Abstract
The western black-legged tick, Ixodes pacificus, is a primary vector of the spirochaete, Borrelia burgdorferi, that causes Lyme disease. We used variation in a 355-bp DNA portion of the mitochondrial cytochrome oxidase III gene to assess the population structure of the tick across its range from British Columbia to southern California and east to Utah. Ixodes pacificus showed considerable haplotype diversity despite low nucleotide diversity. Maximum parsimony and isolation-by-distance analyses revealed little genetic structure except between a geographically isolated Utah locality and all other localities. Loss of mtDNA polymorphism in Utah ticks is consistent with a post-Pleistocene founder event. The pattern of genetic differentiation in the continuous part of the range of Ixodes pacificus reinforces recent recognition of the difficulties involved in using genetic frequency data to infer gene flow and migration.
Similar content being viewed by others
Introduction
The western black-legged tick, Ixodes pacificus, is one of the most medically important ticks in western North America. A primary vector of the Lyme disease spirochaete, Borrelia burgdorferi, it may also transmit the rickettsial pathogen Ehrlichia equi (Lane et al., 1991; Richter et al., 1996). The genetic structure of populations of I. pacificus is potentially important in understanding the epidemiology and evolutionary dynamics of the diseases it vectors (Tabachnick & Black, 1995).
Current gene flow in I. pacificus is probably the result of a complex interaction of factors such as host specificity, the type of host parasitized, and ecological requirements (Hilburn & Sattler, 1986). As with most ticks, I. pacificus is dependent on its hosts for long-range movement (Hilburn & Sattler, 1986). Gene flow potential in I. pacificus should be high, because it parasitizes mobile vertebrates and has a broad host range which includes 80 species of reptiles, birds, and mammals (Lane et al., 1991). However, ecological restrictions may counterbalance this potential. The larval, nymphal and adult life stages of I. pacificus each feed once and then drop from the host to moult or lay eggs under leaf litter (Peavey & Lane, 1996). Effective gene flow is likely to occur only when there is sufficient rainfall, ground cover and soil humidity to allow completion of these critical life stages.
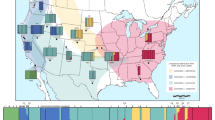
Ixodes pacificus is distributed down the west coast of North America from British Columbia to northern Baja California, with disjunct populations occurring east to Utah (Fig. 1) (Dennis et al., 1998). Ixodes pacificus varies geographically with respect to its host preferences (Arthur & Snow, 1968; Arnason, 1992), its internal transcribed spacers (ITS1 and ITS2) in nuclear ribosomal DNA sequences (Wesson et al., 1993), and its allozymes (Kain et al., 1997).
Map of western North America showing range of Ixodes pacificus by stippled shading. Collecting localities are indicated with dots, with code as in Table 1.
Allozyme data provided an equivocal picture of the population structure and biogeography of I. pacificus (Kain et al., 1997). Eleven of 12 loci sampled across the range of I. pacificus, including Utah, exhibited little genetic differentiation. In contrast, the twelfth locus, glucose-6-phosphate isomerase (GPI), showed a pattern of genetic differentiation that suggested either an adaptive cline or secondary contact along a broad zone in central and southern California. This zone demarcated a northern group of populations (WVC, CNY, CHB; Fig. 1) from a southern group (MDO, SCI, MR, UTAH). Kain et al. (1997) recognized that either a rapid range expansion or high rates of gene flow could explain the pattern of allozyme variation. However, they proposed that the most plausible explanation for the pattern of allozyme variation in I. pacificus was that it had a high rate of gene flow.
The purpose of this study was to use a portion of the mitochondrial DNA (mtDNA) cytochrome oxidase III gene to survey the extent and pattern of population variation across the range of I. pacificus. Theoretically, mtDNA has a fourfold smaller effective population size than nuclear genes because of its uniparental inheritance and haploid nature (Simon et al., 1994). Additionally, a higher mutation rate provides more lineages for genetic drift and gene flow to sort (Simon et al., 1994). Mitochondrial DNA was used successfully to assess the phylogeography of the related tick Ixodes scapularis, the vector of B. burgdorferi in the eastern United States (Norris et al., 1996). In the current study on I. pacificus, populations were chosen to allow assessment of congruence of mtDNA variation with allozyme data, both with respect to the GPI locus and the lack of allozyme differentiation in isolated populations.
