller HJ, Wieschaus E . armadillo, bozooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J Cell Biol 1996; 134:149–63.
Izumi Y, Hirose T, Tamai Y, Hirai SI, Nagashima Y, Fujimoto T, Tabuse Y, kemphues KJ, Ohno S . An atypical PKC directly associates and colocalizes at the epithelial tight junction with Asip, a mammalian homologue of Caenorhabditis elegans polarity protein Par-3. J Cell Biol 1998; 143:95–106.
Balda MS, Matter K . Tight junctions. J Cell Sci 1998; 111:541–7. Chomczynski P, Sacchi N, Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162:156–9.
Chomczynski P, Sacchi N, Single-step method of RNA isolation by acid uanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162:156–9.
Sambrook J, Fritsch EF, Maniatis T . Molecular cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 1989.
Kozak M . Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem 1991; 266:19867–70.
Joberty G, Petersen C, Gao L, Macara IG . The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol 2000; 2:531–9.
Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T . A mammalian Par-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signaling and cell polarity. Nat Cell Biol 2000; 2:540–7.
Wang G, Huang CH, Zhao Y, Cai L, Wang Y, Xiu SJ, Jiang ZW, Yang S, Zhao XT, Huang W, Gu JR . Genetic aberration in primary hepatocellular carcinoma: correlation between p53 gene mutation and loss-of-heterozygosity on chromosome 16q21-q23 and 9p21-p23. Cell Res 2000; 10:311–23.
Lin D, Gish GD, Songyang Z, Pawson T . The carboxyl terminus of B class ephrins constitutes a PDZ domain binding motif. J Biol Chem 1999; 274:3726–33.
Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S . Atypical protein kinase C cooperates with Par-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 1998; 125:3607–14.
Wodarz A, Ramrath A, Grimm A, Knust E . Drosophila atypical protein kinase C associates with Bazooka and control polarity of epithelia and neuroblasts. J Cell Biol 2000; 150:1361–74.
Qiu RG, Abo A, Martin GS . A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCz signaling and cell transformation. Curr Biol 2000; 10:697–707.
Suzuki A, Yamanaka T, Hirose T, Manabe N, Mizumo K, Shimizu M, Akimoto K, Izumi Y, Ohnishi T, Ohno S . Atypical protein kinase C is involved in the evolutionarily conserved PAR protein complex and plays a critical role in establishing epithelial-specific junctional structures. J Cell Biol 2001; 152:1183–96.
Acknowledgements
This work was supported by research grants from the Special Grant for Human Genomics Program of Chinese Academy of Sciences and the Special Funds for Major State Basic Research of China (Grant G1999053905) to YH Xu.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
FANG, C., XU, Y. Down-regulated expression of atypical PKC-binding domain deleted asip isoforms in human hepatocellular carcinomas. Cell Res 11, 223–229 (2001). https://doi.org/10.1038/sj.cr.7290090
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290090
Keywords
This article is cited by
-
The polarity protein PARD3 and cancer
Oncogene (2021)
-
High expression of PARD3 predicts poor prognosis in hepatocellular carcinoma
Scientific Reports (2021)
-
Activation of TNFR2 sensitizes macrophages for TNFR1-mediated necroptosis
Cell Death & Disease (2016)
-
Renal atrophy after ischemia–reperfusion injury depends on massive tubular apoptosis induced by TNFα in the later phase
Medical Molecular Morphology (2014)
-
Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma
Oncogene (2009)