Materials and methods
Samples and DNA
Collecting localities are shown in Fig. 1 and detailed in Table 1. Except for CHB (California, Alameda Co., Anthony Chabot Regional Park), localities correspond to a subset of those used by Kain et al. (1997). Host-seeking adult ticks were collected from vegetation with flannel tick-drags. Ticks were either stored in 95% ethanol or allowed partially to engorge on rabbits (to induce enzyme systems), then frozen at −70°C. Ethanol-preserved specimens were placed individually in 1.5 mL microcentrifuge tubes and centrifuged under vacuum for 15 min to remove excess alcohol. Ticks were then put in 0.5 mL microcentrifuge tubes, frozen in liquid nitrogen, and then dry-homogenized using a flamed 200 μL pipette tip as a pestle. Fifty μL of TE (0.100 M Tris-HCl, pH 8.3, 0.1 mmol EDTA) was added to the dry sample and the solution was boiled for 15 min. Frozen partially engorged ticks were processed using the Qiagen Qiamp Tissue Kit according to the manufacturer’s Tissue protocol. Homogenates and extracted DNA were stored at −20°C until further use.
We employed the polymerase chain reaction (PCR), using universal COIII oligonucleotide primers (modified from Simon et al., 1994) to obtain a partial COIII DNA sequence. The resulting PCR product was cloned using a puc18/SmaI vector and sequenced (GenBank accession no. AF082986). Next, PCR primers specific to I. pacificus were designed from the cloned sequence and used to amplify a 403-bp target (primers: IpA TTC ATA GAA GAC TAT CAC C, IpB TTC AAA GCC AAA ATG ATG AG). From this, 355 bp of sequence was used to appraise geographical variation. PCR products were sequenced directly using either biotinylated solid phase manual sequencing (29 individuals) (Hultman et al., 1989) or automated DNA sequencing methods (51 individuals) (ABI 377, Applied Biosystems, Perkin-Elmer, Inc., Foster City, CA). Sample sizes ranged from 10 to 18 ticks per population.
Data analyses
Sequences were aligned using either the ESEE sequence editing program (Cabot & Beckenbach, 1989) or the Sequence Navigator DNA analysis software (Applied Biosystems Division). Character state changes and nucleotide composition were calculated with PAUP 3.1 (Swofford, 1993). The amino acid sequence was determined using MacClade 3.0.3 (Maddison & Maddison, 1992). Estimates of nucleotide diversity were calculated using HAPLO2 software (Lynch & Crease, 1990) and estimates of haplotype frequency were computed using ARLEQUIN 1.1 software (Schneider et al., 1997).
Relationships among unique haplotypes were assessed by maximum parsimony (MP) using PAUP 3.1 with a heuristic search algorithm and the random stepwise addition option treating all characters as unordered multistate (Swofford, 1993). Ixodes scapularis (GeneBank accession no. AF083466) was used for outgroup comparison. To obtain a single unrooted parsimony network, a 50% majority rule consensus tree was computed.
The null hypothesis of panmixia was tested using an exact test of the differentiation of haplotypes among populations, using ARLEQUIN 1.1 (Schneider et al., 1997). The exact test of population differentiation of haplotypes tests the hypothesis that the observed distribution of frequencies is less likely than the distribution expected under panmixia (Schneider et al., 1997). Probabilities were estimated by permutation analysis using 1000 randomly permuted r × k contingency tables (r=populations and k=different haplotypes) of haplotype frequencies.
Gene flow was calculated using three estimators. The pseudo-maximum likelihood estimator (PMLE) was used to compute the scale parameter θ (Rannala & Hartigan, 1996). For comparison, two FST analogues, GCA (Cockerham & Weir, 1993) and NST (Lynch & Crease, 1990) were used to estimate gene flow also. Gene flow can be estimated from the equation θ=Nm for the continuous generation, island model of population structure, or θ=2Nm for the discrete generation model (Rannala & Hartigan, 1996). Under the continuous generation model of population structure, the PMLE of θ does not assume population equilibrium and incorporates both immigration and birth rates into its estimate based on the relationship θ=immigration rate/birth rate (Rannala & Hartigan, 1996). To estimate Nm under a discrete generation model (Lynch & Crease, 1990), GCA and NST can be used to calculate gene flow for the haploid island model of population structure using the equation Nm=1/(2FST) − 1/2, where GCA and NST=FST. NST partitions nucleotide diversity within and between populations when multiple populations are compared (Lynch & Crease, 1990). NST weights diversity estimates by haplotype genetic distances, whereas the other estimators assume equal genetic distance among haplotypes (Lynch & Crease, 1990). For the computation of θ, we used equations 16 and 17 in Rannala & Hartigan (1996). FSTAT (Goudet, 1995) and HAPLO2 (Lynch & Crease, 1990) software programs were used to calculate GCA and NST, respectively.
An isolation-by-distance analysis (IBD) was performed to examine further the influence of gene flow on the partitioning of genetic variation in I. pacificus (Slatkin & Maddison, 1990). The log10 of pairwise Nm values were regressed against log10 of pairwise distance values (Slatkin & Maddison, 1990).
Results
In total, 80 I. pacificus and one I. scapularis haplotypes were sequenced. Within I. pacificus, 36 nucleotide sites out of 355 bp varied among all haplotypes (Fig. 2) and 2% was the maximum difference between any pair of haplotypes. Thirty variable sites were at the third codon position, five were at first codon positions, and one was at a second codon position. The transition to transversion ratio (1.05:1) was relatively low compared to insects (Simon et al., 1994). Out of a total of four amino acid replacements, three were produced by first codon substitutions resulting in conserved amino acid replacements [pos. 239, Val/Leu, pos. 356, Ileu/Leu (both nonpolar); pos. 209, Asn/Tyr (polar neutral)] (Fig. 2). One second codon position substitution resulted in a nonconserved (polar neutral to nonpolar) amino acid replacement (pos. 84, Ser/Phe). This pos. 84 substitution was found in one tick from the Oregon population, in all ticks from the Utah population, and in the outgroup I. scapularis (Genbank accession no. AF083466).
Unique haplotypes, variable nucleotide positions, relative haplotype frequencies, sample sizes, and nucleotide and haplotype diversity estimates within Ixodes pacificus. Numbers in top row of left side of figure indicate nucleotide positions relative to a cloned sequence (GenBank accession no. AF082986). Numbers in the body of the right side of figure indicate relative haplotype frequencies within each population. Sample sizes and nucleotide diversity estimates (±2 SE) are arrayed along bottom of the right-hand side of figure. HT, haplotype code; N, sample size per population and total number of that haplotype. Locality codes as in Table 1. Note: haplotype frequencies may not equal unity because of rounding errors.
Average nucleotide diversity within populations ranged from 0.0005 (UTAH) to 0.0086 (MR) and the mean within-population nucleotide diversity was 0.0057 (Fig. 2). Average among-population nucleotide diversity was 0.0028 (Fig. 2). A total of 38 unique haplotypes was found within and among populations (Fig. 2). Haplotype diversity within populations ranged from 0.173 for Utah to 0.905 for MR (Fig. 2). Overall haplotype diversity was 0.930. All populations except Utah shared at least one haplotype with another population. Ticks from Utah only carried unique haplotypes (37 and 38). Thirty-four haplotypes were unique to various localities (Fig. 2).
The exact test of differentiation of haplotype frequency was significant over all populations (P<0.001). The pairwise differentiation test was significant in 12 out of 21 comparisons (Table 2). The UTAH population was significantly differentiated from all other populations. In contrast, no apparent geographical pattern to the differentiation was found in the continuous portion of the range.
The PMLE provided an estimate of θ=20.72. Under the continuous generation model, Nm=20.72 (SE=9.67) and, under the discrete generation model, Nm=10.36. The NST value was 0.329 (SE=0.217) and the resulting Nm=1.02. The GCA estimator provided a value of 0.167 which gives a Nm=2.49.
The IBD results indicate that there is no relationship between the magnitude of gene flow and geographical distances (Fig. 3). The R2 value was 0.065 and was nonsignificant (P=0.28) and the regression slope was −0.76.
Strict consensus MP analysis performed on 2000 equally parsimonious trees resulted in an almost completely unresolved polytomous tree (not shown). A parsimony network derived from the 50% majority rule consensus tree is shown. An unrooted tree is presented (Fig. 4) because tree topology was not affected by removing I. scapularis as an outgroup. This was probably because of the large number of nucleotide substitutions separating I. scapularis and I. pacificus relative to the small number of substitutions separating ticks within I. pacificus. With the exception of the Utah locality, the tree revealed little geographical structure. Relationships of many haplotypes were reconstructed as a basal polytomy, whereas others clustered into weakly supported clades or clades that combined geographically distant haplotypes.
Unrooted parsimony network of 38 unique Ixodes pacificus haplotypes derived from the 50% majority rule consensus tree of 2000 equally parsimonious trees derived from a heuristic search. Locality and haplotype codes as in Table 1 and Fig. 2, respectively, with number of individuals in parentheses. Circle sizes are approximately proportional to haplotype frequencies across the range. Short hatching indicates nucleotide substitutions and asterisks indicate nucleotide substitution that results in the nonconserved amino acid replacement. Numbers at branches indicate consensus tree percentage support for that branch.
Discussion
Utah and Santa Cruz Island populations
The isolation of the Utah population is supported by the fact that the mtDNA of I. pacificus from this population was distinguishable from that of all other tick populations evaluated. Furthermore, there was reduced haplotype diversity (0.173) in Utah ticks compared to other populations (0.762) (Fig. 2). The Utah locality can be characterized as a montane island with sufficient moisture and ground cover to support I. pacificus (Dennis et al., 1998). It is ecologically isolated from the western portion of the range by a large expanse of arid high desert habitat that is unsuitable to I. pacificus (Dennis et al., 1998). Such a distributional pattern could result from either Pleistocene range fragmentation or a more recent founding event (Hewitt, 1996). The loss of mtDNA polymorphism in the Utah population, coupled with the fact that haplotypes in this population were not unusually diverged (Fig. 4), suggests a post-Pleistocene founder event leading to a genetic bottleneck, rather than Pleistocene vicariance (Hewitt, 1996). Ixodes pacificus has the potential for both natural and human-mediated long-range dispersal because it parasitizes birds, large mobile mammals, livestock, and domestic pets (Lane et al., 1991).
Allozyme data did not distinguish the Utah population (Kain et al., 1997). Such a result could be explained by high rates of gene flow, non-neutrality of alleles, or retention of ancestral polymorphisms (Avise, 1994). These alternative hypotheses are difficult to choose between. Our mtDNA data suggest that glucose-6-phosphate isomerase alleles may be influenced by selection (see below). Because gene flow models assume demographic equilibrium, we cannot distinguish gene flow from retention of polymorphisms if the population is not in equilibrium (Whitlock & McCauley, 1999). If either a founder event, range fragmentation, or restriction of gene flow have occurred, allozymes, as markers of gene flow, are not as sensitive as mtDNA (Avise, 1994).
In contrast to the distinctness of the Utah population, there was no evidence that ticks from Santa Cruz Island were differentiated from mainland populations. Allozyme data, likewise, show no differences between Santa Cruz Island and mainland ticks (Kain et al., 1997). We suggest that this is most probably because of the known high incidence of recent anthropogenic effects, particularly as movement of pets and livestock between the mainland and island (Miller, 1985).
Western portion of range
No discernible geographical patterns of differentiation in mtDNA were exhibited by I. pacificus within the western portion of its range, and high haplotype diversity contrasted with low nucleotide diversity. Although estimates of gene flow were moderate to high, the data do not comfortably fit this scenario, because there were so many unique haplotypes restricted to single localities and no significant isolation-by-distance effect (Fig. 3). Likewise, recent, rapid range expansion seems unlikely in the presence of such a large number of unique haplotypes. Another explanation is unusually rapid local evolutionary rates, perhaps following an earlier rapid range expansion that spread the haplotypes (1, 2, 15) that are now widely distributed. The failure to settle unambiguously on a single hypothesis may be because of a number of factors, including unsuitability of mtDNA as a gene target, sampling biases, or gene flow models that are biased or whose assumptions are violated. Gene flow models assume equilibrium between the effective movement of genes into a population and genetic drift (Whitlock & McCauley, 1999). If such equilibrium exists, then geographical genetic structure will result even in the presence of high rates of gene flow (Whitlock & McCauley, 1999).
The general informativeness of mtDNA for this level of analysis was demonstrated in a survey of the closely related I. scapularis (Norris et al., 1996). Here 12S and 16S mtDNA sequences exhibited a broad geographical pattern where mtDNA in this species was divided into two distinct clades (Norris et al., 1996). Because third positions in COIII sequences are generally faster-evolving than the faster bases in 12S and 16S sequences (Simon et al., 1994), it is reasonable to expect that the mtDNA region sequenced in this study should be informative in I. pacificus.
In fact, mtDNA data agreed with the lack of a discernable pattern in 11 out of 12 allozyme loci (Kain et al., 1997), and it is likely that this pattern characterizes a significant part of the genetic structure of I. pacificus. Because mtDNA showed no pattern suggesting secondary contact, it is most plausible that the pattern in the twelfth allozyme locus, GPI, is caused by selection on that locus.
As for sampling biases, we note that we sampled ticks over a relatively broad area at each locality. Even if we had unduly concentrated our sampling efforts, we should have found high homogeneity within samples rather than the many rare alleles, which we actually observed. It is conceivable, however, that our sample sizes were insufficient to detect rare, but shared, haplotypes, which would lead to the appearance of many unshared haplotypes.
Inadequacy of gene flow models remains a reasonable factor in the apparent lack of pattern presented by our data. Gene flow estimates in this study varied 20-fold and ranged from moderate, Nm=1.02 (NST) to very high, Nm=20.72 (PMLE, continuous generation model). An average estimate generated from allozyme data gives a Nm=5.70 (Kain et al., 1997). On the whole, it is difficult to determine which estimator is most appropriate to this study (Whitlock & McCauley, 1999). Indirect measures of gene flow and migration that are extrapolated from gene frequency data are variants of FST, a standardized measure of genetic variance among populations. The assumptions underlying these extrapolations include (i) low rates of mutation, (ii) a stepping stone pattern of migration, (iii) spatial and temporal demographic stability, (iv) nonoverlapping generations (for some estimators), and (v) equilibrium between gene flow and genetic drift (Whitlock & McCauley, 1999). We consider these assumptions in succession below.
As a violation of the first assumption, high rates of mutation could produce a large number of unique haplotypes in the face of moderate to high levels of gene flow (Schneider, 1996). Gene flow models assume relatively low rates of mutation, which may apply to nuclear genes, but which may not apply to mtDNA genes (Whitlock & McCauley, 1999). A high rate of mutation in the COIII gene in I. pacificus may be evidenced by the unusually low transition to transversion ratio (1.05).
For the second assumption, that there is a stepping stone pattern of migration, the isolation-by-distance model employed here uses a one- or two-dimensional stepping stone model. If spatial structure of gene flow deviates from this model, then an isolation-by-distance effect may not be detected (Whitlock & McCauley, 1999). Likewise, if the stepping stone model applies on a local level, but not at larger distances, then an isolation-by-distance effect would appear to be absent (Whitlock & McCauley, 1999).
The third assumption of gene flow models is that there is spatial and demographic stability. Deviations from this model have not been extensively explored, although there are some indications that there could be significant levels of gene flow with the presence of many unique haplotypes and the lack of an isolation-by-distance effect. Nürnberger & Harrison (1995) in a study on mtDNA in gyrinid water beetles, found that demographic instability appeared to have led to high levels of haplotype heterogeneity in the presence of high rates of gene flow. These beetles were localized in a series of isolated ponds with high extinction rates followed by founder events (Nürnberger & Harrison, 1995). Their study concluded that significant inadequacies remain in the ability of current population genetic models to analyse the relationship between population structure and gene flow.
For the fourth assumption, three of four estimators used in this study assume discrete generations. However, there is some evidence that I. pacificus has overlapping generations (Peavey & Lane, 1996) and therefore the fourth, and highest, estimate of migration rate (Nm=20.72) might be most accurate.
Finally, if there is violation of the fifth assumption of equilibrium between gene flow and genetic drift, then estimates of gene flow could be inflated because of the lack of such equilibrium. Recent and rapid range expansion usually results in a pattern of little population genetic structure (Slatkin & Hudson, 1991), but under certain conditions can produce patterns of high within-population haplotype diversity within populations, and high estimates of gene flow (Patton et al., 1996). The original gene pool would have to be highly polymorphic, with colonists expanding into new territory and carrying a biased subsample of haplotypes that included both rare, localized and more common alleles. The presence of a few ancestral haplotypes within most populations would inflate gene flow estimates even if current rates were low (Patton et al., 1996).
In conclusion, we were able to find mtDNA haplotype differentiation between a population of Utah ticks and all populations of I. pacificus where allozyme data were not able to do so. We found no differentiation of mtDNA haplotypes within the continuous portion of the range. This probably reveals failures in current gene flow models to reflect adequately the complexity of evolutionary processes underlying the geographical differentiation of mtDNA haplotypes in I. pacificus. Because many systematic analyses are begun with little or no ecological or genetic data available on the organism being investigated, it will be crucial to continue to develop realistic gene flow models that can be applied in empirical studies such as ours.
References
Arnason, C. S. (1992). Biology of the Western Black-legged Tick, Ixodes pacificus, (Cooley and Kohls, 1943): A Potential Vector of Lyme Disease in South Coastal British Columbia. M.S. Thesis, Simon Fraser University, B.C., Canada.
Arthur, D. R. and Snow, K. R. (1968). Ixodes pacificus Cooley and Kohls, 1943: its life-history and occurrence. Parasitology. 58: 893–906.
Avise, J. C. (1994). Molecular Markers, Natural History and Evolution. Chapman & Hall, New York.
Cabot, E. L. and Beckenbach, A. T. (1989). Simultaneous editing of multiple nucleic acid and protein sequences with ESEE. Comp Appl Biosci. 5: 233–234.
Cockerham, C. C. and Weir, B. (1993). Estimation of gene flow from F-statistics. Evolution. 47: 855–863.
Dennis, D. T., Nekomoto, T. S., Victor, J. C., Paul, W. S. and Piesman, J. (1998). Reported distribution of Ixodes scapularis and Ixodes pacificus (Acari: Ixodidae) in the United States. J Med Entomol. 35: 629–638.
Goudet, J. (1995). Fstat, Version 1.2: a computer program to calculate F-statistics. J Hered. 86: 485–486.
Hewitt, G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc. 58: 247–276.
Hilburn, L. R. and Sattler, P. W. (1986). Are tick populations really less variable and should they be?. Hered. 57: 113–117.
Hultman, T., Stahl, S., Hornes, E. and Uhlen, M. (1989). Direct solid phase sequencing of genomic and plasmid DNA using magnetic beads as solid support. Nucl Acids Res. 17: 4937–4946.
Kain, D. E., Sperling, F. A. H. and Lane, R. S. (1997). Population genetic structure of Ixodes pacificus (Acari: Ixodidae) using allozymes. J Med Entomol. 34: 441–450.
Lane, R. S., Piesman, J. and Burgdorfer, W. (1991). Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Ann Rev Ent. 36: 587–609.
Lynch, M. and Crease, T. R. (1990). The analysis of population survey data on DNA sequence variation. Mol Biol Evol. 7: 377–394.
Maddison, W. P. and Maddison, D. R. (1992). MacClade. Sinauer Associates, Sunderland, MA.
Miller, D. R. (1985). The California Channel Islands — past, present, and future: an entomological perspective. In: Menke, A. S. and Miller, D. R. (eds) Entomology of the Channel Islands, pp. 3–27. Santa Barbara Museum of Natural History, Santa Barbara, CA.
Norris, D. E., Klompen, J. S. H., Keirans, J. E., Black W. C. Iv (1996). Population genetics of Ixodes scapularis (Acari: Ixodidae) based on mitochondrial 16S and 12S genes. J Med Entomol. 33: 78–89.
Nürnberger, B. and Harrison, R. G. (1995). Spatial population structure in the whirligig beetle Dineutus assimilis: evolutionary inferences based on mitochondrial DNA and field data. Evolution. 49: 266–275.
Patton, J. L., Da Silva, M. N. F. and Malcolm, J. R. (1996). Hierarchical genetic structure and gene flow in three sympatric species of Amazonian rodents. Mol Ecol. 5: 229–238.
Peavey, C. A. and Lane, R. S. (1996). Field and laboratory studies on the timing of oviposition and hatching of the western black-legged tick, Ixodes pacificus (Acari: Ixodidae). Exp Appl Acarol. 20: 695–711.
Rannala, B. and Hartigan, J. A. (1996). Estimating gene flow in island populations. Genet Res. 67: 147–158.
Richter, P. J. J. R., Kimsey, R. B., Madigan, J. E., Barlough, J. E., Dumler, J. S. and Brooks, D. L. (1996). Ixodes pacificus (Acari: Ixodidae) as a vector of Ehrlichia equi (Rickettsiales: Ehrlichieae). J Med Entomol. 33: 1–5.
Schneider, C. J. (1996). Distinguishing between primary and secondary intergradation among morphologically differentiated populations of Anolis marmoratus. Mol Ecol. 5: 239–249.
Schneider, S., Kueffer, J. -M., Roessli, D. and Excoffier, L. (1997). Arlequin, Version 11: A software for population genetic data analysis Genetics and Biometry Laboratory. University of Geneva, Switzerland.
Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H. and Flook, P. (1994). Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 87: 651–701.
Slatkin, M. and Hudson, R. R. (1991). Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 129: 555–562.
Slatkin, M. and Maddison, W. P. (1990). Detecting isolation by distance using phylogenies of genes. Genetics. 126: 249–260.
Swofford, D. L. (1993). PAUP: Phylogenetic Analysis Using Parsimony, Version 3.1. Illinois Natural History Survey, Champaign, IL.
Tabachnick, W. J. and Black, W. C. I. V. (1995). Making a case for molecular population genetic studies of arthropod vectors. Parasitol Today. 11: 27–30.
Wesson, D. M., Mclain, D. K., Oliver, J. H. Jr, Piesman, J. and Collins, F. H. (1993). Investigation of the validity of species status of Ixodes dammini (Acari: Ixodidae) using rDNA. Proc Natl Acad Sci USA. 90: 10,221–10,225.
Whitlock, M. C. and Mccauley, D. E. (1999). Indirect measures of gene flow and migration:FST(1/(4Nm + 1). Heredity. 82: 117–125.
Acknowledgements
We thank the following people for their invaluable assistance in collecting I. pacificus: D. Kindree (West Vancouver), C. S. Arnason (Simon Fraser University), R. A. Gresbrink (Oregon State Health Division, retired), S. G. Bennett and J. P. Webb, Jr. (Orange County Vector Control District), K. Padgett (University of California at Berkeley), K. MacBarron (San Diego County Department of Environmental Health), L. Laughrin (University of California Natural Reserve System, Santa Cruz Island Reserve), J. R. Clover (California Department of Health Services), P. Coy (California Department of Transportation). We thank M. Lynch and B. Rannala for their assistance with data analyses. This project was partially funded by Sigma Xi, and the Walker Fund of UC Berkeley to DEK, a Hatch Grant to FAHS, and by grant AI22501 from the National Institutes of Health and California Agricultural Experiment Station Project 5719-AH to RSL.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kain, D., Sperling, F., Daly, H. et al. Mitochondrial DNA sequence variation in Ixodes pacificus (Acari: Ixodidae). Heredity 83, 378–386 (1999). https://doi.org/10.1038/sj.hdy.6886110
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6886110
Keywords
This article is cited by
-
Epidemiologic profile of hard ticks and molecular characterization of Rhipicephalus microplus infesting cattle in central part of Khyber Pakhtunkhwa, Pakistan
Parasitology Research (2022)
-
Genetics of Thrips palmi (Thysanoptera: Thripidae)
Journal of Pest Science (2020)
-
Analyses of mitochondrial genes reveal two sympatric but genetically divergent lineages of Rhipicephalus appendiculatus in Kenya
Parasites & Vectors (2016)
-
Identification and characterization of Rhipicephalus (Boophilus) microplus and Haemaphysalis bispinosa ticks (Acari: Ixodidae) of North East India by ITS2 and 16S rDNA sequences and morphological analysis
Experimental and Applied Acarology (2014)
-
Molecular analysis of Ixodes granulatus, a possible vector tick for Borrelia burgdorferi sensu lato in Taiwan
Experimental and Applied Acarology (2009)